Abstract
DNA replication origins are necessary for the duplication of genomes. In addition, plasmid-based expression systems require DNA replication origins to maintain plasmids efficiently. The yeast autonomously replicating sequence (ARS) assay has been a valuable tool in dissecting replication origin structure and function. However, the dearth of information on origins in diverse yeasts limits the availability of efficient replication origin modules to only a handful of species and restricts our understanding of origin function and evolution. To enable rapid study of origins, we have developed a sequencing-based suite of methods for comprehensively mapping and characterizing ARSs within a yeast genome. Our approach finely maps genomic inserts capable of supporting plasmid replication and uses massively parallel deep mutational scanning to define molecular determinants of ARS function with single-nucleotide resolution. In addition to providing unprecedented detail into origin structure, our data have allowed us to design short, synthetic DNA sequences that retain maximal ARS function. These methods can be readily applied to understand and modulate ARS function in diverse systems.
The application of genomic tools to classical genetic techniques has led to a rapid expansion of our understanding of biological processes at a systematic, global level. Microbial systems such as yeast are particularly well-suited for the merging of these methods due to our ability to perform experiments on a large scale. Recent advances have transformed the study of basic principles such as genetic interactions (Costanzo et al. 2011) and structural features of DNA elements (Sharon et al. 2012). In this study, we have developed genomic tools for comprehensively mapping and dissecting replication origins in yeast using simple screening techniques that can be readily extended to other DNA elements.
Origins of DNA replication act as sites of initiation of DNA replication via recruitment of the origin recognition complex (ORC) and other proteins necessary for the duplication of the genome in every cell cycle (Sclafani and Holzen 2007). While origins on the whole are an essential part of every genome, functional redundancy of these noncoding elements subjects them to a poorly understood set of evolutionary forces. A comprehensive understanding of origin location and structure across multiple species would shed light on the interaction between DNA replication dynamics and genome structure as well as on the coevolution of origin sequences and origin-interacting proteins.
Yeast origins promote replication and maintenance of episomal plasmids as cis-acting autonomously replicating sequences (ARSs) (Stinchcomb et al. 1979). This property is essential for plasmid-based expression systems (Parent et al. 1985; Böer et al. 2007) and has been widely used to map origins and dissect their functional domains uncovering a diversity of molecular determinants of ARS function among different yeast species. Examples include Saccharomyces cerevisiae, which uses a short (11–17 bp) T-rich ARS Consensus Sequence (ACS) motif for ARS function (Broach et al. 1983); Kluyveromyces lactis, whose ACS motif is 50 bp (Liachko et al. 2010); and fission yeast Schizosaccharomyces pombe, whose origins do not seem to have a consensus motif, instead using a more promiscuous AT-binding property of ORC (Chuang and Kelly 1999). Recent work also suggests a diversity of origin sequences among Schizosaccharomyces yeasts (Xu et al. 2012). In some cases, species that are relatively closely related use different consensus sequences (Liachko et al. 2011; Di Rienzi et al. 2012).
Yeast replication origins can also be characterized through experiments focused on the dynamics of chromosome replication (Raghuraman et al. 2001; Yabuki et al. 2002; Smith and Whitehouse 2012). While these studies are useful in detailing the temporal order of events during genome replication, they fall short of generating a complete map of potential origin sites, due to low resolution and variability in origin usage and efficiency in different cells within a population. Deletion of all known active origin sites on a chromosome does not completely abrogate replication (Dershowitz et al. 2007), suggesting the presence of cryptic origins whose chromosomal replication initiation signal is too weak to be detected in population-based assays. Thus, ARS mapping and functional dissection remain the most precise tools for understanding the molecular determinants of yeast origin function. However, a lack of methods to comprehensively identify and dissect ARSs has slowed progress because origins are typically studied in small numbers. Thus, despite decades of study, >30% of suspected S. cerevisiae origins remain unconfirmed (Nieduszynski et al. 2007; Siow et al. 2012).
We have developed an approach that couples high-throughput ARS screening with deep sequencing to map ARSs, delineate their functional regions, and measure the effect of all possible point mutations on individual ARS fragments using massively parallel mutational scanning. Our approach yields the most comprehensive, high-resolution S. cerevisiae ARS data set to date and can be readily applied to other yeast strains and species. Using our data, we have designed and tested a 100-bp ARS fragment that is able to maintain episomal plasmids with much greater stability than wild type and acts as an improved replication origin in its native genomic context. Such improved replication origin modules can be useful for regulating genomic replication as well as to increase the stability of plasmids in diverse strains and species of yeast.
Results
High-throughput mapping of genomic ARS locations
To obtain a complete map of ARS locations in S. cerevisiae, we generated a >12× genomic library of overlapping restriction fragments cloned into a shuttle vector that lacks ARS function but contains a URA3 selectable marker. Transformation-competent ura3 yeast were transformed with this library and selected for growth on medium lacking uracil. Colony formation requires the replication and propagation of the plasmid and allows the recovery of ARS-bearing plasmids (Fig. 1A). Plasmid inserts were amplified using vector-specific primers and identified en masse using paired-end deep sequencing.
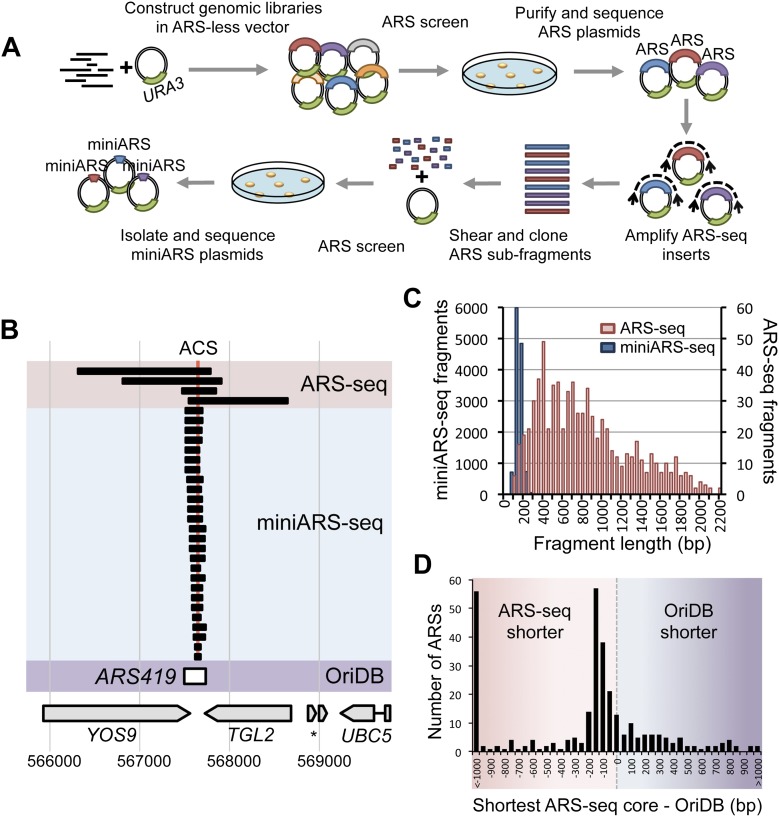
Figure 1.
High-throughput, high-resolution mapping of ARSs using ARS-seq and miniARS-seq. (A) Genomic libraries were constructed in a URA3 vector lacking an ARS. Yeast were transformed with these libraries for selection of ARS-containing plasmids. ARS plasmids were isolated from pooled yeast colonies and sequenced using insert-flanking primers (ARS-seq, top row). Inserts from the ARS-seq plasmid pools were amplified using vector-specific primers. Randomly sheared and size-selected fragments were cloned into an ARS-less vector and rescreened for ARS function (miniARS-seq, bottom row). (B) A sample locus (at ARS419) comparing results of ARS-seq (red highlight), miniARS-seq (blue highlight), and OriDB annotation (purple highlight). The best match of the ACS motif is indicated at the top (red vertical line). Corresponding coordinates on chromosome 4 and annotated nearby genes are shown at the bottom. (C) Size distributions of ARS-seq and miniARS-seq fragments listed in Supplemental Tables 1 and 2. (D) Distribution of the size differences between OriDB-annotated confirmed ARSs and the corresponding shortest ARS-seq/miniARS-seq fragments or inferred functional cores (see Methods).
Our screen, which we have named “ARS-seq,” yielded 720 distinct DNA fragments (median length: 702 bp) representing 366 unique genomic loci (227 loci were overlapped by multiple fragments) (Supplemental Table 1). To better define the false-positive (FP) and false-negative (FN) rates associated with ARS-seq, we manually cloned selected DNA fragments (Supplemental Analysis) and tested them for ARS activity by transforming yeast and monitoring colony formation on selective medium. The majority of our ARS-seq data (263 ARSs) overlapped with regions previously annotated as confirmed ARSs in the OriDB database (Siow et al. 2012), and a further 58 ARSs overlapped with regions annotated as “likely ARSs.” Manual validation drastically increased the data set's accuracy and coverage and identified a further 48 unique ARS loci. Our findings suggest FP and FN rates of ∼12% for ARS-seq (Supplemental Fig. 1; Supplemental Analysis). Our data are also consistent with a recently published partially overlapping set of manually validated ARS candidates (Müller and Nieduszynski 2012), further underscoring the value of an unbiased screening tool to map genomic DNA elements comprehensively.
Delineating essential ARS regions using miniARS-seq
While the average length of ARS fragments identified by ARS-seq was 702 bp, it is known that the regions required for ARS function in S. cerevisiae (as well as in several other yeasts) can be <100 bp (Sclafani and Holzen 2007; Liachko et al. 2010, 2011). We developed the miniARS-seq method to identify the minimal functional regions of ARSs en masse. We PCR-amplified all ARS inserts from the plasmid pools isolated from the ARS-seq experiment. We used DNase I to randomly shear the inserts and used gel purification to isolate fragments in the 100-bp to 200-bp range. These short subfragments of ARSs were cloned into the ARS-less URA3 vector and used to transform yeast to select for fragments that retain ARS activity (Methods). ARS plasmids were extracted from yeast in high numbers, and the inserts were sequenced to map overlapping short ARS fragments (Fig. 1A).
After stringent filtering to remove FPs (for details on data filtering, see the Supplemental Analysis), our experiments yielded a total of 12,338 unique genomic miniARS fragments (median length: 147 bp) representing 181 unique ARS regions (Supplemental Table 2). The recovery of miniARS-seq fragments correlated with coverage in the original ARS-seq screen (Supplemental Fig. 2), as well as the score of the ACS match within the ARS. We used information from overlapping fragments to infer functional cores of miniARS-seq contigs, which further delineated minimal functional ARS regions (median length: 92 bp) (Fig. 1B,C). Subsequent manual validation suggested a false-positive rate of 3.9% prior to validation (Supplemental Tables 3, 4; Supplemental Analysis). Combining the data from a single ARS-seq and miniARS-seq experiment generated an ARS map with much higher resolution than the combined ARS/origin data set curated by OriDB (Fig. 1D). While it is known that elements flanking the ACS are necessary for ARS function, our data also suggest that in most cases essential flanking elements reside on the 3′ side of the T-rich strand of the ACS (Supplemental Analysis). To test the sufficiency of a 3′-extended ACS for ARS function, we manually validated four unique ARS fragments, each <35 bp in length, that contain an ACS and a flanking B1 element and found three-fourths to retain ARS function, although strongly attenuated in one case (Supplemental Analysis).
Deep mutational scanning of ARS1
The contribution of specific nucleotides to ARS function can be elucidated by mutagenesis experiments. However, such experiments are limited by throughput. To test the functional consequences of all single substitution mutations on a given ARS in a massively parallel fashion, we developed mutARS-seq—a deep mutational scanning approach coupled with high-throughput sequencing (Patwardhan et al. 2009; Fowler et al. 2010; Haberle and Lenhard 2012), which we applied to a 100-bp fragment of the well-studied ARS1 for method validation. This fragment contains the ACS as well as the B1 and B2 elements essential for ARS1 function and is sufficient to support plasmid replication. We used a randomly mutagenized oligonucleotide to generate a library of ARS1 mutant variants in an ARS-less vector. The resulting library contained >22,000 ARS1 inserts. Every position within each individual ARS1 insert had a 2% chance of bearing a mutation. These variant plasmids were used to transform yeast in large numbers, and the resulting library was directly competed in liquid culture. The abundance of each variant at different times in the competition was measured by 101-bp paired-end deep sequencing and was used to calculate a relative fitness value for each mutant allele (Fig. 2A). The resulting library yielded data for all 300 possible single substitutions, deletions at most positions, and combinations of these mutations.
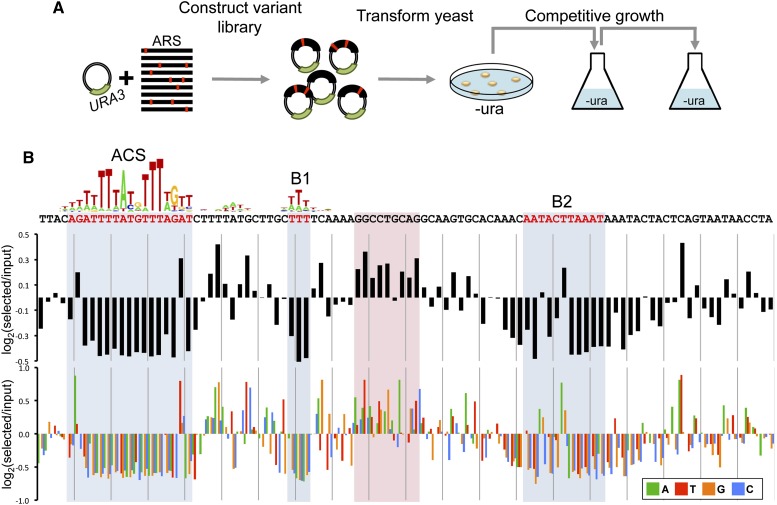
Figure 2.
Comprehensive mutational scanning of a 100-bp fragment of ARS1 using mutARS-seq. (A) A library of randomly mutagenized ARS1 fragments was cloned into a URA3 vector. Yeast were transformed with these libraries and competed in liquid batch growth. (B) The log2 of the enrichment ratio is shown for all substitution mutations within the ARS1 fragment (top: average; bottom: individual nucleotide substitutions). (Blue) Previously described ACS, B1, and B2 elements. (Red box) A region of nucleotides that repress wild-type ARS1 function. The data shown are the composite of multiple samples as described in the Supplemental Analysis.
Our data showed strong depletion of plasmids with nucleotide substitutions and deletions at the ACS, B1, and B2 domains of the ARS1 fragment (Fig. 2B; Supplemental Fig. 6). In addition, several positions showed a preference for mutations over wild-type nucleotides, the most striking example being a 9-bp region between the B1 and B2 elements and two positions within the core ACS (Fig. 2B). Closer inspection of the sequence preference surrounding the B2 element revealed that the optimal combination of nucleotides at this locus would re-create a perfect 11-bp ACS match on the reverse strand, as previously predicted (Wilmes and Bell 2002). Analysis of variants carrying two substitutions revealed synergistic relationships between nucleotides in the ACS, B1, and B2 elements, while most other substitution combinations showed weak epistatic (nonadditive) effects (Supplemental Analysis). Analysis of alleles bearing deletions further demonstrated the functional importance of the ACS, B1, and B2 elements. Surprisingly, there was a strong functional benefit to variants with single-base deletions between the ACS and the B2 element (Supplemental Fig. 6). Note that these beneficial deletions also occur between the core ACS and the TTT of the B1 element. However, this does not contradict the presumably rigid distance between the core ACS and the B1 element since there is an “extra” T following the B1's TTT which in the case of such a deletion would join the last two T's of the original B1 to give the same TTT. Our findings show that mutARS-seq is effective at measuring effects of mutations across the ARS as well as probing for positional effects and spacing constraints.
Using mutARS-seq data to optimize ARS1 function
The data from mutARS-seq can be used to optimize the function of ARS1. We constructed a synthetic DNA fragment consisting of nucleotides found to be the most beneficial for ARS1 function at each position. This 100-bp sequence (ARS1max) (Supplemental Fig. 8) contained 53 mutations relative to the wild-type ARS1 sequence (Supplemental Fig. 8a) and promoted faster growth in selective medium than either its wild-type equivalent or ARS1hi1 (the best-performing allele identified by mutARS-seq) (Supplemental Fig. 8), indicating increased ARS activity (Fig. 3A). DNA fragments bearing ARS1 and ARS1max were cloned into CEN plasmids, and ARS activity was quantified using the minichromosome maintenance assay, which measures plasmid loss per generation in nonselective growth conditions (Donato et al. 2006). The 100-bp ARS1max sequence had a drastically reduced plasmid loss rate relative to the original ARS1, indicating a level of ARS efficiency comparable to a 7-kb fragment of the efficient ARS121 (systematic name: ARS1021) (Fig. 3B; Supplemental Fig. 8b; Walker et al. 1990). Extending the length of the ARS fragments to 2.5 kb significantly increased function of the wild-type ARS1, but had very little effect on ARS1max, suggesting that the 100-bp ARS1max has reached maximal ARS efficiency (Fig. 3B). However, the 2.5-kb DNA fragment bearing ARS1max still showed a lower plasmid loss rate than the 2.5-kb ARS1 fragment. We also found ARS1max to be largely resistant to the effects of mcm4-chaos3, an oncogenic allele of MCM4—a component of the replicative DNA helicase (Fig. 3B; Shima et al. 2007). Since the sequence of ARS1max contains a reverse complement ACS match at the position where the B2 element is located in ARS1, it is possible that ARS1max is no longer dependent on the original ACS for function. We tested this hypothesis by mutating a critical TT dinucleotide in the original ACS (Supplemental Fig. 9). Our results show that mutating the original ACS abolishes the ARS function of ARS1, but does not abolish the ARS function of ARS1max. This finding suggests that ARS1max may be a dimeric ARS (Bolon and Bielinsky 2006).
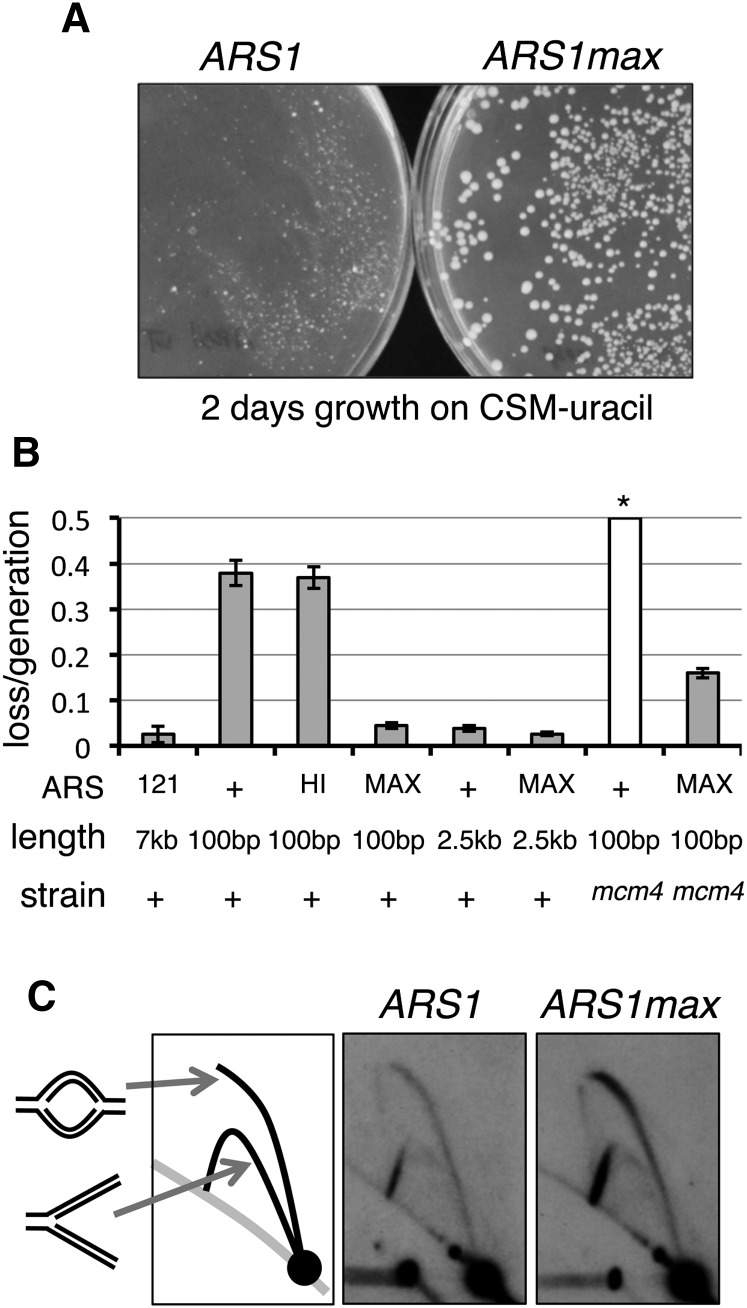
Figure 3.
Optimized synthetic ARS function. (A) ARS1 and ARS1max sequences were cloned into a URA3 vector. Yeast were transformed with resultant plasmids and grown for 2 d on medium lacking uracil. Colony size is representative of ARS function. (B) Plasmid loss assays were performed on CEN vectors containing ARSs indicated: (+) ARS1; (HI) ARS1hi1; (MAX) ARS1max1.1; (121) ARS121. A control vector YCP121, bearing a 7-kb ARS121 fragment is shown (121) as an example of very strong ARS function. (*) The loss rate of wild-type ARS1 in mcm4-chaos3 cells (mcm4) was too high to be measured. (C) Genomic DNA replication was assayed in strains with ARS1 and ARS1max integrated into the native chromosomal locus. The signal ratio of the top (bubble) arcs to the bottom (Y) arcs is indicative of replication initiation at the assayed locus.
To assay ARS1max function in a genomic context, we made a strain in which ARS1 was replaced with ARS1max. DNA two-dimensional (2D) gel analysis (Brewer and Fangman 1987) showed an increase in origin firing at the ARS1 locus in the ARS1max strain (Fig. 3C). Our data show that while in the wild-type ARS1 strain 5.9%–6.2% of the selected replicating intermediate signal comes from the Y-arcs, in the ARS1max strain 2.1%–2.5% of the signal comes from the Y-arcs (Supplemental Fig. 10). While there is no definitive way of converting these measurements into an actual number of firing events, combined, our results indicate that synthetic ARS1max is much more efficient than the wild-type ARS1 both in plasmid and genomic contexts.
Discussion
Combining straightforward genetic tools with large-scale genomic analysis drastically increases scientific productivity and can lead to discoveries inaccessible by traditional means. In yeast, the ARS assay is a simple approach that provides the ability to dissect with fine resolution DNA sequences that are sufficient to initiate DNA replication. While genomic loci are subject to regulation that may differ from plasmid-based sequences, understanding ARSs, the smallest functional unit of replication origins, can serve as a platform for further understanding these sequences in their complex chromosomal context.
While the first budding yeast ARS was discovered more than three decades ago (Stinchcomb et al. 1979), a complete map of ARS locations is not yet available despite laborious efforts to manually validate large numbers of ARSs (Donato et al. 2006; Nieduszynski et al. 2006; Müller and Nieduszynski 2012). Our methodology combines ARS screening with next-generation sequencing to enable the generation of a comprehensive ARS map with the average resolution of <200 bp. The ARS assay is useful for the study of replication origins in yeast species diverged >500 million yr, so we expect that these methods will be portable to a variety of strains and species. Complete ARS maps allow for evolutionary analysis of origin structure and genomic distribution, while simultaneously providing data sets for the study of mechanistic features of origin selection. Generating such maps for the other sequenced yeast species will add a new dimension to our understanding of how replication origins affect genome biology.
In addition to delineating all ARS sequences within a genome, understanding the functional contribution of each nucleotide within an ARS is an important goal. Traditional mutagenesis techniques require the laborious task of testing individual mutant alleles for phenotypic effects. These approaches are usually limited in throughput and sensitivity, lowering the number of alleles that can be tested and the amount of information that can be gathered from each mutation. Our deep mutational scanning approach (mutARS-seq) allows the mapping of the effects of all mutations at all positions in a given ARS sequence. The synthesis of ARS1max underscores the value of such data, providing enough knowledge of ARS1 structure to allow deliberate modulation of its activity. This method can be easily applied to other origins, either to elucidate mechanistic features of ARS function in other species or to understand structure–function relationships in different origins within the same species. In addition, approaches similar to ones presented here can be adapted for the mapping and dissection of other DNA elements, such as promoters, genes, and centromeres.
Methods
Reagents
All strains and primers are listed in Supplemental Table 5. Genomic DNA was isolated using the YeaStar Genomic DNA Kit (Zymo Research). PCR purification and purification of digested plasmids were performed using the DNA Clean and Concentrator-5 Kit (Zymo Research). All enzymes used were from New England Biolabs unless otherwise noted. All primers were purchased from IDT, unless otherwise noted (for a complete list of oligos, see Supplemental Table 5). The transformation host yeast strain used in all experiments was W303-1A. All yeast growth was performed at 30°C; all bacterial growth was performed at 37°C. Bacterial transformants were selected on standard LB medium with ampicillin (100 μg/mL). Yeast transformants were selected on standard synthetic complete medium lacking uracil.
Illumina library construction
Illumina adapter-containing primers that anneal within the vector sequences outside of the insert were used to amplify the relevant DNA, using the high-fidelity enzyme Phusion HF (15 cycles of amplification). Gel extraction was used to isolate fragments of appropriate length for each experiment, followed by purification with the DNA Clean and Concentrator-5 kit (Zymo Research). ARS-seq and miniARS-seq experiments were sequenced using primers OCA275 and OCA276, or IL575 and OCA276.
Vector construction
The vectors used for this study were derivatives of subcloning vector pRS406. An MscI site or an NruI site were inserted into the BamHI site of pRS406 to create pIL19 and pIL22, respectively. Full vector sequences are available upon request.
Construction and screening of ARS-seq libraries
Genomic DNA from a ρ zero derivative of FY4 (prototroph, S288C strain background) was purified using the YeaStar Genomic DNA Kit. Genomic DNA was digested to completion with one of four four-cutter restriction enzymes—MboI, AluI, HaeIII, or RsaI. To prevent insert concatamerization, fragmented DNA was treated with Antarctic Phosphatase and purified with the DNA Clean and Concentrator-5 Kit prior to ligation. DNA digested with MboI was ligated into the BamHI site of pRS406. DNA digested with AluI, HaeIII, and RsaI was ligated into the MscI site of pIL19 and the NruI site of pIL22 (each insert DNA pool was cloned into both vectors separately). To maximize cloning efficiency, each ligation reaction was purified using DNA Clean and Concentrator -5 columns and digested with the vector cloning restriction enzyme (BamHI, MscI, and NruI for pRS406, pIL19, and pIL22, respectively).
Ligation products were used to transform Alpha-Select Gold Efficiency competent Escherichia coli cells (Bioline). Cloning efficiency and insert sizes were checked using colony PCR on random E. coli colonies. Plasmid DNA was purified using the Wizard Plus SV Miniprep Kit (Promega). Total library coverage was ∼12× genome size (∼3× for each restriction enzyme pool). Library transformations of yeast were conducted using a standard lithium acetate method and plated onto complete synthetic agar medium lacking uracil. The host strain for transformations was W303-1A. Yeast colonies were grown for 3 d at 30°C. To enrich for ARS plasmids and to eliminate nontransformed cells, yeast colonies were replica-plated onto fresh -Ura plates and grown for two more days at 30°C. Plasmids were extracted from pooled yeast colonies by glass bead disruption, followed by DNA purification using Wizard Plus SV Miniprep Kit columns. To remove genomic yeast DNA and to facilitate the purification of individual plasmids for further testing, the extracted total DNA was used to transform E. coli, resultant ampicillin-resistant colonies were scraped and pooled, and plasmids were extracted. Forty-eight ARS clones were individually purified, Sanger-sequenced (primers IL429 and IL430), and used to retransform yeast to confirm function.
Construction and screening of miniARS-seq libraries
ARS inserts were amplified from purified ARS-seq plasmid pools using primers IL594 and OCA272. DNA was sheared using DNase I (Roche) and resolved on 2% agarose gels. Fragments corresponding to 100–200 bp in size were cut out of the gel and purified using the GenElute minus EtBr (Sigma-Aldrich) and the DNA Clean and Concentrate-5 kits. Ends of sheared DNA fragments were made blunt using the Klenow fragment of DNA polymerase I, dephosphorylated with Antarctic Phosphatase, and purified with the DNA Clean and Concentrator-5 Kit. Insert DNA was ligated into vectors pIL19 and pIL22 separately, as above. Library coverage was calculated using colony PCR. Approximately 100,000 clones bearing inserts were screened in total. Yeast transformations and plasmid extractions were performed as above. Plasmids extracted from the first round of miniARS-seq were used to transform yeast again to remove false positives that passed through the first screen. ARS clones were individually purified, Sanger-sequenced (primers IL429 and IL430), and used to retransform yeast to confirm function.
Construction and screening of mutARS-seq libraries
Oligo mutARS1_trilink was synthesized by Trilink Biotechnologies. The central 100 bp was randomly mutagenized at a frequency of 2% at each position. The outer 12 bp at each end of the oligo was not mutagenized and contained homology for amplification primers and an MscI restriction site for cloning. Primers IL585 and IL586 were used to amplify the oligo mutARS1_trilink. The resulting fragment was digested with MscI, phosphatase-treated, and ligated into the NruI site of pIL22 as above. Colony PCR on sets of random colonies was used to estimate library coverage; 22,000–24,000 insert-bearing colonies were pooled together for use in the screen. Sanger sequencing of a subset of clones was used for quality control.
The plasmid library was used to transform yeast as above. Yeast were grown for 3 d at 30°C, at which point plates were scraped and inoculated into a 1-L culture of medium lacking uracil. Samples were taken after 12 and 20 h. Total DNA was purified and used as template for primers IL594 and one of the barcoded primers IL576, IL589, IL590, and IL593. Fragments of appropriate length were gel-purified and sequenced using primers IL591 and IL592.
Manual validation of ARS function
DNA segments to be tested were amplified by PCR with primers containing appropriate restriction sites (BamHI, MscI, or NruI). The resulting fragments were cloned into either pRS406, pIL19, or pIL22 (depending on restriction sites present within the insert), Sanger-sequenced, and used to transform yeast. Growth on synthetic complete medium lacking uracil was assayed after 3 d at 30°C.
Construction and characterization of ARS1max plasmids and strains
Synthetic ARS1max sequences were constructed by PCR fusing overlapping primers ARS1max_F and ARS1max_R. (The ARS1max mutant noted in Supplemental Fig. 9 was cloned using primers ARS1maxGG_F and ARS1max_R.) The resulting insert was digested with MscI and cloned into pIL22 and pIL07 (Liachko et al. 2010)—a centromeric vector used for plasmid loss assays. Plasmid loss assays were performed as described (Donato et al. 2006). Long ARS1max fragments were constructed using standard PCR fusion techniques (the mutant ARS1 noted in Supplemental Fig. 9 was constructed using primers IL721 and IL722) and either cloned into appropriate vectors or integrated, replacing the ARS1 sequence using a standard pop-in/pop-out method in the FY3 strain. The integrated ARS1max strain was backcrossed to FY3 (resulting in strain ILY506). Genomic 2D gel experiments were performed as described (Brewer and Fangman 1987). Analysis of replication intermediates on 2D gels was performed using the Quantity One v.4.6.9 software on the Personal Molecular Imager (Bio-Rad) (Supplemental Fig. 10).
ARS-seq data analysis
For more detailed descriptions of computational data analysis, see the Supplemental Analysis. ARS-seq sequencing reads were mapped using Bowtie version 0.12.7 to the October 2003 version of the yeast genome (Saccer1) to correspond with the coordinate system used by OriDB. Custom scripts for filtering and other analyses were written in Python. All ACS positions and scores were determined by the GIMSAN and SADMAMA motif analysis tools (Keich et al. 2008; Ng and Keich 2008).
Assigning the mapped read pairs to fragments generated by the four-cutter restriction enzymes used to fragment the insert DNA yielded 926 unique contiguous fragments. Quality score filtering and removing fragments that had a combined read count of 1 left 720 fragments (Fig. 1C; Supplemental Table 1), which assembled into 366 contigs. To improve the resolution of ARS-seq, we inferred functional cores of ARSs using a dynamic programming approach that requires each core to be at least 50 bp long and keeps track of the resulting read depths of all core segments. ARS candidates selected for manual validation are described in the Supplemental Analysis.
miniARS-seq data analysis
For more detailed descriptions of computational data analysis, see the Supplemental Analysis. Mapping of the reads was done using Bowtie and the Saccer1 version of the S. cerevisiae genome as above. The above filtering resulted in 12,338 unique miniARS fragments that were assembled into 181 unique contigs (the average contig consisted of 68 overlapping fragments) (Supplemental Table 2). Inferred functional core segments were assigned in a slightly different way than for ARS-seq, by defining the endpoints as the 0.05 quantile position of endpoints from all fragments within a contig on both sides closest to the center of the contig subject to the constraint that the resulting contig is at least 50 bp long. Manual validation targets are discussed in the Supplemental Analysis.
mutARS-seq data analysis
More detailed descriptions of computational data analysis can be found in the Supplemental Analysis. Reads were mapped using Bowtie2 (Langmead and Salzberg 2012). The reference sequence used was S288C background ARS416 (chr4, 462510-462609). Custom Python scripts were used to combine overlapping reads into a single variant sequence taking into account quality score information at each position. The enrichment value for each variant was calculated by dividing the variant counts against the counts of the wild-type allele and taking the base 2 logarithm.
Data access
The sequencing data from this study have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRA051407 (ARS-seq), SRA051408 (miniARS-seq), and SRA051406 (mutARS-seq).
Acknowledgments
I.L. was supported by F32 GM090561 and T32 HG00035. M.J.D. is a Rita Allen Scholar. This project was supported in part by grants from the National Center for Research Resources (5P41RR011823-17) and the National Institute of General Medical Sciences (8 P41 GM103533-17) from the National Institutes of Health. We thank Bonita Brewer for help with 2D gel experiments and analysis. We also thank Jay Shendure, Choli Lee, and Jacob Kitzman for help with sequencing; Douglas Fowler, Carlos Araya, and Sara Di Rienzi for help with data analysis; as well as Bonita Brewer, M.K. Raghuraman, Stanley Fields, Rupali Patwardhan, Kimberly Lindstrom, and Aaron Miller for helpful discussions and critical reading of the manuscript.
Author contributions: I.L. designed and performed the experiments, analyzed data, and wrote the paper. R.A.Y. performed experiments and analyzed data. U.K. analyzed data and wrote the paper. M.J.D. designed experiments, analyzed data, and wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.144659.112.
Freely available online through the Genome Research Open Access option.
References
- Böer E, Steinborn G, Kunze G, Gellissen G 2007. Yeast expression platforms. Appl Microbiol Biotechnol 77: 513–523 [DOI] [PubMed] [Google Scholar]
- Bolon Y-T, Bielinsky A-K 2006. The spatial arrangement of ORC binding modules determines the functionality of replication origins in budding yeast. Nucleic Acids Res 34: 5069–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Broach JR, Li YY, Feldman J, Jayaram M, Abraham J, Nasmyth KA, Hicks JB 1983. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol 47: 1165–1173 [DOI] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ 1999. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci 96: 2656–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Myers CL, Andrews B, Boone C 2011. Charting the genetic interaction map of a cell. Curr Opin Biotechnol 22: 66–74 [DOI] [PubMed] [Google Scholar]
- Dershowitz A, Snyder M, Sbia M, Skurnick JH, Ong LY, Newlon CS 2007. Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol Cell Biol 27: 4652–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Lindstrom KC, Mann T, Noble WS, Raghuraman MK, Brewer BJ 2012. Maintaining replication origins in the face of genomic change. Genome Res 22: 1940–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato JJ, Chung SCC, Tye BK 2006. Genome-wide hierarchy of replication origin usage in Saccharomyces cerevisiae. PLoS Genet 2: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S 2010. High-resolution mapping of protein sequence-function relationships. Nat Methods 7: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Lenhard B 2012. Dissecting genomic regulatory elements in vivo. Nat Biotechnol 30: 504–506 [DOI] [PubMed] [Google Scholar]
- Keich U, Gao H, Garretson JS, Bhaskar A, Liachko I, Donato J, Tye BK 2008. Computational detection of significant variation in binding affinity across two sets of sequences with application to the analysis of replication origins in yeast. BMC Bioinformatics 9: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Bhaskar A, Lee C, Chung SCC, Tye B-K, Keich U 2010. A comprehensive genome-wide map of autonomously replicating sequences in a naive genome. PLoS Genet 6: e1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Tanaka E, Cox K, Chung SCC, Yang L, Seher A, Hallas L, Cha E, Kang G, Pace H, et al. 2011. Novel features of ARS selection in budding yeast Lachancea kluyveri. BMC Genomics 12: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CA, Nieduszynski CA 2012. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res 22: 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P, Keich U 2008. GIMSAN: A Gibbs motif finder with significance analysis. Bioinformatics 24: 2256–2257 [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Knox Y, Donaldson AD 2006. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 20: 1874–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S-I, Ak P, Benham CJ, Donaldson AD 2007. OriDB: A DNA replication origin database. Nucleic Acids Res 35: D40–D46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent SA, Fenimore CM, Bostian KA 1985. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast 1: 83–138 [DOI] [PubMed] [Google Scholar]
- Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J 2009. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol 27: 1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E 2012. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 30: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC 2007. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet 39: 93–98 [DOI] [PubMed] [Google Scholar]
- Siow CC, Nieduszynska SR, Müller CA, Nieduszynski CA 2012. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res 40: D682–D686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483: 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282: 39–43 [DOI] [PubMed] [Google Scholar]
- Walker SS, Francesconi SC, Eisenberg S 1990. A DNA replication enhancer in Saccharomyces cerevisiae. Proc Natl Acad Sci 87: 4665–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bell SP 2002. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc Natl Acad Sci 99: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yanagisawa Y, Tsankov AM, Hart C, Aoki K, Kommajosyula N, Steinmann KE, Bochicchio J, Russ C, Regev A, et al. 2012. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol 13: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]