Abstract
Spontaneous DNA damage may occur nonrandomly in the genome, especially when genome maintenance mechanisms are undermined. We developed single-strand DNA (ssDNA)–associated protein immunoprecipitation followed by sequencing (SPI-seq) to map genomic hotspots of DNA damage. We demonstrated this method with Rad52, a homologous recombination repair protein, which binds to ssDNA formed at DNA lesions. SPI-seq faithfully detected, in fission yeast, Rad52 enrichment at artificially induced double-strand breaks (DSBs) as well as endogenously programmed DSBs for mating-type switching. Applying Rad52 SPI-seq to fission yeast mutants defective in DNA helicase Pfh1 or histone H3K56 deacetylase Hst4, led to global views of DNA lesion hotspots emerging in these mutants. We also found serendipitously that histone dosage aberration can activate retrotransposon Tf2 and cause the accumulation of a Tf2 cDNA species bound by Rad52. SPI-seq should be widely applicable for mapping sites of DNA damage and uncovering the causes of genome instability.
Some regions of the eukaryotic genomes are more vulnerable than others when facing challenges to DNA integrity. In the human genome, special loci called fragile sites become prone to breakage when cells are treated with aphidicolin, a DNA polymerase inhibitor (Glover et al. 1984). In the budding yeast Saccharomyces cerevisiae, lowering DNA polymerase activity led to heightened levels of translocations at two hotspots, each composed of a pair of yeast retrotransposons (Ty elements) (Lemoine et al. 2005). It was recently shown that these and several other Ty elements are translocation hotspots even in cells with normal DNA polymerase activity (Chan and Kolodner 2011, 2012). The discoveries of at-risk genomic sites correlate with the findings that specific mechanisms have evolved for their protection. For example, in S. cerevisiae, DNA helicases Rrm3 and Pif1 promote replication through hard-to-replicate sites and thus prevent fork stalling that may potentially cause DNA damage (Ivessa et al. 2003; Paeschke et al. 2011).
A complete understanding of the extent of genome fragility and the molecular machineries involved in protecting at-risk sites requires systematic mapping of genomic hotspots of DNA damage. This is made possible by the advent of high-throughput technologies such as microarray and second-generation sequencing. In particular, chromatin immunoprecipitation (ChIP) followed by microarray (ChIP-chip) analyses of a commonly used double-strand break (DSB) marker, phosphorylated histone H2A(X), have revealed in wild-type budding yeast and fission yeast cells the genome-wide distribution of sites prone to triggering phosphorylation of H2A(X) by primary DNA damage checkpoint kinases (Rozenzhak et al. 2010; Szilard et al. 2010).
In both budding yeast and fission yeast, Rad52 (also known as Rad22 in the fission yeast Schizosaccharomyces pombe) is essential for DSB repair and homologous recombination (Mortensen et al. 2009). Nuclear foci formed by fluorescent protein-tagged Rad52 have been widely used to assess the level of DNA damage in live cell imaging analysis (Lisby et al. 2001; Du et al. 2003). Unlike phospho-H2A(X), which is not found at the immediate vicinity of DSBs due to end resection (Shroff et al. 2004), Rad52 is recruited to single-strand DNA (ssDNA) exposed by resection, in a replication protein A (RPA)–dependent manner (Lisby et al. 2004). Besides DSBs, Rad52 may bind to other ssDNA-containing DNA structures, such as single-strand gaps, which can also initiate recombination (Lettier et al. 2006). Thus, mapping Rad52 binding sites is expected to be a complementary approach to phospho-H2A(X) location analysis. In this report, we present a method called SPI-seq (pronounced “Spy-seq,” for ssDNA-associated protein immunoprecipitation followed by sequencing) and its application to genome-wide analysis of Rad52 binding sites in fission yeast. Importantly, SPI-seq reveals not only the locations, but also the strand specificity of Rad52 binding, thereby providing extra information on the nature of DNA lesions that lead to repair protein recruitment.
Results
Rad52 enrichment patterns at DSBs
We developed the SPI-seq procedure for two reasons. First, because Rad52 preferentially associates with ssDNA, the adaptor ligation method used in standard ChIP-seq protocols, which involves end repair and dA-tailing of double-strand DNA (dsDNA), is not suitable for Rad52 binding site analysis. Moreover, ssDNA binding by Rad52 often has strand specificity, which, if known, may help deducing the initiating event that leads to ssDNA exposure and Rad52 recruitment.
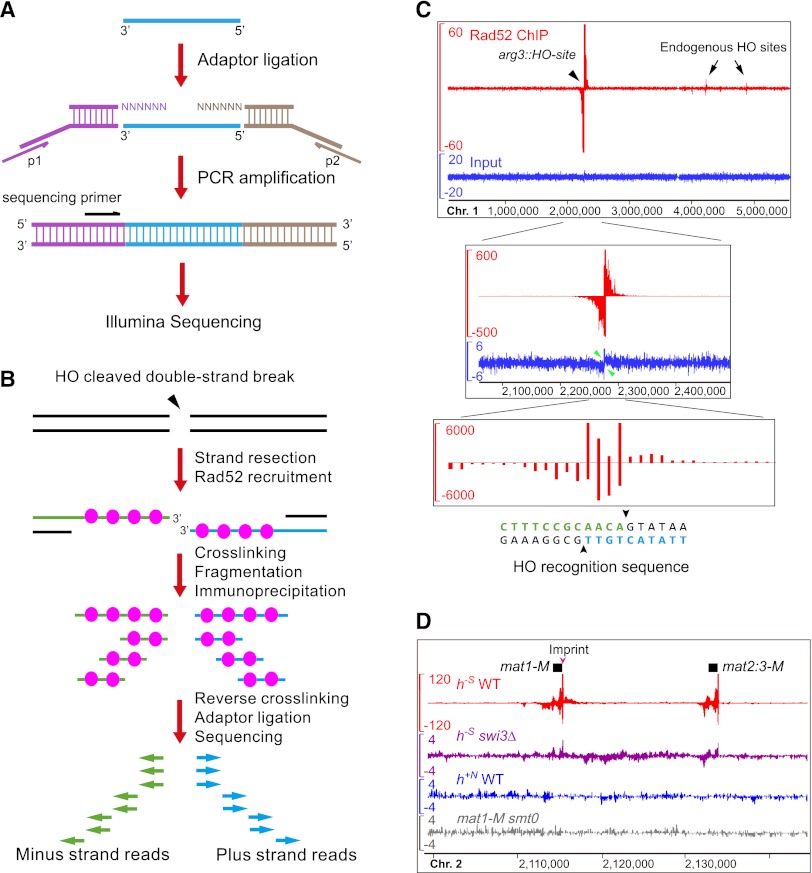
The key design of SPI-seq is an adaptor-ligation scheme based on methods used for tagging the 3′ end of single-strand cDNA (Shibata et al. 2001) and the 5′ end of single-strand RNA (Clepet et al. 2004). In this scheme (Fig. 1A), prior to adaptor addition, all DNA fragments are heat denatured to become single-stranded. Two adaptors, one with a 5′ overhang composed of six random nucleotides (N6), and the other with a 3′ N6 overhang, are ligated to the ends of the ssDNA using T4 DNA ligase. This design preserves the strand polarity information because the 3′ end of ssDNA can only be ligated to the adaptor with the 3′ overhang, and the 5′ end can only be ligated to the adaptor with the 5′ overhang. We incorporated the Illumina single-end sequencing primer sequence into the adaptor tagging the 3′-end. As DSB resection generates 3′-ended ssDNA, sequencing from the 3′ end allows capturing the sequences immediately adjacent to the DSB.
Figure 1.
SPI-seq assay detected Rad52 bound at artificially induced and naturally occurring double-strand breaks (DSBs). (A) An adaptor-ligation scheme specially designed for single-strand DNA (ssDNA). (B) The expected outcome of SPI-seq when applied to an HO-induced DSB. Because of our adaptor-ligation design, the chromatin immunoprecipitation (ChIP)–enriched sequencing reads should map to the strands opposite to the ones bound by Rad52. (C) SPI-seq analysis of HO-induced DSBs. The upper and middle panels show the same read density profiles generated by kernel density estimation. (Upper panel) The whole chromosome 1. The y-axis scale of the ChIP signal track is limited to 60 to allow the visualization of the weak signals at the endogenous HO sites. (Middle panel) Close-up view of arg3∷HO-site and its surrounding region. The ChIP signal track shows the full height of the ChIP signal at arg3∷HO-site. Green arrowheads point to the HO-site-flanking regions where strand-specific signal loss occurred in the input, presumably due to resection. (Lower panel) ChIP-enriched reads mapped to individual nucleotide positions at the HO cleavage site, without kernel smoothing. (D) SPI-seq signals at mating-type loci. To visualize reads mapped to the duplicated mating-type cassette, nonuniquely aligned reads were randomly assigned. The units on the y-axes in C and D are reads per 10 million.
As a proof-of-principle test, we first analyzed Rad52 enrichment at an artificial DSB. Expressing the budding yeast HO endonuclease in the presence of a 24-nucleotide (nt) HO recognition sequence inserted at the arg3 locus (arg3∷HO-site) on fission yeast chromosome 1 resulted in a persistent DSB and eventual cell death (Du et al. 2003, 2006). The fact that a mutation of the HO recognition sequence renders the cell resistant to continuous HO expression suggests that no other sequences can be efficiently cut by HO in the fission yeast genome (Li et al. 2012). Based on our SPI-seq design, we expected that ChIP-enriched sequencing reads mapped to the right side of the DSB should be in forward orientation (displayed as plus strand reads on a genome browser), whereas reads mapped to the left side of the DSB should be in reverse orientation (displayed as minus strand reads on a genome browser) (Fig. 1B).
By use of strains expressing epitope-tagged Rad52, we performed SPI-seq on cells suffering HO-induced DSBs. As a control, DNA from the input was processed using the same procedure. Sequencing reads from the input were found evenly distributed along all three chromosomes on both strands, suggesting that our adaptor ligation scheme did not introduce any gross bias (Fig. 1C; Supplemental Fig. S1A). In contrast, reads from the ChIP sample showed strong and specific enrichment signals at arg3∷HO-site. As expected, Rad52-bound DNA exhibited clear strand polarity, with plus-strand SPI-seq signals on the right and minus-strand signals on the left. The extensive spreading of the ChIP signal probably resulted from continuous DNA resection during the many hours between HO cutting and sample collection. At the single-nucleotide resolution, the plus-strand and minus-strand ChIP-enriched reads overlapped by 4 nt, matching precisely the 3′-overhangs generated by HO (Fig. 1C, bottom panel).
In the HO-expressing strain, besides arg3∷HO-site, we unexpectedly found four other sites with detectable Rad52 binding, two on chromosome 1 and two on chromosome 3 (Fig. 1C, Supplemental Fig. S1A,B). The SPI-seq signal patterns at these sites match that of a DSB, indicating there are endogenous sequences cleavable by HO, albeit only inefficiently. The high-resolution, strand-specific SPI-seq data allowed us to pinpoint the locations of these endogenous HO cleavage sites, even though the enriched signals were as low as 0.36% of that at arg3∷HO-site (Supplemental Fig. S1C). In all four cases, the cleavage site sequences conform to the consensus HO recognition sequence (Supplemental Fig. S1D; Nickoloff et al. 1990). The low efficiency of HO cutting at these sites may be due to either less-than-ideal recognition sequence or prohibitive chromatin structure, as it is known that in the budding yeast, HO cannot cleave the recognition sequences at the silent donor loci HML and HMR (Strathern et al. 1982).
Homothallic fission yeast cells undergo mating-type switching when a strand discontinuity (called an “imprint”) at the mat1 locus causes the formation of a one-ended DSB during replication (Arcangioli et al. 2007). This DSB then initiates recombination between mat1 and the homologous sequences at one of the silent donor loci, mat2-P or mat3-M. The reference genome of S. pombe is that of a heterothallic h−S strain. In an h−S strain, there is only one donor locus, mat2:3-M, which harbors the same “minus” cassette as the one at mat1, thus its mating type stays unchanged despite constant DSB formation and recombination (Beach and Klar 1984). When we applied SPI-seq to an h−S strain, we observed, as expected, Rad52 binding at both mat1-M and mat2:3-M (Fig. 1D; Supplemental Fig. S2A). On the right side of the mat1 DSB, as observed at the HO-induced DSBs, the ChIP signal showed plus-strand polarity, presumably due to resection-exposed ssDNA. On the left side of the mat1 DSB, we observed the enrichment of not only minus-strand reads, as we saw on the left side of HO-induced DSBs, but also plus-strand reads, likely reflecting the association of Rad52 with recombination intermediates, such as the displaced strand of the extended displacement loop (D-loop) or the newly synthesized DNA unwound from the extended D-loop (Supplemental Fig. S2B).
The establishment of the mat1 imprint requires both cis-acting DNA elements (smt) and trans-acting switch (swi) genes (Arcangioli et al. 2007). Consistent with the essential role of the imprint for inducing the DSB and recombination, Rad52 binding at the mating loci was not observed in an smt0 strain and was only barely noticeable in an h−S swi3Δ strain (Fig. 1D). Interestingly, no Rad52 binding at the mating loci was observed in heterothallic h+N strains (Fig. 1D). It was reported that Leupold's strain 975, from which h+N laboratory strains originated, had a very low imprinting level (Beach and Klar 1984). The exact cause of the imprinting defect of strain 975 is unknown.
Except for the mating-type region in h−S strains, we did not detect any significant Rad52 binding in the wild-type genome. We hypothesized that we might discover sites vulnerable to DNA damage by applying SPI-seq to mutants with heightened levels of genomic instability. In the following sections, we present two such applications.
Rad52 enrichment patterns in pfh1 helicase mutants
In fission yeast, Pfh1 is the sole homolog of budding yeast Rrm3 and Pif1 and is essential for maintaining both the nuclear genome and the mitochondrial genome (Pinter et al. 2008). As shown recently by two-dimensional gel analysis, in the nucleus, Pfh1 prevents replication fork stalling at hard-to-replicate sites, including RNA polymerase III (Pol III) transcribed tRNA and 5S rRNA genes (Sabouri et al. 2012; Steinacher et al. 2012). To catalog sites protected by Pfh1 in a genome-wide manner, we analyzed Rad52 distribution in two cold-sensitive mutants of pfh1, pfh1-R20 and pfh1-R23 (Tanaka et al. 2002; Ryu et al. 2004). In the R20 mutant, at the restrictive temperature of 20°C, dozens of Rad52 binding peaks were found throughout the genome, with the strongest ones near the centromeres of chromosomes 2 and 3 (Fig. 2; Supplemental Fig. S3). The R23 mutant did not have as many conspicuous Rad52 peaks. Instead, we found a dramatic enrichment of Rad52 at the mating-type region (Supplemental Fig. S3), probably as a result of failed attempts to repair the mat1 DSB (the pfh1 strains used in the analysis were h−S). Consistent with the SPI-seq results, we found that in the homothallic h90 strain background, R23 but not R20 showed strong mating-type switching defect (Supplemental Fig. S4). This phenotype of R23 resembles that of class II swi mutants (Arcangioli et al. 2007), suggesting the possibility that pfh1 may act together with the class II swi genes in resolving the recombination intermediates during mating-type switching.
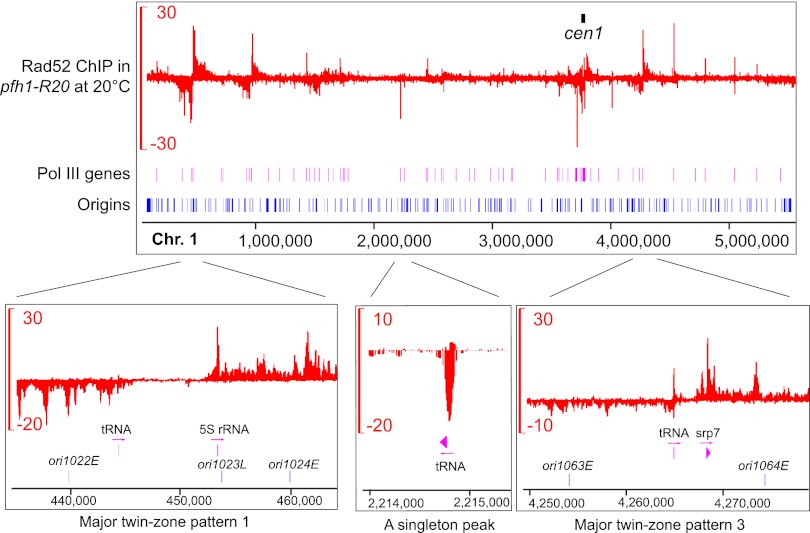
Figure 2.
Rad52 enrichment patterns in pfh1-R20. Rad52 SPI-seq read distribution along chromosome 1 in pfh1-R20 at 20°C. The units on the y-axes are reads per 10 million. The positions and naming of replication origins are as described by Hayashi et al. (2007).
The SPI-seq results indicate that there are a large number of DNA lesion hotspots in pfh1-R20 at the restrictive temperature. Thus, we carried out in-depth analysis of the Rad52 binding patterns in this mutant. Apart from those found at the centromeres and the mating-type region, the Rad52 enrichment signals in pfh1-R20 can be classified into two types (Fig. 2; Supplemental Fig. S3). Characteristic of the first type are two closely spaced Rad52-binding zones on opposite strands, with minus-strand signal on the left and plus-strand signal on the right. There are 11 sites displaying this type of signals, which we named the twin-zone patterns, and based on signal intensity, they are further classified as major twin-zone patterns or minor twin-zone patterns. Within each major twin-zone pattern, a gap devoid of Rad52 enrichment, with a size ranging from 3 kb to 22 kb, can be always found between the two enrichment zones. Belonging to the second type are isolated peaks with signals mainly on one strand, and we call them singleton peaks. The ChIP signals of the seven major twin-zone patterns spread tens of kbs on either side, whereas the minor twin-zone patterns and singleton peaks tend to be much narrower.
To understand the nature of DNA lesion hotspots in pfh1-R20, we visually examined the relationship between Rad52 binding sites and other genomic features. Remarkably, Rad52 binding sites are almost always associated with Pol III–transcribed genes (for brevity, hereafter referred to as Pol III genes). In S. pombe, there are a total of 206 Pol III genes scattered throughout the nuclear genome, including 171 tRNA genes, 33 5S rRNA genes, the U6 snRNA gene snu6, and the 7SL RNA gene srp7 (Hamada et al. 2001; Marck et al. 2006). Within each major twin-zone pattern, we could find two Pol III genes flanking the Rad52-free gap (Fig. 2; Supplemental Table S1). At the minor twin-zone patterns and singleton peaks, a Pol III gene can be often found close to the positions where Rad52 ChIP signal reaches a maximum (Fig. 2; Supplemental Fig. S3). In addition, the levels of Rad52 enrichment at centromere-flanking regions largely correlate with the numbers of the Pol III genes near these regions (Supplemental Fig. S5). Thus, DNA lesion hotspots in pfh1-R20 are intimately related to Pol III genes, consistent with the known role of Pfh1 in overcoming replication block at Pol III genes (Sabouri et al. 2012; Steinacher et al. 2012).
We hypothesized that the extensive spreading of ChIP signal at major twin-zone patterns may result from two Pol III genes acting synergistically together. To test this idea, we deleted the left-side Pol III gene at major twin-zone pattern 1. As predicted, the deletion not only abolished Rad52 enrichment on the left side but also strongly reduced the intensity and the spreading of ChIP signals on the right side, essentially converting the major twin-zone pattern to a singleton peak (Supplemental Fig. S6).
The narrower Rad52 distribution at singleton peaks and minor twin-zone patterns rendered them amenable for software-based peak detection. Thus, we searched for peaks outside of centromere-flanking regions, the mating-type region, and regions containing major twin-zone patterns with the software MACS (Zhang et al. 2008). Using a P-value cutoff of 10−7, we found 53 peaks with specific Rad52 enrichment in pfh1-R20 (see Supplemental Methods). Consistent with our visual impression, 39 (73%) of these peaks overlap with Pol III genes, including 32 singleton peaks and seven peaks associated with minor twin-zone patterns, and 11 out of the 14 remaining peaks are also adjacent to Pol III genes (Supplemental Table S2). Permutation tests showed a significant association of these peaks with Pol III genes (P < 0.001), but not with sn/sno RNA genes, highly transcribed protein-coding genes, or predicted G-quadruplexes (Fig. 3A). The peaks are not associated with the COC loci, where TFIIIC but not Pol III binds (Noma et al. 2006). We did observe a significant association of the peaks with replication origins, but this may be a secondary consequence of the association of Rad52 peaks with Pol III genes, as a great majority of peak-associated origins are adjacent to Pol III genes (Fig. 3B).
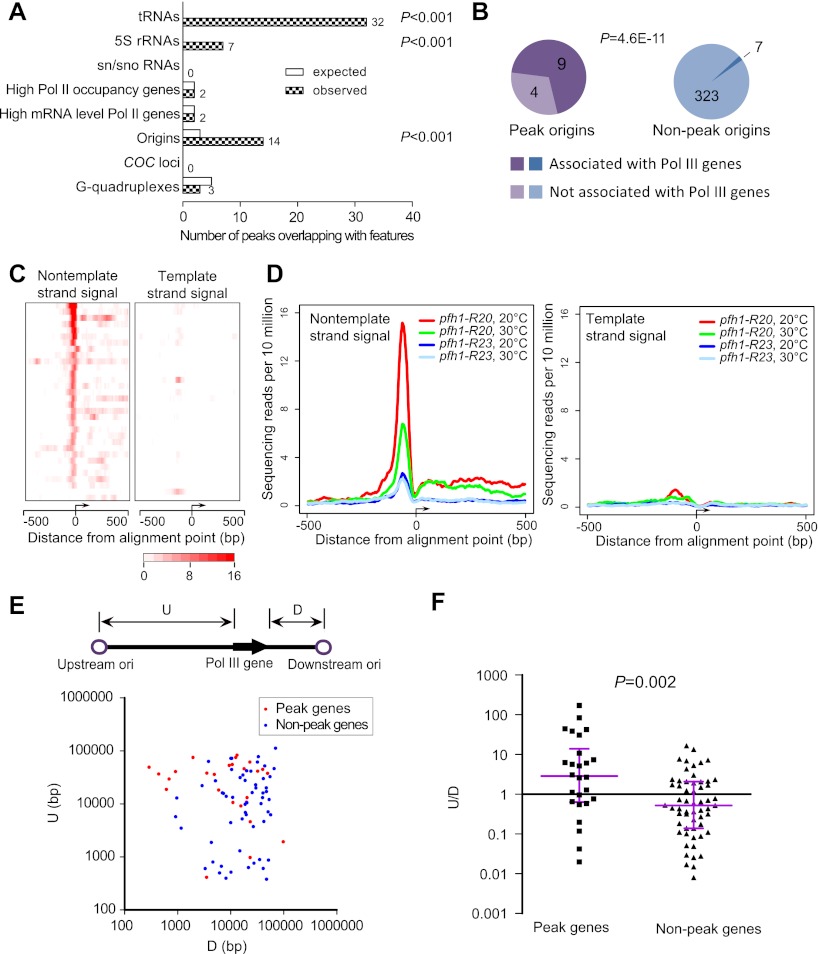
Figure 3.
Analysis of the narrow Rad52 peaks in pfh1-R20. (A) Rad52 peaks identified by MACS were analyzed to detect their associations with known genomic features. The numbers of observed peaks overlapping with genomic features are listed. For details, see Supplemental Methods. (B) Origins that overlap with Rad52 peaks are enriched with those close to Pol III genes. An origin is deemed associated with a Pol III gene if it is within 300 bp from a Pol III gene. P-value was determined by Fisher's exact test. (C) Heatmap representations of the SPI-seq signals at the 32 singleton peaks overlapping with Pol III genes. The peaks are aligned according to the Pol III gene transcription orientation, with the first nucleotide of the mature RNAs as the alignment point. (D) Average plots of Rad52 SPI-seq signals at the 32 singleton peaks overlapping with Pol III genes. The peaks are aligned as in C. (E) A scatter plot of the distances between Pol III genes and their neighboring origins. The distance from the 5′ end of a Pol III gene to its upstream origin was denoted as its “U” value, while the distance from the 3′ end to the downstream origin as its “D” value. Peak genes refer to the Pol III genes overlapping with the singleton peaks. To reduce errors caused by imprecise origin mapping, we did not include the genes whose distance to the closest origin is <250 bp. (F) The ratios of U/D for peak and nonpeak Pol III genes were plotted. P-value was obtained by comparing the log-transformed ratios of the two groups using Welch's t-test.
For the 39 peaks overlapping with Pol III genes, the peak polarity highly correlates with the transcription direction of the Pol III genes. The SPI-seq signals of 36 peaks are mainly on the nontemplate strands (P = 3.61 × 10−8, McNemar's test) (Supplemental Table S2). To directly visualize this correlation, we displayed as heatmaps the ChIP signals of the 32 singleton peaks overlapping with Pol III genes (Fig. 3C). Averaging the signals from these peaks also revealed the same trend (Fig. 3D). We suspect this correlation is related to the observations that in Pfh1-depleted cells tRNA genes became polar fork barriers that caused stronger elevation of replication pausing and recombination when replication and transcription move in opposite directions (Sabouri et al. 2012; Steinacher et al. 2012).
There are a total of 109 Pol III genes in the genomic regions searched by MACS, and more than half of them neither overlap nor adjoin Rad52 peaks. As these nonpeak genes are occupied by Pol III to the same extent as the peak genes (Noma et al. 2006), lack of Rad52 enrichment cannot be attributed to lower Pol III occupancy. When we compared gene–origin distances for the nonpeak genes versus singleton-peak genes, we found that the latter tend to be closer to the downstream replication origins, whereas the former tend to be closer to the upstream origins (Fig. 3E,F). Thus, the selective binding of Rad52 at peak genes may also be due to the polar fork blocking effect of Pol III genes (see Supplemental Discussion).
The confined and largely unimodal distribution of SPI-seq signals at the singleton peaks allowed us to assess where in a Pol III gene Rad52 enrichment preferentially occurs. As seen in the heatmap and the average plot (Fig. 3C,D), the highest levels of Rad52 enrichment were observed at a position just 5′ to the mature RNA sequence, where Pol III initiation complex binds (Hamada et al. 2001). Thus, we propose that it is the Pol III initiation complex or the poised polymerase complex, rather than the transcribing Pol III machinery, that poses the strongest threat to incoming replication fork when Pfh1 function is compromised. This model is consistent with the report that in S. cerevisiae, fork pausing at tRNA genes is due to the presence of the transcription initiation complex rather than the act of transcription (Ivessa et al. 2003).
Replication-associated Rad52 enrichment patterns in the hst4 mutant
Acetylation at histone H3 lysine 56 (H3K56Ac) preferentially occurs during S phase and in response to DNA damage in yeasts and human (Costelloe and Lowndes 2010). In S. pombe, H3K56 acetylation requires the acetyltransferase Rtt109 (Xhemalce et al. 2007). Deacetylation of H3K56Ac is mediated by the sirtuin-family deacetylase Hst4. Loss of Hst4 caused growth defect and hypersensitivity to genotoxic agents (Haldar and Kamakaka 2008). To better understand the consequences of H3K56 hyperacetylation, we examined the Rad52 enrichment profile in hst4Δ by SPI-seq.
In hst4Δ, Rad52 SPI-seq signal appeared as dozens of distinct peaks on each chromosome (Fig. 4A), with half-peak widths in the range of kbs to tens of kbs, significantly wider than the singleton peaks seen in pfh1-R20. Visual inspection revealed that peak locations correlate with replication origins in a specific manner; namely, the summit of a plus-strand peak is often found in the left vicinity of the nearest origin, whereas the summit of a minus-strand peak is usually seen in the right vicinity of the nearest origin. Furthermore, the high-intensity peaks seem to reside preferentially within the longer inter-origin intervals. Computational analysis confirmed the visual impression (Fig. 4B). In average plots, Rad52 enrichment is more pronounced for longer inter-origin intervals, within which the ChIP signal reaches its height near one of the flanking origins—the minus-strand peaks near the left origin and the plus-strand peaks near the right origin. The Rad52-bound DNA in hst4Δ was most likely single-stranded, as we detected similar enrichment patterns of RPA in this mutant (Supplemental Fig. S7).
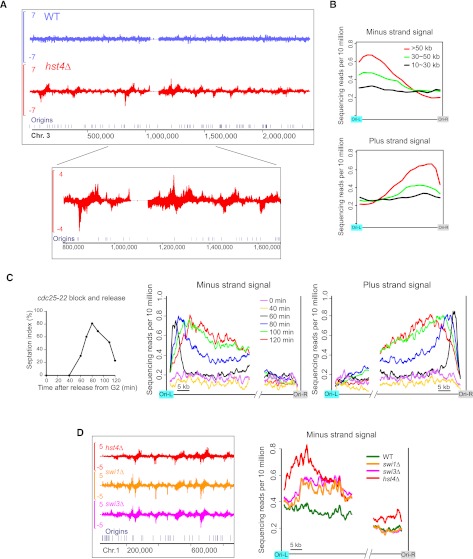
Figure 4.
Replication-coupled Rad52 enrichment patterns in hst4Δ. (A) Rad52 SPI-seq read distribution along chromosome 3. (B) Averaged SPI-seq signal within inter-origin regions of different sizes. Inter-origin regions were classified into three groups according to their sizes. We divided each inter-origin region to 100 bins of equal sizes. The average read number within each bin was calculated, and a sliding window with a window size of 10 bins and a step size of five bins was applied to smooth the curves. (C) Rad52 SPI-seq analysis of hst4Δ cells synchronously released into cell cycle from a G2 arrest. The septation index (percentage of septated cells) shown on the left was measured with Calcofluor staining. The sequencing reads from inter-origin regions >50 kb were used to draw the average plots shown on the right. We first calculated the average read number at each nucleotide position within the first 35 kb and the last 15 kb of the inter-origin regions, and then applied a sliding window with a window size of 1 kb and a step size of 0.1 kb to smooth the curves. (D) Rad52 SPI-seq analysis of swi1Δ and swi3Δ. Average plots were drawn as in C. In A and D, a bandwidth of 500 bp was used for kernel density estimation, and the units on the y-axes of the browser views are reads per 10 million.
To examine whether Rad52 binding is influenced by cell cycle phase, we synchronized cells in G2 using the cdc25-22 temperature-sensitive allele and monitored Rad52 enrichment at different time points after the release from G2 arrest (Fig. 4C). In S. pombe, the onset of septation (formation of the division septum) coincides with the beginning of S phase. Thus, we used the septation index as an indicator of cell cycle progression, which shows that cells began to enter S phase sometime between 40 and 60 min after G2 release. No Rad52 enrichment was detected at pre-S-phase time points (0–40 min), and Rad52 enrichment patterns emerging at early S-phase time points (60–80 min) were sharper and closer to the replication origins than those found at later time points (100–120 min), suggesting that Rad52 binding occurs during replication and extends away from the origin as replication progresses.
The replication-associated Rad52 accumulation in hst4Δ prompted us to examine two other mutants with a replication-related defect, swi1Δ and swi3Δ. Fission yeast Swi1 and Swi3 proteins (orthologs of Tof1 and Csm3 in budding yeast, and Timeless and Tipin in mammals, respectively) form a fork protection complex, and their mutants accumulate Rad52 foci during unperturbed replication, probably as a consequence of ssDNA gaps formed near replication forks (Noguchi et al. 2004). In swi1Δ and swi3Δ, we found similar Rad52 peak patterns as those in hst4Δ (Fig. 4D). Thus, fork-associated ssDNA may be a common cause of Rad52 accumulation in these mutants (see Supplemental Discussion). The Rad52 peaks were at positions closer to the origins in hst4Δ than in swi1Δ and swi3Δ, implying that forks may suffer earlier or more frequent problems in hst4Δ.
Epistasis analysis showed that the Rad52 enrichment pattern in hst4Δ is due to H3K56 hyperacetylation mediated by Rtt109, as deleting rtt109 eliminated the Rad52 pattern (Supplemental Fig. S8A), and so did H3K56R mutation (Supplemental Fig. S8B). The K56A and K56Q mutations that mimic the charge state of acetylated lysine had the same effect, suggesting that the functional consequence of K56 acetylation is not merely due to charge neutralization (Supplemental Fig. S8B).
In S. cerevisiae, genetic analysis suggested that Rtt101, Mms22, and Mms1, which are components of a potential E3 ubiquitin ligase complex, act downstream from H3K56Ac (Collins et al. 2007). In S. pombe, Mms22 (also known as Mus7) and Mms1 function together in genome stability maintenance (Dovey and Russell 2007; Yokoyama et al. 2007; Dovey et al. 2009). It was recently shown that mms1Δ can suppress the growth defect of hst4Δ (Vejrup-Hansen et al. 2011). We found that mms22Δ suppressed the Rad52 enrichment pattern of hst4Δ (Supplemental Fig. S8C), suggesting that in S. pombe, the Mms22–Mms1 complex also acts as a downstream effector of H3K56Ac. Consistent with this model, we found that the synthetic lethality/sickness of the hst4Δ cds1Δ double mutant can be suppressed by either rtt109Δ or mms22Δ (Supplemental Fig. S8D).
Histone dosage aberration causes retrotransposon Tf2 activation and the accumulation of a Tf2 cDNA species bound by Rad52
In fission yeast, there are three pairs of histone H3-H4 genes (hht1-hhf1, hht2-hhf2, and hht3-hhf3). To generate the histone H3K56 point mutations shown in Supplemental Figure S8B, we employed a strain in which two of the gene pairs hht1-hhf1 and hht3-hhf3 were deleted (hereafter referred to as Δ1Δ3) (Mellone et al. 2003). Consistent with the observations that the Δ1Δ3 strain lacks any overt growth defect or chromosome instability (Mellone et al. 2003), we did not observe in this strain any notable Rad52 enrichment patterns when uniquely mapped reads were displayed on a genome browser (Fig. 5A). However, when nonuniquely mapped reads were randomly assigned and visualized, we were surprised to see strong strand-specific enrichment of Rad52 at the 13 Tf2 retrotransposons, which share almost identical DNA sequences (Fig. 5A). Tf2 belongs to the Ty3/Gypsy family of long terminal repeat (LTR)–containing retrotransposons and is the only type of transposable elements capable of remobilization in standard S. pombe strains (Bowen et al. 2003). ChIP-enriched reads can also be aligned to the Tf-fragment 1 element, and a number of solo LTRs, probably due to their sequence similarity to the full-length Tf2. Thus, our SPI-seq analysis fortuitously identified a novel connection between histone genes and Tf2 retrotransposon.
Figure 5.
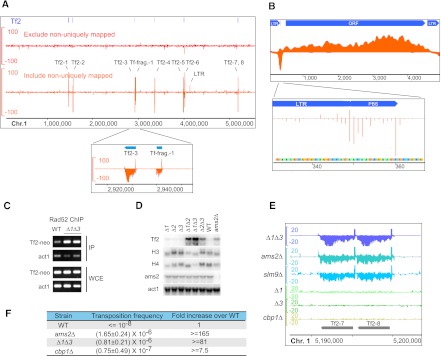
Rad52 enrichment on retrotransposon cDNA in histone dosage mutants. (A) Rad52 SPI-seq read distribution along chromosome 1 in Δ1Δ3. Enrichment at Tf2 retrotransposons was detected when nonuniquely aligned reads were randomly assigned and visualized. (B) Rad52 SPI-seq reads from Δ1Δ3 were aligned to a full-length Tf2 sequence (GenBank L10324.1). (Bottom panel) The unaveraged read numbers at primer-binding site (PBS). (C) ChIP-PCR detection of Rad52 enrichment on Tf2-12-neoAI. (D) Northern blotting analysis of Tf2 transcription in H3-H4 gene deletion mutants and ams2Δ. (E) Rad52 SPI-seq analysis of histone deletion mutants and mutants with higher-than-normal Tf2 expression. The tandem-linked Tf2-7 and Tf2-8 and their surrounding regions are shown. (F) Tf2 transposition frequency determined using Tf2-12-neoAI. The standard deviation was obtained from four independent cultures. The units on the y-axes of the browser views in A and E are reads per 10 million.
If Rad52 binding occurred on the Tf2 genomic DNA in the Δ1Δ3 strain, we would expect to see some ChIP signal at Tf2-flanking regions. However, virtually no enriched reads could be detected on the Tf2-flanking genomic DNA (Fig. 5A). Thus, Rad52 most likely bound to Tf2 cDNA. Furthermore, within the internal region between the two LTRs, SPI-seq signals were found exclusively on the nontemplate strand except for the primer-binding site (PBS), where a strong peak was seen on the template strand (Fig. 5B). These observations, together with the fact that the signals on the LTRs were significantly lower than those in the internal region, led us to propose that the Rad52-bound cDNA may represent a reverse transcription intermediate, in which the LTR region is double-stranded and the rest of the cDNA remains single-stranded (Supplemental Fig. S9).
To verify the SPI-seq results, we performed ChIP-PCR analysis using strains in which one Tf2 is tagged with a neo marker (Sehgal et al. 2007). Indeed, in Δ1Δ3, we detected Rad52 enrichment on the neo marker gene (Fig. 5C).
In budding yeast, histone gene deletions cause the increase of retrotransposon Ty1 transcription (Nyswaner et al. 2008). To test the possibility that Tf2 transcription in Δ1Δ3 may be altered, we performed Northern blotting analysis (Fig. 5D). Tf2 transcripts were hardly detectable in strains in which none or only one of the three pairs of H3-H4 genes was deleted, yet their abundance increased greatly in all three mutants with two pairs of H3-H4 genes deleted, especially Δ1Δ3.
The GATA-type transcription factor Ams2 is required for S-phase activation of H3 and H4 genes in fission yeast (Takayama and Takahashi 2007). Thus, it was no surprise that we observed up-regulation of Tf2 transcription in ams2Δ, too (Fig. 5D). Together, these findings suggest that Tf2 silencing in fission yeast requires normal levels of histone expression.
To see whether Tf2 derepression is always associated with Rad52 binding to Tf2 cDNA, we carried out SPI-seq analysis on ams2Δ, as well as two other mutants defective in Tf2 silencing, the HIRA histone chaperone mutant slm9Δ and the CENP-B mutant cbp1Δ (Greenall et al. 2006; Cam et al. 2008). In both ams2Δ and slm9Δ, we observed Rad52 enrichment on Tf2, with patterns similar to those in Δ1Δ3 (Fig. 5E). Surprisingly, no Rad52 enrichment on Tf2 was observed for cbp1Δ (Fig. 5E). ChIP-qPCR analysis confirmed the SPI-seq results (Supplemental Fig. S10A). The lack of Rad52 binding to Tf2 cDNA in cbp1Δ correlated with a reduced accumulation of Tf2 cDNA, even though the level of Tf2 RNA was similar to that of the other mutants (Supplemental Fig. S10B,C). The reason for the unique phenotype of cbp1Δ remains to be determined.
Pronounced binding of Rad52 to Tf2 cDNA is associated with a dramatic increase of transposition, as we found that the Tf2 transposition frequencies in Δ1Δ3 and ams2Δ were more than 80-fold higher than that in the wild type, and more than 10-fold higher than that in cbp1Δ (Fig. 5F). Tf2 mobilization is often mediated by integrase-independent recombination between its cDNA and pre-existing genomic copies of Tf2 (Hoff et al. 1998; Sehgal et al. 2007). We speculate that the binding of Rad52 to an immature form of Tf2 cDNA may contribute to this main mode of Tf2 transposition.
Discussion
We have developed a method for revealing genome-wide locations of DNA repair proteins associated with ssDNA. By use of this method, we identified specific Rad52 accumulation patterns in several different mutants and thus generated high-resolution global views of DNA lesion hotspots emerging upon the impairment of genome maintenance factors.
Comparison with other methods that can reveal DNA lesion hotspots
There are two types of assays that can reveal DNA lesion hotspots. The first type of assays maps the breakpoint junctions of chromosomal rearrangements. Because rearrangements are repair products of DNA lesions, these methods indirectly infer the locations of the initiating lesions. In budding yeast, such an approach has led to the discovery of the Ty element–associated fragile sites (Lemoine et al. 2005). One important method in this category is the gross chromosomal rearrangement (GCR) assay developed by the Kolodner laboratory, which is based on the selection against two markers near the left end of chromosome V in budding yeast (Chen and Kolodner 1999). The rearrangement breakpoints in hundreds of GCR-containing strains have been mapped, although doing such mapping one strain at a time is a time- and cost-consuming endeavor (Putnam et al. 2005; Chan and Kolodner 2012).
A limitation of the growth selection-based rearrangement detection systems is the difficulty of capturing rearrangement events that compromise cell growth. In addition, the locations of the selection markers may affect the types of rearrangement that can be recovered. For example, rrm3 mutant has the same GCR rate as the wild type despite the known role of Rrm3 in preventing replication stalling at hard-to-replicate sites, possibly due to the lack of such sites in the 12-kb region monitored by the GCR assay (Schmidt and Kolodner 2006). Recently, deep sequencing-based methods have been employed to simultaneously identify tens of thousands of translocation junctions without growth selection (Chiarle et al. 2011; Klein et al. 2011). These approaches rely on an artificial DSB to induce and map translocations and thus are still not bias free.
Our SPI-seq method belongs to the second type of approaches, which directly monitors the physical locations of DNA damage and thus does not depend on DNA repair outcomes. Microarray-based mapping of ssDNA has been achieved through random priming without denaturation (Feng et al. 2006) or benzoyl naphthoyl DEAE (BND) cellulose enrichment (Buhler et al. 2007). DSBs have been mapped by microarray using an end-labeling procedure (Feng et al. 2011). These DNA-targeted assays are prone to in vitro artifacts and noises due to DNA denaturation and breakage. New protocols that lyse cells and label DNA in agarose plugs minimize but may not completely eliminate these problems (Feng et al. 2011).
Unlike assays directly targeting DNA, SPI-seq and other ChIP-based methods indirectly capture lesion-associated DNA by enriching proteins bound at lesion sites. These methods are relatively insensitive to in vitro manipulations that may perturb DNA conformations, as the protein–DNA interaction is fixed by formaldehyde prior to cell lysis. Because lesion-associated proteins vary in their binding specificity and distribution patterns, ChIP data reflect not only the locations of the DNA lesions but also the properties of the protein being enriched. For example, phospho-H2A(X) is subject to many factors influencing the distribution and activities of the ATM/ATR checkpoint kinases, and thus it remains uncertain whether all sites enriched for phospho-H2A(X) correspond to DSBs.
Due to superior resolution and lower cost, ChIP-seq is replacing ChIP-chip to become the method of choice for location analysis of DNA-binding proteins. ssDNA-binding proteins pose a challenge for ChIP-seq because routine sequencing library preparation procedures cannot ligate adaptors to ssDNA. The strand polarity of ssDNA offers an extra dimension of spatial information that, to our knowledge, cannot be captured by any existing approaches. Our SPI-seq method is tailor-designed for ssDNA-binding proteins, and thus can report the true locations of ssDNA, and more importantly, reveal strand polarity information. Because SPI-seq does not directly monitor ssDNA, in cases where signals are detected on both strands (e.g., the mat1 locus), SPI-seq data alone cannot distinguish between binding to ssDNA of both polarities and binding to dsDNA.
Potential applications
SPI-seq can be easily adopted for proteins other than Rad52 and RPA, and nonyeast organisms. For example, in human cells, repair proteins such as RAD51, RAD52, BRCA2, and FANCD2, as well as checkpoint signaling proteins like ATR and ATRIP, accumulate on ssDNA upon DSB formation (Bekker-Jensen et al. 2006). As long as suitable antibodies are available for ChIP, the genome-wide locations of any of these proteins can be analyzed using SPI-seq. In addition, the adaptor-ligation scheme we designed can also be applied to ChIP-enriched dsDNA, thus allowing other DNA lesion markers such as phospho-H2A(X) to be profiled by the same procedure. Besides mapping spontaneous DNA lesion hotspots, SPI-seq should also be useful for cataloging in vivo cleavage sites of genome-editing nucleases, such as zinc-finger nucleases (ZFNs) and transcription activator like effector nucleases (TALENs).
Methods
Strains and culturing conditions
The strains used in this study are listed in Supplemental Table S3. Cells used for ChIP assay were cultured in EMM (Edinburgh Minimal Medium) with necessary supplements at 30°C unless otherwise noted. For the HO experiment, cells were shifted to thiamine-free medium for 17 h before crosslinking by formaldehyde. For pfh1 mutants, cells were cultured at the restrictive temperature for 18 h before crosslinking.
SPI-seq
Formaldehyde crosslinking was performed for 1 h at 4°C. Cells were lysed by glass bead beating. Chromatin was fragmented by sonication. Antibody-enriched chromatin fragments were reverse-crosslinked overnight at 65°C. ChIP DNA or input DNA was predenatured at 95°C and quenched on ice. The DNA was mixed with two pre-annealed adaptors added at final concentrations of 1.5 μM each. Adaptor 1 was composed of oligo A (5′-GCTCTTCCGATCXXXXCNNNNNN-NH2-3′) and oligo B (5′-p-GXXXXGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT-NH2-3′). XXXX in the oligo sequence denotes 4-nucleotide multiplex indexes. Adaptor 2 was composed of oligo C (5′-NNNNNNGTTCAGAGTTCTGCGACAGGAGAG-NH2-3′) and oligo D (5′-CAAGCAGAAGACGGCATACGACCTCTCCTGTCGCAGAACTCTGAAC-3′). The ligation reaction was conducted using the Quick Ligation Kit (New England Biolabs) overnight at 16°C. The ligation product was then separated on a Low Range Ultra agarose gel (Bio-Rad). DNA in the 250- to 500-bp range was retrieved with the GFX PCR DNA and Gel Band Purification Kit (GE Healthcare). The gel-purified DNA was amplified by PCR using primers P1 (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA-3′) and P2 (5′-CAAGCAGAAGACGGCATACGA-3′) for 25 cycles. The PCR product was processed by gel-based size selection, again retaining DNA in the 250- to 500-bp range. Equal amounts of DNA tagged with different multiplex indexes were mixed together and sequenced on an Illumina GA-II instrument using the sequencing primer 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATC-3′. Sequencing reads were aligned to the genome using SOAP2 (Li et al. 2009). The read alignment output was converted to a read density profile using kernel density estimation (Valouev et al. 2008) and visualized with Integrated Genome Browser (Nicol et al. 2009). Unless otherwise noted, the kernel density bandwidth was set at 30 bp.
Data access
The sequencing data described here are deposited at the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE39166.
Acknowledgments
We thank M.-Q. Dong and P. Russell for comments and suggestions on the manuscript. We thank S. MacNeill, P. Espenshade, and H. Levin for providing strains and plasmids. This work was supported by grants from the Chinese Ministry of Science and Technology and the Beijing Municipal Government to L.-L.D., and grants from Grant-in-Aid for Young Scientists (B) from JSPS KAKENHI (grant nos. 22770176, 20770005) to Y.T.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.146357.112.
References
- Arcangioli B, Roseaulin L, Holmes A 2007. Mating-type switching in S. pombe. In Topics in current genetics, Vol. 17, pp. 251–283. Springer, Berlin/Heidelberg [Google Scholar]
- Beach DH, Klar AJ 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J 3: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL 2003. Retrotransposons and their recognition of pol II promoters: A comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13: 1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SIS 2008. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451: 431–436 [DOI] [PubMed] [Google Scholar]
- Chan JE, Kolodner RD 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet 7: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JE, Kolodner RD 2012. Rapid analysis of Saccharomyces cerevisiae genome rearrangements by multiplex ligation-dependent probe amplification. PLoS Genet 8: e1002539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23: 81–85 [DOI] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho Y-J, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. 2011. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clepet C, Le Clainche I, Caboche M 2004. Improved full-length cDNA production based on RNA tagging by T4 DNA ligase. Nucleic Acids Res 32: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Costelloe T, Lowndes NF 2010. Chromatin assembly and signalling the end of DNA repair requires acetylation of histone H3 on lysine 56. Subcell Biochem 50: 43–54 [DOI] [PubMed] [Google Scholar]
- Dovey CL, Russell P 2007. Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe. Genetics 177: 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey CL, Aslanian A, Sofueva S, Yates JR III, Russell P 2009. Mms1-Mms22 complex protects genome integrity in Schizosaccharomyces pombe. DNA Repair 8: 1390–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L-L, Nakamura TM, Moser BA, Russell P 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol Cell Biol 23: 6150–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L-L, Nakamura TM, Russell P 2006. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Di Rienzi SC, Raghuraman MK, Brewer BJ 2011. Replication stress-induced chromosome breakage is correlated with replication fork progression and is preceded by single-stranded DNA formation. G3 1: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover TW, Berger C, Coyle J, Echo B 1984. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet 67: 136–142 [DOI] [PubMed] [Google Scholar]
- Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK 2006. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J Biol Chem 281: 8732–8739 [DOI] [PubMed] [Google Scholar]
- Haldar D, Kamakaka RT 2008. Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot Cell 7: 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Huang Y, Lowe TM, Maraia RJ 2001. Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol Cell Biol 21: 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Tazumi M, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H 2007. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff EF, Levin HL, Boeke JD 1998. Schizosaccharomyces pombe retrotransposon Tf2 mobilizes primarily through homologous cDNA recombination. Mol Cell Biol 18: 6839–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598 [DOI] [PubMed] [Google Scholar]
- Lettier G, Feng Q, De Mayolo AA, Erdeniz N, Reid RJD, Lisby M, Mortensen UH, Rothstein R 2006. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet 2: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J 2009. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Li P, Li J, Li M, Dou K, Zhang M-J, Suo F, Du L-L 2012. Multiple end joining mechanisms repair a chromosomal DNA break in fission yeast. DNA Repair 11: 120–130 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H 2006. The RNA polymerase III-dependent family of genes in hemiascomycetes: Comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res 34: 1816–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC 2003. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol 13: 1748–1757 [DOI] [PubMed] [Google Scholar]
- Mortensen UH, Lisby M, Rothstein R 2009. Rad52. Curr Biol 19: R676–R677 [DOI] [PubMed] [Google Scholar]
- Nickoloff JA, Singer JD, Heffron F 1990. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol Cell Biol 10: 1174–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE 2009. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, Yates JR III, Russell P 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24: 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SIS 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872 [DOI] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Aubert SD, Zakian VA 2008. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol 28: 6594–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Pennaneach V, Kolodner RD 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol Cell Biol 25: 7226–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenzhak S, Mejía-Ramírez E, Williams JS, Schaffer L, Hammond JA, Head SR, Russell P 2010. Rad3ATR decorates critical chromosomal domains with γH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet 6: e1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu G-H, Tanaka H, Kim D-H, Kim J-H, Bae S-H, Kwon Y-N, Rhee JS, MacNeill SA, Seo Y-S 2004. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res 32: 4205–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA 2012. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev 26: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD 2006. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc Natl Acad Sci 103: 18196–18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Lee C-YS, Espenshade PJ 2007. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet 3: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Carninci P, Watahiki A, Shiraki T, Konno H, Muramatsu M, Hayashizaki Y 2001. Cloning full-length, cap-trapper-selected cDNAs by using the single-strand linker ligation method. Biotechniques 30: 1250–1254 [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC 2012. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev 26: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Klar AJ, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31: 183–192 [DOI] [PubMed] [Google Scholar]
- Szilard RK, Jacques P-E, Laramée L, Cheng B, Galicia S, Bataille AR, Yeung M, Mendez M, Bergeron M, Robert F, et al. 2010. Systematic identification of fragile sites via genome-wide location analysis of γ-H2AX. Nat Struct Mol Biol 17: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Takahashi K 2007. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res 35: 3223–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ryu G-H, Seo Y-S, Tanaka K, Okayama H, MacNeill SA, Yuasa Y 2002. The fission yeast pfh1(+) gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res 30: 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejrup-Hansen R, Mizuno K, Miyabe I, Fleck O, Holmberg C, Murray JM, Carr AM, Nielsen O 2011. Schizosaccharomyces pombe Mms1 channels repair of perturbed replication into Rhp51 independent homologous recombination. DNA Repair 10: 283–295 [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Miller KM, Driscoll R, Masumoto H, Jackson SP, Kouzarides T, Verreault A, Arcangioli B 2007. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J Biol Chem 282: 15040–15047 [DOI] [PubMed] [Google Scholar]
- Yokoyama M, Inoue H, Ishii C, Murakami Y 2007. The novel gene mus7+ is involved in the repair of replication-associated DNA damage in fission yeast. DNA Repair 6: 770–780 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]