Polyglutamine-binding protein 1 (PQBP1) has been associated with familial X-linked neurodegenerative disorders, grouped together as Renpenning syndrome. Here, Wang et al. identify PQBP1 as an effector of alternative splicing, influencing multiple mRNA targets in human cells. Depletion of PQBP1 in mouse cortical neurons results in reduced dendritic outgrowth and aberrant alternative splicing. The investigators conclude that aberrant alternative splicing contributes to the disease pathology of PQBP1-linked neurological disorders.
Keywords: PQBP1, alternative splicing, neurite outgrowth, neurological disorders
Abstract
Polyglutamine-binding protein 1 (PQBP1) is a highly conserved protein associated with neurodegenerative disorders. Here, we identify PQBP1 as an alternative messenger RNA (mRNA) splicing (AS) effector capable of influencing splicing of multiple mRNA targets. PQBP1 is associated with many splicing factors, including the key U2 small nuclear ribonucleoprotein (snRNP) component SF3B1 (subunit 1 of the splicing factor 3B [SF3B] protein complex). Loss of functional PQBP1 reduced SF3B1 substrate mRNA association and led to significant changes in AS patterns. Depletion of PQBP1 in primary mouse neurons reduced dendritic outgrowth and altered AS of mRNAs enriched for functions in neuron projection development. Disease-linked PQBP1 mutants were deficient in splicing factor associations and could not complement neurite outgrowth defects. Our results indicate that PQBP1 can affect the AS of multiple mRNAs and indicate specific affected targets whose splice site determination may contribute to the disease phenotype in PQBP1-linked neurological disorders.
Alternative splicing (AS) is a major source for biological diversity and a crucial determinant of cell identity and fate. To date, >90% of human precursor messenger RNAs (pre-mRNAs) are reported to undergo AS, with immense variation across tissue types and developmental stages (Wang et al. 2008). The regulation of AS is carried out by the interaction of RNA-binding proteins (RBPs), notably hnRNPs and SR proteins, with cis-elements in the pre-mRNAs (Black 2003). The dynamics of RBP cis-element recognition are further modulated by a wide spectrum of spliceosome-associated proteins and are responsive to signal transduction networks (Varani and Nagai 1998; Wahl et al. 2009). Tissue-specific AS regulation is especially prominent in the nervous system (Castle et al. 2008), where neurons must promptly respond to complex stimuli and perform downstream functions with flexibility and precision (Dredge et al. 2001). Accordingly, neuron-specific AS networks modulate such diverse functions, including neurotransmission, receptor activity, and ion channel function, and thus impact higher brain functions, including cognition, coordination, and learning (Grabowski and Black 2001).
Aberrant AS is a primary cause for many human diseases (Wang and Cooper 2007). Diverse neurological disorders are closely linked to aberrant splicing and can be classified as cis-acting and trans-acting splicing defects (Licatalosi and Darnell 2006). Cis-acting defects originate from mutations in the disease-associated gene that change sequences important for the correct splicing of the pre-mRNA. A representative example is frontotemporal dementia with Parkinsonism linked to chromosome 17, which arises from mutations causing missplicing of the mRNA for the microtubule-associated protein tau (Licatalosi and Darnell 2006). Trans-acting defects arise from mutations or misregulation of splicing regulatory factors. One example is retinitis pigmentosa, characterized by a progressive loss of rod photoreceptor cells, which results from mutations in HPRP3, PRP31, and PRPC8—factors crucial for the assembly and functioning of U small nuclear ribonucleoproteins (snRNPs) in pre-mRNA splicing (McKie et al. 2001; Vithana et al. 2001; Chakarova et al. 2002). Although increasing numbers of splicing-related disorders have been identified, the molecular link between AS regulation and the disease phenotype remains to be elucidated.
Polyglutamine-binding protein 1 (PQBP1) is a highly conserved protein with mutations associated with ∼10 familial X-linked mental retardation (XLMR) diseases (Kalscheuer et al. 2003; Stevenson et al. 2005). Patients with these diseases share similar symptoms, such as intellectual deficiency, microcephaly, and short stature (Kalscheuer et al. 2003; Stevenson et al. 2005). Although these diseases were separately discovered and originally thought to be unrelated, they were recently grouped together as “Renpenning syndrome” because of the common symptomatic manifestation and mutations in PQBP1 (Stevenson et al. 2005).
The molecular function of PQBP1 remains unclear. PQBP1 is linked to transcription regulation based on its interaction with RNA polymerase II and other transcription factors (Waragai et al. 1999; Okazawa et al. 2002). Recent studies also reported that PQBP1 was dynamically associated with stress granules and interacted with dynactin components, indicating a possible role for PQBP1 in RNA transport or cytoplasmic RNA processing (Kunde et al. 2011). Also, PQBP1 has been implicated in pre-mRNA splicing: PQBP1 interacts with known splicing factors such as WBP11 and U5-15KD (Komuro et al. 1999; Waragai et al. 2000) and is part of the early spliceosome (Makarova et al. 2004). Consistent with this view, a disease-associated mutant of PQBP1 decreased splicing efficiency (Tapia et al. 2010).
We previously identified PQBP1 in a screen for factors that affect the AS of the mRNA for Bcl-X, a mammalian apoptosis regulator (Moore et al. 2010). Here, we show that PQBP1 affects the AS of numerous other pre-mRNAs in human cells. In addition, we used whole-transcriptome sequencing to identify a global set of PQBP1 AS targets in primary mouse neurons, which revealed that PQBP1 coordinates important neuronal functions and maintains normal neurite development. Collectively, our findings indicate that aberrant AS contributes to the disease pathology of PQBP1-associated neurological disorders.
Results
PQBP1 affects the AS of mRNAs encoding apoptotic factors
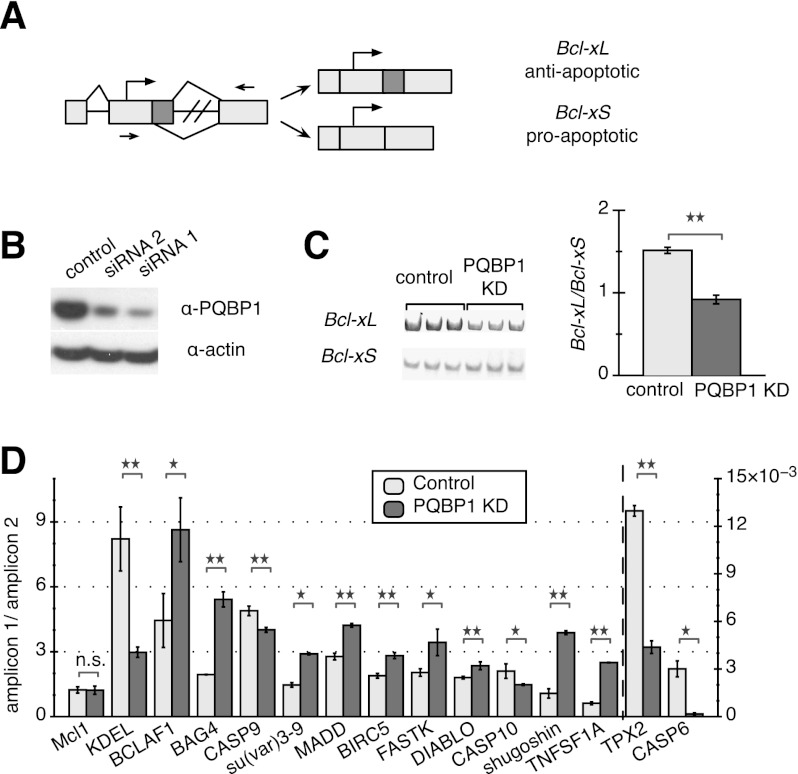
We previously identified PQBP1 as a potential effector for Bcl-x splicing in HeLa cells in a genome-wide siRNA screen using a fluorescence-based minigene assay (Fig. 1A; Moore et al. 2010). To verify the effect of PQBP1 on the AS of Bcl-x, we examined the splicing of endogenous Bcl-x after depletion of PQBP1 with two different siRNAs (Fig. 1B) using RT–PCR primers flanking the alternatively spliced region (Fig. 1A). As expected, depletion of PQBP1 favored a shift from the anti-apoptotic isoform Bcl-xL toward the proapoptotic isoform Bcl-xS via increased use of an alternative 5′ splice site in the second exon (Fig. 1A,C; Supplemental Fig. S1A,B). These results confirmed that PQBP1 affects the AS of Bcl-x in HeLa cells.
Figure 1.
AS targets of PQBP1 in HeLa cells. (A) Schematic of Bcl-x. Light rectangles are exons, with dark color marking the alternatively spliced region, and black lines are introns. Arrows show translation start sites, and arrowheads mark quantitative PCR primer sets. (B) Western blot of HeLa cell lysates where PQBP1 is knocked down by siRNA. The control sample was treated with a siRNA targeting firefly luciferase mRNA. Antibodies are listed on the right. Knockdown (KD) with two different siRNAs is shown. (C) Gel analysis of RT–PCR of Bcl-x splicing isoforms for control and PQBP1 knockdown samples is shown (left panel) with quantification (right panel). Mean of three independent measurements ± SD are shown. Changes were tested by one-way ANOVA. (**) Statistically significant with P-value < 0.01. See also Supplemental Figure S1, A and B. (D) Quantification of real-time PCR data on 14 mRNAs that showed significant AS changes between control and PQBP1 knockdown samples, with Mcl1 (not a target of PQBP1) as the negative control. The Y-axis depicts the ratio between amplicons for constitutive and alternative exonic regions; TPX-2 and CASP6 follow the Y-axis on the right side, while the rest follow the Y-axis on the left side. Light columns show the ratio of two amplicons from the control sample, and dark columns show the ratio of two amplicons from the PQBP1 knockdown sample. Mean of three independent measurements ± SD are shown. The difference of the ratio between control and PQBP1 knockdown samples was analyzed by one-way ANOVA test. (*) Statistically significant with P-value < 0.03; (**) statistically significant with P-value < 0.01; (n.s.) not significant. See also Supplemental Figure S1C.
We now show that PQBP1 affects additional AS events. We analyzed 31 mRNAs encoding apoptosis-related factors with annotated alternatively spliced isoforms. We designed primers to amplify the alternatively spliced exonic region and constitutive exonic region of each mRNA, and the ratio of the two amplicons was compared between the control and PQBP1 knockdown samples by RT–PCR. Approximately half of these mRNAs showed significant changes in AS when PQBP1 was knocked down (Fig. 1D; Supplemental Fig. S1C). These results provided further evidence that PQBP1 affects the AS of many mRNAs.
PQBP1 is associated with splicing factors
To further clarify PQBP1's role in AS regulation, we systematically determined the spectrum of proteins that are associated with PQBP1. In addition, to understand the aberrant function of disease-related PQBP1 mutants, we compared the associated protein profile of wild-type PQBP1 and two mutant variants identified in Renpenning syndrome patients.
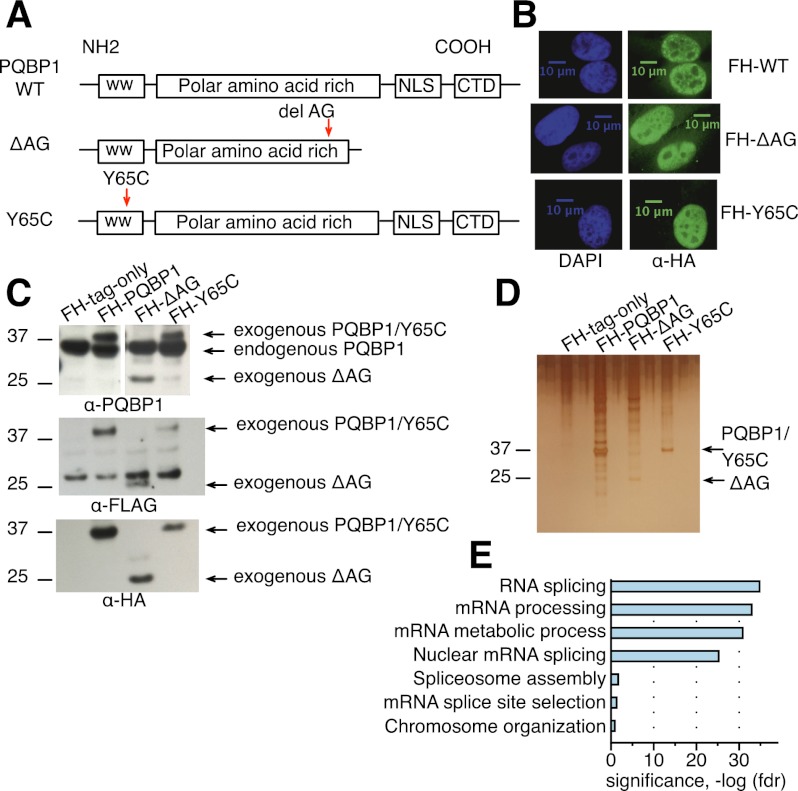
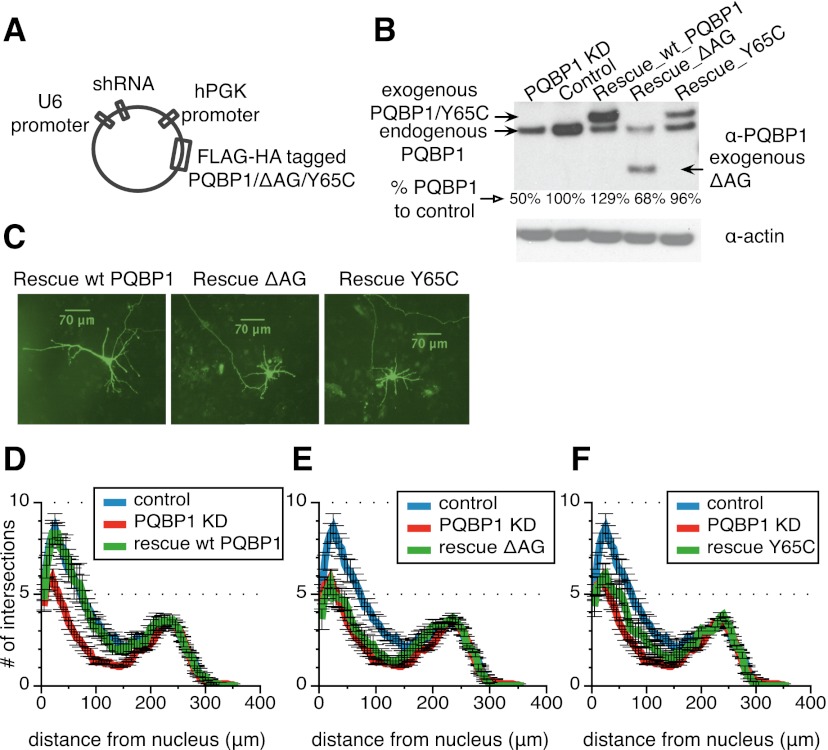
PQBP1 protein contains a WW domain, a polar amino acid-rich domain, and a C-terminal domain with a nuclear localization signal (NLS) (Fig. 2A; Waragai et al. 1999). The mutations in PQBP1 identified from Renpenning syndrome patients can be classified into two categories: frameshifting mutations in the AG dinucleotide hexamer in the polar amino acid-rich domain and missense mutations in the WW domain (Musante et al. 2010). Here, we examined two representative disease-linked variants: one with an AG dinucleotide deletion (ΔAG) in the polar amino acid-rich domain, resulting in a truncated protein, and one with a point mutation (Y65C) in the WW domain (Fig. 2A; Kalscheuer et al. 2003). PQBP1 localizes predominantly in the nucleus (Fig. 2B; Waragai et al. 1999; Komuro et al. 1999; Kalscheuer et al. 2003). Interestingly, both ΔAG and Y65C mutants localize to the nucleus (Fig. 2B), although ΔAG loses a potential NLS (Fig. 2A), indicating that there must be an additional unidentified NLS or that it enters together with another protein.
Figure 2.
The spectrum of proteins associated with PQBP1 and disease-linked mutants. (A) Schematics of the protein domain organization of wild-type (WT) PQBP1 and two disease-linked mutants. Boxes are protein domains, with names shown in the center. (WW) WW domain; (CTD) C-terminal domain. Red arrowheads show the mutation sites in the disease-linked PQBP1 variants. (B) Immunofluorescence staining of HeLa cells stably expressing Flag-HA-tagged wild-type PQBP1, ΔAG, or Y65C. Cells were fixed and stained with antibodies or dyes listed at the bottom of each panel column. (C) Western blot of HeLa cell lines stably expressing empty Flag-HA vector, Flag-HA-tagged wild-type PQBP1, Flag-HA-tagged ΔAG, and Flag-HA-tagged Y65C. The antibodies used are indicated at the bottom of each panel. (D) Silver staining of proteins copurified with Flag-HA-tagged empty plasmid, Flag-HA-tagged wild-type PQBP1, Flag-HA-tagged ΔAG, or Flag-HA-tagged Y65C. (E) GO term enrichments of proteins that were pulled down with wild-type PQBP1. Statistics were calculated with human genes as the background. The X-axis is the corrected P-value (false discovery rate [FDR]) in negative log for enrichment.
Proteins associated with wild-type PQBP1 were strongly enriched for known or potential splicing factors and spliceosome components (Fig. 2E; Supplemental Table S1). HeLa cells expressing stably integrated Flag-HA-tagged full-length PQBP1, ΔAG, or Y65C were generated by stable integration of retroviral constructs and shown by Western blotting to express exogenous proteins at a level comparable with endogenous PQBP1 (Fig. 2C). Tandem affinity purification with Flag and HA antibodies followed by mass spectroscopy was performed to identify PQBP1- or mutant-associated proteins (Fig. 2D; Supplemental Table S1). WBP11, a previously known PQBP1 interactor (Komuro et al. 1999), was detected in all three replicate purifications of wild-type PQBP1, confirming the validity of our approach (Supplemental Table S1). U5-15KD, another proposed interactor of PQBP1 (Waragai et al. 2000), was not identified, possibly because of cell specificity or because the interaction is not strong enough to be detected in our tandem affinity purification. ΔAG showed significantly reduced splicing factor association compared with wild-type PQBP1, while Y65C lost associations with all splicing-related factors (Supplemental Table S1). These results established an association, either direct or indirect, of PQBP1 to the splicing machinery and identified the WW domain as important for linking PQBP1 to splicing complexes. This finding is consistent with the idea that disease-linked mutations may disrupt splicing-related functions of PQBP1.
PQBP1 influences SF3B1's (subunit 1 of the splicing factor 3B [SF3B] protein complex) recognition of splicing sites
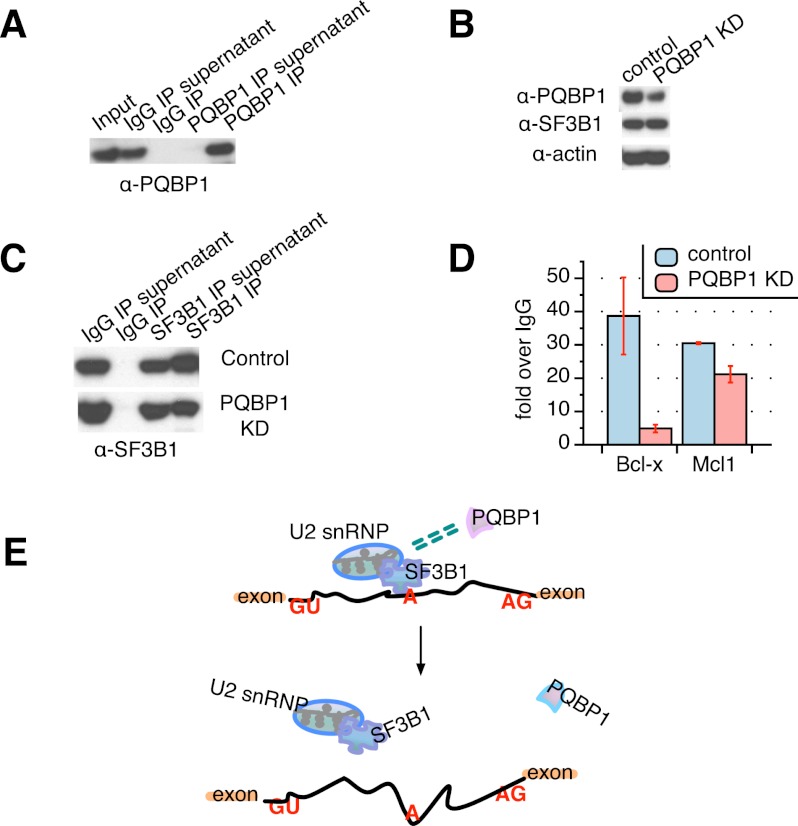
To explore the mechanism of PQBP1 in AS regulation, we first examined whether PQBP1 directly binds pre-mRNAs whose splicing it affects. Although previous studies showed PQBP1 bound to polyrG resin in vitro (Komuro et al. 1999), PQBP1 protein itself has no known RNA-binding domains (Fig. 2A; Lunde et al. 2007). We used the CLIP (cross-linking and immunoprecipitation) method to determine whether PQBP1 directly binds its targets in vivo (Ule et al. 2003). Live HeLa cells were exposed to UV light to cross-link RNA–protein complexes, and then PQBP1 was immunopurified from cell lysates (Fig. 3A). RT–PCR quantification of Bcl-x and nine other target mRNAs (Fig. 1D) showed no enrichment in PQBP1 immunoprecipitations compared with an irrelevant immunoglobulin G (IgG) control. These results suggest that PQBP1 does not bind AS target mRNAs directly, with the caveat that it may fail to be effectively cross-linked under the conditions where other known splicing factors are observed to bind.
Figure 3.
SF3B1 function is affected by PQBP1. (A) Western blot of PQBP1 CLIP together with an irrelevant IgG control, probed with PQBP1 antibody. PQBP1 was not detected in depleted supernatants or the IgG pull-down, indicating efficient and specific antibody binding of PQBP1. (B) Western blot of SF3B1 CLIP from PQBP1 knockdown (KD) samples and samples treated with siRNA targeting firefly luciferase mRNA as control. Shown are total cell lysates of control (lane 1) and PQBP1 knockdown (lane 2) HeLa cells probed with antibodies indicated at the left. (C) Western blot of SF3B1 CLIP from PQBP1 knockdown samples and samples treated with siRNA targeting firefly luciferase mRNA as control. Lanes 1 and 2 are the IgG controls, and lanes 3 and 4 are SF3B1 CLIP stained by SF3B1 antibody for control and PQBP1 knockdown samples. (D) Quantification of Bcl-x and Mcl1 mRNA enrichment in the SF3B1–RNA complex as from SF3B1 CLIP in control and PQBP1 knockdown samples, respectively, by RT–PCR. Data values normalized to the IgG control and mean of three independent measurements ± SD are shown. (E) Proposed model mechanism of PQBP1 AS regulation. A schematic of part of a hypothetical mRNA is shown. Orange rectangles are exons, and the black line is the intron. “GU” marks the 5′ splice site, “A” marks the branch site recognized by the U2 snRNP with help from components like SF3B1, and “AG” marks the 3′ splice site. Green dashed double lines indicate the association between PQBP1 and SF3B1. Through an association with SF3B1 and other splicing factors, PQBP1 influences the recruitment of U2 snRNP to specific target sites, thus affecting splicing decisions on a subset of mRNAs. Loss or mutations of PQBP1 interrupt the splicing factor association and the recognition of specific splice sites. See the Discussion.
PQBP1, on the other hand, affects the association of the spliceosome factor SF3B1 with a target pre-mRNA. Among the proteins that are associated with PQBP1 are SF3B protein complex members (Supplemental Table S1), which form part of the U2 snRNP. The function of the SF3B complex is critical for the assembly of the spliceosome complex A as well as the successful execution of the splicing reaction (Wahl et al. 2009). Furthermore, SF3B1 was reported to regulate the AS of Bcl-x through direct interaction with a cis-element in the Bcl-x mRNA (Massiello et al. 2006), and we observed that CLIP with SF3B1 showed a 40-fold enrichment of Bcl-x binding over an IgG control (Fig. 3D, blue column over Bcl-x).
To test whether the recognition of Bcl-x by SF3B1 is influenced by PQBP1, we knocked down PQBP1 in HeLa cells and reanalyzed the enrichment of Bcl-x in the SF3B1–RNA complex (Fig. 3B,C). PQBP1 depletion led to an eightfold decrease of Bcl-x mRNA levels in SF3B1 CLIP compared with a control knockdown (Fig. 3D), although the level of SF3B1 protein remained unchanged (Fig. 3B), and the steady-state level of Bcl-x mRNA did not change upon PQBP1 knockdown (Supplemental Fig. S2).
To further test whether PQBP1's impact on SF3B1–RNA recognition is specific to the targets of PQBP1, we analyzed the mRNA of Mcl1 in the SF3B1–RNA complex. Like Bcl-X, Mcl1 is a mammalian apoptosis regulator and has two splicing isoforms: anti-apoptotic Mcl1-L and proapoptotic Mcl1-S (Akgul et al. 2004). However, while knockdown of SF3B1 significantly shifted the splicing of Mcl1 toward Mcl1-S (Moore et al. 2010), PQBP1 knockdown had little effect (Fig. 1D; Moore et al. 2010). We therefore hypothesized that the knockdown of PQBP1 should have relatively minor influence on Mcl1 mRNA enrichment in the SF3B1–RNA complex. Indeed, CLIP data showed only a 1.4-fold decrease of Mcl1 mRNA enrichment in the SF3B1–RNA complex in the PQBP1 knockdown sample compared with the control (Fig. 3D). Additionally, the decrease was mostly likely due to a Mcl1 mRNA steady-state level change of 1.5-fold (Supplemental Fig. S2). Therefore, PQBP1 influenced the recognition of SF3B1 to one mRNA whose AS is affected by PQBP1 but not for an mRNA where AS was regulated by SF3B1 but not PQBP1. These results do not rule out possibilities that PQBP1 may affect mRNA AS through other interactions or that other splicing factors may assist in the process.
Loss of PQBP1 causes defects in neurite outgrowth
Previous work found that PQBP1 is most highly expressed in the CNS of embryonic or newborn rodents, with the peak around birth (Qi et al. 2005). Considering the tissue-specific expression pattern of PQBP1 as well as its disease association, we examined the role of PQBP1 in AS modulation in embryonic rodent neurons.
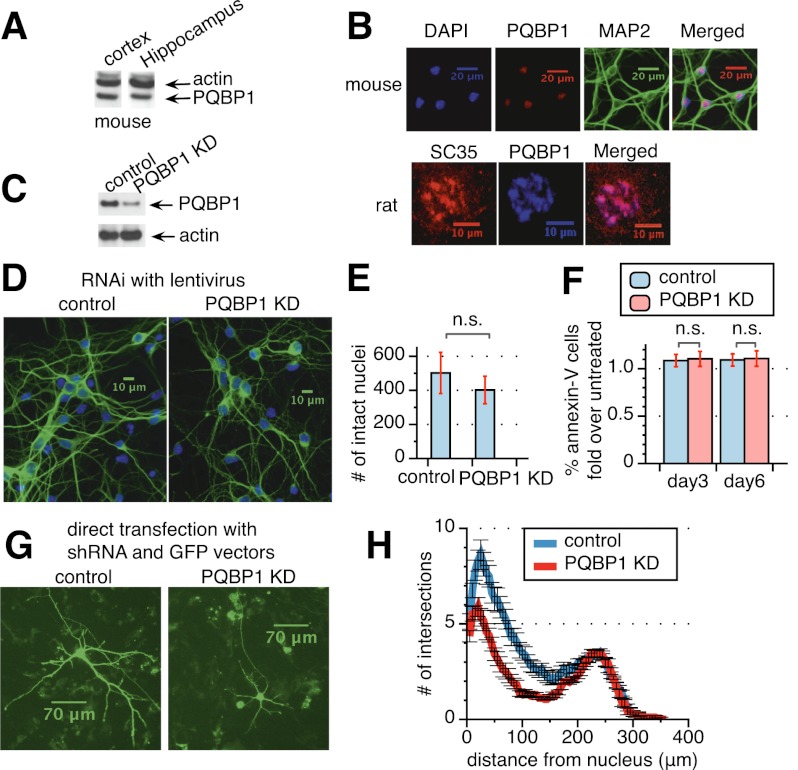
PQBP1 is highly expressed in embryonic mouse cortical and hippocampal neurons, as shown by Western blots (Fig. 4A). In addition, PQBP1 localizes predominantly in the nucleus of neurons (Fig. 4B, top panels), as shown previously in other cell types (Waragai et al. 1999; Komuro et al. 1999; Kalscheuer et al. 2003). A closer examination demonstrated that PQBP1 colocalizes with SC35—a splicing factor in nuclear speckles, a nuclear domain enriched for splicing regulatory factors (Fig. 4B, bottom panels; Supplemental Fig. S3A; Spector and Lamond 2011).
Figure 4.
PQBP1 affects dendritic outgrowth and branching in mouse cortical neurons. (A) Western blot of PQBP1 expression in mouse embryonic cortical and hippocampal neurons, compared with endogenous β-actin expression. (B) Immunofluorescence staining of mouse embryonic primary cortical neurons (the four top panels) and rat embryonic primary cortical neurons (the three bottom panels). Neurons were fixed and stained with antibodies or dyes listed at the top of each panel. See also Supplemental Figure S3A. (C) Western blot of mouse embryonic cortical neurons infected with virus from a nontargeting control shRNA or a PQBP1 targeting shRNA. See also Supplemental Figure S3B. (D) Immunofluorescence staining of mouse primary embryonic cortical neurons 5 d after infection with virus from a nontargeting control shRNA or a PQBP1 targeting shRNA. (Green) MAP2 staining; (blue) DAPI staining. See also Supplemental Figure S3C. (E) Comparison of the number of intact nuclei between neuron samples 5 d after infection with virus from a nontargeting control shRNA or a PQBP1 targeting shRNA. Neurons were fixed and stained with DAPI. Healthy and intact nuclei were counted for each sample. Mean of eight independent measurements ± SD is shown. The difference in the number of intact nuclei between control and PQBP1 knockdown samples was analyzed by one-way ANOVA test. (n.s.) Not significant. (F) Comparison of the percentage of Annexin V-stained neurons between samples infected with virus from a nontargeting control shRNA or a PQBP1 targeting shRNA. The number of neurons with Annexin V staining and the percentage of Annexin V-positive neurons were calculated and normalized to neuron samples that were not treated by virus. The Y-axis depicts the fold of the percentage of Annexin V-positive neurons in control or PQBP1 knockdown samples to samples that were untreated. Neurons on day 3 and day 6 after infection/plating were studied. The mean of six independent measurements ± SD is shown. The difference in the percentage of Annexin V-positive neurons between control and PQBP1 knockdown samples was analyzed by one-way ANOVA test. (n.s.) Not significant. (G) Immunofluorescence of individual neurons that were transiently transfected with either a nontargeting control shRNA or a PQBP1 targeting shRNA together with a GFP vector. Neurons were grown on a layer of feeder glial cells and examined 5 d after transfection. (H) Sholl analysis of dendritic branching for neurons transiently transfected with a nontargeting control shRNA or PQBP1 targeting shRNA together with a GFP vector. Concentric circles were drawn at intervals of 5 μm. The number of dendritic branch intersections with each concentric circle was counted (mean ± SE, n = 16–17). The X-axis depicts the distance of the concentric circle from the neuron soma center, and the Y-axis depicts the number of intersections the corresponding circle encounters with dendrites.
To understand the importance of PQBP1 in neuron development, we knocked down PQBP1 in mouse embryonic primary cortical neurons with shRNAs delivered by lentivirus (Fig. 4C). Neurons were first examined by immunostaining of the microtubule-associated protein 2 (MAP2), a marker for dendrites. Interestingly, upon PQBP1 knockdown, the number of projections and length of neurites were decreased, and the connection between neurons degenerated considerably (Fig. 4D). A second shRNA against PQBP1 led to a similar defect in neurite morphology (Supplemental Fig. S3B,C). To determine whether this defect is simply due to cell apoptosis, we compared the number of DAPI-stained intact nuclei after fixation as well as the percentage of Annexin V-stained cells between control and PQBP1 knockdown neurons (Fig. 4E,F). No significant differences were observed, indicating that the defects observed from PQBP1 knockdown are not from neuron apoptosis.
PQBP1 knockdown neurons showed substantially diminished dendrite branching and dendrite length (Fig. 4G,H). To quantify the changes in neurite outgrowth, we transiently transfected neurons with a nontargeting control shRNA or a PQBP1 targeting shRNA together with a GFP vector (Fig. 4G). Sholl analysis was then performed on neurons that were GFP-highlighted for control and PQBP1 knockdown (Fig. 4H; Sholl 1953).
To confirm that the dendritic outgrowth defect observed was a direct effect of PQBP1 knockdown, we examined whether the defect could be rescued by re-expressing wild-type PQBP1 in comparison with the disease-associated PQBP1 mutants. shRNA-resistant, Flag-HA-tagged cDNAs encoding human PQBP1, ΔAG, and Y65C were coexpressed in mouse cortical neurons with shRNAs targeting endogenous mouse PQBP1 (Fig. 5A; Supplemental Fig. S3D). Western blots confirmed the knockdown of endogenous mouse PQBP1 and the concomitant expression of exogenous dual-tagged human PQBP1 and variants (Fig. 5B). Total PQBP1 expression in all three rescue samples was compatible with the level of PQBP1 in the control sample, with the exception of ΔAG, which reached only 68% of the control (Fig. 5B). To quantitatively compare neurite outgrowth between mutant PQBP1 and wild-type rescue, we transfected mouse cortical neurons with the rescue constructs (Fig. 5C) and performed Sholl analysis after 5 d. A significant restoration of dendrite density and length was observed in wild-type PQBP1-rescued neurons, while ΔAG and Y65C showed less improvement (Fig. 5D,E,F). In the case of ΔAG, it is possible that insufficient expression of ΔAG may contribute to the deficiency of rescue.
Figure 5.
Wild-type (WT) PQBP1 but not the mutants rescues the neuron morphological defects. (A) Schematic of the pLKO.1-derived lentiviral constructs used to generate viruses for the rescue experiments. See also Supplemental Figure S3D. (B) Western blot of neuron samples that were infected with virus from an unmodified PQBP1 targeting shRNA, a nontargeting control shRNA, or PQBP1 targeting shRNA constructs coexpressing exogenous wild-type PQBP1, ΔAG, or Y65C. Antibodies used are listed at the right. Quantification of total PQBP1 level for each sample compared with control is shown below each lane. (C) Immunofluorescence of individual neurons that were transiently transfected with PQBP1 targeting shRNA constructs coexpressing exogenous wild-type PQBP1, ΔAG, or Y65C together with a GFP vector. Neurons were grown on a layer of feeder glial cells. (D–F) Sholl analysis of dendritic branching for neurons that were transiently transfected with a nontargeting control shRNA, an unmodified PQBP1 targeting shRNA, or PQBP1 targeting shRNA constructs coexpressing wild-type PQBP1 (D), ΔAG (E), or Y65C (F) (mean ± SE; n = 10–17).
Combining the results above, we conclude that PQBP1 is important in maintaining neurite projection and outgrowth in neuron development. Moreover, deviation in neurite structure may be part of the defects brought about by the disease-associated mutants of PQBP1.
Coupling of neuron projection and PQBP1 function
Dendrite structure is the crucial basis for neuron network formation and information processing. To clarify the molecular function of PQBP1 in maintaining neurite outgrowth, we systematically profiled the AS targets of PQBP1 in mouse embryonic cortical neurons by RNA sequencing (RNA-seq).
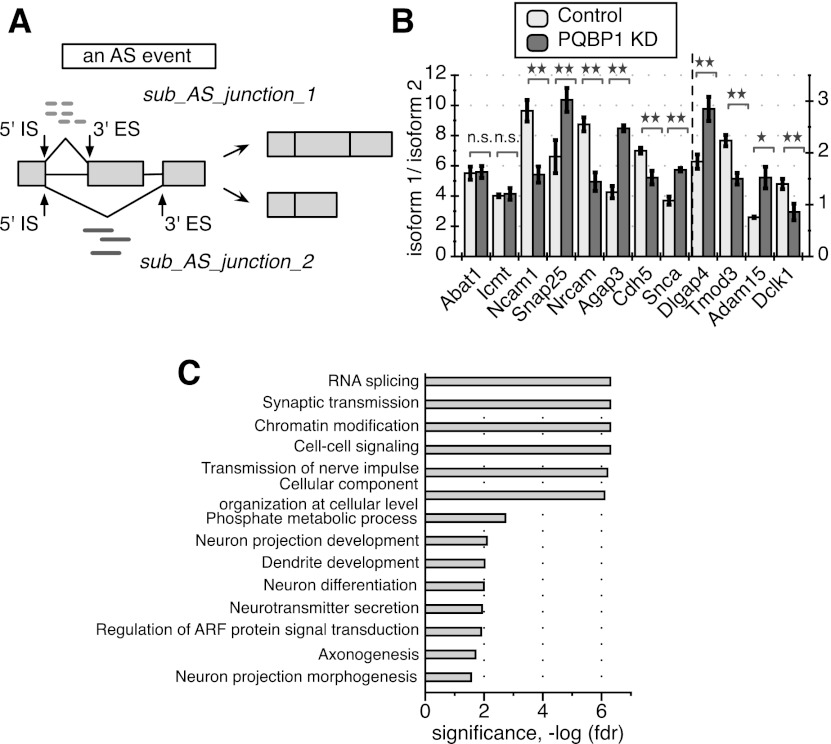
We considered RNA-seq reads that were directly mapped over splice junctions as the most straightforward evidence to quantify the splicing changes. For each splice junction, a 5′ initiation site (5′ IS) is defined as the final nucleotide position of the upstream exon, and the 3′ end site (3′ ES) is defined as the first nucleotide position of the downstream exon (Fig. 6A). An AS event is then defined as a set of junctions that share the same 5′ IS or the same 3′ ES, with each junction in the set referred to as a sub-AS junction (Fig. 6A). This definition covers all six primary AS patterns except for intron retention, which was not considered in our analysis (Supplemental Fig. S4A; Black 2003). An AS event under an experimental condition is represented by the distribution of read counts mapped to each sub-AS junction within the AS event; i.e., the “usage” of each sub-AS junction under that condition. We developed methods to identify AS events that experienced significant differential “usage” of any sub-AS junction between control and PQBP1 knockdown conditions (for details, see the Supplemental Material) and defined these AS events as AS targets of PQBP1. In total, 457 targets were identified in the process (Supplemental Fig. S4B; Supplemental Table S2).
Figure 6.
PQBP1's AS targets in mouse embryonic cortical neurons. (A) A schematic of a defined AS event and quantification of splicing changes. A hypothetical alternatively spliced mRNA is shown, with light rectangles representing exons and black lines between exons representing introns. The middle exon is the alternatively spliced exon. The 5′ IS is the start of a splicing junction, and the 3′ ES is the end of the junction. The AS event shown here includes two sub-AS junctions that share the same 5′ IS. Light dashed short lines mark RNA-seq reads that are mapped to sub-AS junction 1, and dark solid short lines mark reads mapped to sub-AS junction 2. The distribution of reads mapped to the two sub-AS junctions was used for AS quantification in our analysis. See also Supplemental Figure S4, Supplemental Table S2, and the Supplemental Material. (B) RT–PCR validation of 10 randomly picked AS events identified as PQBP1 targets and two randomly picked AS events identified as non-PQBP1 targets in the computational analysis. Abat1 and Icmt were predicted non-PQBP1 targets. The Y-axis depicts the ratio between isoform 1 and isoform 2. Dlgap4, Tmod3, Adam15, and Dclk1 follow the Y-axis on the right side, while the rest follow the Y-axis on the left side. The mean of three independent measurements ± SD is shown. The differences between control and PQBP1 knockdown (KD) samples were analyzed by one-way ANOVA test. (*) Statistically significant with P-value < 0.05; (**) statistically significant with P-value < 0.01; (n.s.) not significant. (C) GO term functional enrichment of AS targets of PQBP1. Statistics were calculated with all genes that are expressed in mouse embryonic cortical neurons (profiled from RNA-seq data) as the background. The GO term enrichment profile here is specific compared with RNA-seq studies for other proteins in neurons (see the text). The X-axis is the corrected P-value (FDR) in negative log for enrichment. See also Supplemental Table S4.
To validate PQBP1 targets identified by RNA-seq, we picked 10 candidate targets together with two candidate nontargets from the analysis and designed primers to amplify each splicing isoform of each gene. We then compared the ratio of the isoform expression levels by RT–PCR for control and PQBP1 knockdown samples. All 10 identified targets exhibited significant AS shift upon PQBP1 knockdown, while the two nontargets showed no discernible changes (Fig. 6B). This result demonstrated the sensitivity and specificity of our computational method to identify AS targets of PQBP1.
The PQBP1 AS targets showed tissue-specific functional enrichment, with strong gene ontology (GO) enrichments for neuron projection development/morphogenesis, dendrite development, and axonogenesis (Fig. 6C; Supplemental Table S4) versus genes that were expressed in mouse embryonic cortical neurons profiled from the RNA-seq data. This observation was in accordance with the neurite outgrowth defect observed when PQBP1 was knocked down (Fig. 4D,G,H). Targets of PQBP1 also showed strong enrichment in RNA splicing, synaptic transmission, chromatin modification, cell–cell signaling, phosphate metabolic process, neurotransmitter transport, and ADP-ribosylation factor (ARF) protein signal transduction (Fig. 6C; Supplemental Table S4). The GO term enrichment profile for PQBP1's AS targets is specific compared with high-throughput studies on other proteins in neurons, especially RNA splicing, chromatin modification, dendrite development-related functions, and ARF signal transduction (Ule et al. 2005; Gehman et al. 2011; Charizanis et al. 2012). Bcl-x AS is not conserved from humans to mice and was not identified as a target in this analysis. Among the 14 AS targets identified in HeLa cells that were related to apoptosis control (Fig. 1D), only the mRNA of BCLAF1 was recognized as an AS target of PQBP1 in mouse neurons, indicating the tissue specificity of PQBP1's regulation in AS.
Considering the physical and potential functional association of PQBP1 and SF3B1, we also knocked down SF3B1 in mouse cortical neurons and examined the AS of the 10 verified targets of PQBP1 (Supplemental Fig. S5A). Interestingly, all 10 targets also showed a significant AS shift upon SF3B1 knockdown (Supplemental Fig. S5B).
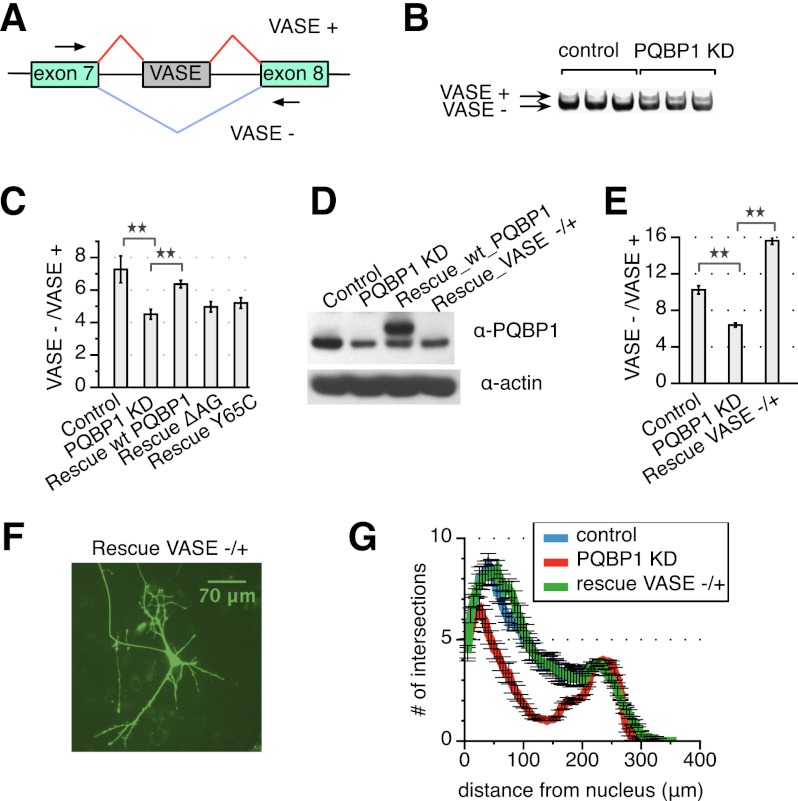
Among the top hits of PQBP1 AS targets was NCAM-140, a major isoform of neural cell adhesion molecules 1 (Ncam1) (Fig. 6B). Specifically, a 30-nucleotide (nt) exon between exons 7 and 8 is alternatively spliced in NCAM-140 and is designated VASE (variable alternatively spliced exon) (Fig. 7A). The VASE-excluding isoform of NCAM-140 (VASE−) promotes neurite outgrowth, while the VASE-including isoform (VASE+) inhibits neurite outgrowth (Doherty et al. 1992). The embryonic brain mainly expresses VASE− with gradual ascendancy until final dominance of VASE+ expression toward adulthood, except in regions such as the olfactory bulb, where reformation of synapses and turnover of neurons continue (Small and Akeson 1990). Thus, the AS of VASE in NCAM-140 is a critical switch for neuronal plasticity. To confirm PQBP1's effect on VASE, we designed primers flanking the VASE exon and examined VASE+ and VASE− isoforms by RT–PCR and electrophoresis (Fig. 7A). VASE+ was increased upon PQBP1 knockdown, while VASE− decreased (Fig. 7B), resulting in a significant decrease in the ratio of VASE−/VASE+ expression in response to PQBP1 shRNA versus a control hairpin (Fig. 6B, Ncam1). These results were confirmed with a second shRNA targeting PQBP1 (Supplemental Fig. S6).
Figure 7.
PQBP1-modulated AS of NCAM-140 influences neurite outgrowth. (A) A schematic of alternatively spliced region of NCAM-140. Green rectangles are exons, and black lines are introns. The gray rectangle marks the VASE. Arrowheads are primers designed for RT–PCR and electrophoresis. (B) NCAM-140 VASE splicing isoforms for neuron samples infected with virus from a nontargeting control shRNA or a PQBP1 targeting shRNA were analyzed by RT–PCR and gel electrophoresis. See also Supplemental Figure S6. (C) RT–PCR quantification for VASE− and VASE+. The ratio of the two isoform levels are compared among neurons infected with virus from a nontargeting control shRNA, an unmodified PQBP1 targeting shRNA, or PQBP1 targeting shRNA constructs coexpressing exogenous wild-type (WT) PQBP1, ΔAG, or Y65C. The mean of three independent measurements ± SD is shown. The differences between samples were analyzed by one-way ANOVA test. (**) Statistically significant with P-value < 0.01. (D) Western blot of neurons infected with virus from a nontargeting shRNA, an unmodified PQBP1 targeting shRNA, a PQBP1 targeting shRNA construct coexpressing exogenous wild-type PQBP1, or a PQBP1 targeting shRNA construct coexpressing exogenous VASE− to restore the ratio of VASE−/VASE+. (E) RT–PCR quantification for VASE− and VASE+ expression levels of neurons infected with virus from a nontargeting shRNA, an unmodified PQBP1 targeting shRNA, or a PQBP1 targeting shRNA construct coexpressing exogenous VASE− to restore the ratio of VASE−/VASE+. The mean of three independent measurements ± SD is shown. The differences between samples were analyzed by one-way ANOVA test. (**) Statistically significant with P-value < 0.01. (F) Immunofluorescence of individual neurons that were transiently transfected with a PQBP1 targeting shRNA construct coexpressing exogenous VASE− to restore the ratio of VASE−/VASE+ together with a GFP vector. Neurons were grown on a layer of feeder glial cells. (G) Sholl analysis of dendritic branching for individual neurons that were transiently transfected with a nontargeting control shRNA, an unmodified PQBP1 targeting shRNA, or a PQBP1 targeting shRNA construct coexpressing exogenous VASE− to restore the ratio of VASE−/VASE+ (mean ± SE, n = 15–17).
As a further confirmation that PQBP1 impacts the AS of NCAM-140, we examined AS of NCAM-140 after PQBP1 knockdown in neurons coexpressing the wild-type PQBP1, ΔAG, and Y65C constructs from Figure 5A. Interestingly, neurons rescued with wild-type PQBP1 restored the ratio of VASE−/VASE+ to levels observed in control samples (Fig. 7C). In contrast, ΔAG and Y65C were less effective in correcting the VASE−/VASE+ ratio (Fig. 7C).
To test whether NCAM-140 is one of the PQBP1 targets responsible for the defects observed in cells lacking PQBP1, we examined the effect of restoring the ratio of VASE− and VASE+. We coexpressed VASE− under the hPGK promoter in the lentiviral shRNA vector as described above (Fig. 5A). Knockdown of PQBP1 was verified by Western blot (Fig. 7D), and the change in the ratio of VASE−/VASE+ was verified by RT–PCR (Fig. 7E). Neurons were transfected with the rescue construct (Fig. 7F), and Sholl analysis on transfected neurons displayed a significant recovery from the neurite outgrowth defect caused by PQBP1 knockdown (Fig. 7G). These results confirm the role of PQBP1 in the AS of NCAM-140 and modulating neurite outgrowth.
Discussion
In this study, we showed that the disease-associated factor PQBP1 plays a role in the AS of mRNAs. We demonstrated that PQBP1 associates with core spliceosome components and has a protein association network enriched for other splicing factors. Following down-regulation of PQBP1, we observed aberrant splicing profiles for many mRNAs in both human cells and primary mouse cortical neurons. Specifically in neurons, mRNA profiling showed preferential misregulation of AS in factors that are important for neurite outgrowth, which is defective upon PQBP1 depletion. Collectively, we showed that PQBP1 has a role in AS and indicate aberrant AS as a possible mechanism underlying the symptoms of neurological disorders associated with PQBP1.
PQBP1 affects AS
Our work presents several lines of evidence that PQBP1 affects the AS of many pre-mRNAs. Previous work has indicated an association of PQBP1 with mRNA processing factors (Waragai et al. 1999, 2000; Okazawa et al. 2002; Komuro et al. 1999). We discovered that PQBP1 colocalizes with SC35 in nuclear speckles (Fig. 4B; Supplemental Fig. S3A), which are nuclear subdomains enriched for many splicing regulatory factors (Spector and Lamond 2011). Moreover, profiling of PQBP1-associated proteins revealed a set of factors highly enriched in splicing regulators and spliceosome components (Fig. 2E; Supplemental Table S1). Among these binding partners, we also identified WBP11, a previously reported interactor of PQBP1 (Supplemental Table S1; Komuro et al. 1999). These results support PQBP1's connection with the splicing machinery.
PQBP1 affects U2 snRNP component SF3B1 function
We observed that PQBP1 did not associate with functional AS target mRNAs in vivo in CLIP experiments, suggesting that it does not bind directly to the AS targets identified in HeLa cells (Fig. 3A), consistent with its apparent lack of known RNA-binding domains (Fig. 2A). SF3B1, which was previously shown to directly bind substrate mRNAs and affect Bcl-x AS (Gozani et al. 1998; Massiello et al. 2006) and was herein identified in PQBP1 affinity purifications (Supplemental Table S1), constituted a prime candidate to work together with PQBP1. Consistent with this hypothesis, depletion of PQBP1 decreased SF3B1's association with Bcl-x mRNA (Fig. 3D). In contrast, the binding of SF3B1 to Mcl1, a target of SF3B1 (but not of PQBP1) was not affected by the knockdown of PQBP1 (Fig. 3D). These findings suggest that PQBP1 affects the association of SF3B1 with a distinct subset of substrate mRNAs. Furthermore, we found that a subset of PQBP1 targets affected in their splicing was similarly influenced by the knockdown of SF3B1 (Supplemental Fig. S5). SF3B1 is a central component of the U2 snRNP. Recruitment of U2 snRNP to the intronic branchpoint sequence (BPS) is a key step in the assembly of the spliceosomal A complex and thus the selection of splice sites (Wahl et al. 2009). We propose a model whereby through an association with SF3B1, PQBP1 influences the recruitment of U2 snRNP to specific target sites, thus affecting splicing decisions on a subset of mRNAs (Fig. 3E). The precise mechanism by which this occurs remains to be explored.
PQBP1 affects the AS of mRNAs in neurons
Depletion of PQBP1 causes a decrease in outgrowth and branching of dendrites in mouse primary cortical neurons (Fig. 4G,H; Supplemental Fig. S3B,C). This morphological defect was not apoptosis-related (Fig. 4E,F). Qualitatively, dendrites close to the nucleus appeared more affected than distal dendrites by knockdown of PQBP1 (Figs. 4H, 5D). The defect in neurite morphology caused by loss of PQBP1 could be rescued by re-expression of wild-type PQBP1 (Fig. 5C,D), confirming the correlation between neurite deformity and the loss of PQBP1 expression. Transcriptome-wide profiling revealed that PQBP1's AS targets were enriched for factors regulating neuron projection development (Fig. 6C).
To further investigate the effect of PQBP1 on neurons, we focused on the VASE-associated isoforms of NCAM-140 encoded by Ncam1, which has been implicated in neurite outgrowth and development. Ncam1 contains five Ig-like domains involved in homophilic binding of Ncam1 either in cis or trans, which in turn controls cell–cell interactions and the induction of neurite outgrowth (Walsh and Doherty 1997). The AS of VASE determines the inclusion or exclusion of the fourth Ig domain and alters the homophilic binding of NCAM-140 (Lahrtz et al. 1997). The VASE− isoform of NCAM-140 stimulates—while VASE+ inhibits—neurite outgrowth (Doherty et al. 1992; Liu et al. 1993; Saffell et al. 1994).
We found that the ratio of VASE−/VASE+ expression corresponded to the level of functional PQBP1 and neurite outgrowth (Figs. 4D, 5C, 7C,E,F). Moreover, restoration of the proper ratio of VASE−/VASE+ “rescued” the abnormal neurite outgrowth associated with the knockdown of PQBP1 (Fig. 7F,G). These results identified NCAM-140 as a critical target connecting PQBP1-mediated AS to the cellular process of neurite projection and outgrowth. However, the ability of enforced VASE−/VASE+ expression to rescue the neurite outgrowth defect caused by PQBP1 down-regulation does not exclude the possibility that other PQBP1 targets contribute to this phenotype. For example, PQBP1 AS targets are also enriched in ARF-related proteins (Fig. 6C). The members of the ARF family of small GTPases were previously reported to affect dendritic branching and spine formation (Hernández-Deviez et al. 2002; Moore et al. 2007).
A possible role for PQBP1 in neurological disorders
Clinical genetics identified mutations in PQBP1 as the direct cause for Renpenning syndrome, a representative XLMR disorder characterized by microcephaly, short stature, multiple body malformations, and mental retardation (Kalscheuer et al. 2003; Stevenson et al. 2005). We examined the function of two disease-associated PQBP1 variants—a truncated protein resulting from a ΔAG deletion and a point mutation containing a Y65C mutation in the WW domain. We discovered that the disease-associated PQBP1 mutants had deficiencies in splicing-related functions. Our proteomics profiling indicates that the association between splicing-related factors and wild-type PQBP1 was lost or altered in the Y65C and ΔAG mutants, respectively (Supplemental Table S1). These results indicate that the association of PQBP1 with splicing factors is important for its function. In line with their compromised association with the splicing machinery, neither ΔAG nor Y65C restored the AS of NCAM-140 VASE when PQBP1 was knocked down in mouse primary neurons (Fig. 7C). Furthermore, both mutants were also unable to rescue the defects in neurite outgrowth upon depletion of PQBP1 (Fig. 5E,F). Defects in dendritic development are consistently found in many other mental retardation disorders, including Fragile-X mental retardation and Rett syndrome (Parrish et al. 2007). Our results suggest that disease-associated PQBP1 variants are deficient in maintaining the proper function of PQBP1 in AS. Moreover, aberrant splicing of specific AS targets may contribute to the disease symptoms in PQBP1-related mental retardation disorders.
In summary, we propose that PQBP1, a protein related to neurological diseases, affects AS through association with the core splicing complex. Aberrant AS of specific targets resulting from misregulation of PQBP1 mutants may be one of the causes of PQBP1-related neurological disorders.
Materials and methods
Plasmids, cell culture, and siRNA transfection
Details of plasmid and stable cell line constructions are included in the Supplemental Material. HeLa and 293T cell lines (American Type Culture Collection) were grown in standard conditions. siRNAs were transfected with Hiperfect (Qiagen) at a final concentration of 40 nM. siRNAs were siGENOME siRNAs (Thermo Scientific) for PQBP1 and firefly luciferase. Transfections were optimized to reach ≥85% protein depletion 72 h post-transfection, as determined by Western blotting.
Mouse cortical culture, plasmid transfection, and lentivirus infection
Mouse embryonic cortical neurons were prepared and seeded as in Kim et al. (2010). For direct neuron transfection, MISSION shRNA lentivirus constructs for PQBP1 (Sigma) and a nontargeting shRNA vector (Addgene Plasmid 1864) were transfected into neurons on the day of seeding with Lipofectamine 2000 (Invitrogen). For lentiviral infection of neurons, 293T cells were transfected with shRNA constructs and packaging vectors. Lentivirus-containing supernatants were collected 48 h post-transfection. Neurons were then incubated with the supernatants for 6 h on the day of seeding. Infection was optimized to reach ≥60% protein depletion 72 h post-infection, as determined by Western blotting.
RNA extraction and RT–PCR
Cellular RNA was extracted by RNeasy minikit (Qiagen). RNAs (1.5–4 μg) were reverse-transcribed by SuperScript First-Strand Synthesis system (Invitrogen). Bcl-x and NCAM-140 VASE splicing isoforms were analyzed by RT–PCR through primers flanking the alternatively spliced region. A linear range for cDNA input was determined by running a twofold dilution series of 28-cycle PCR reactions. Products were run on 5% Tris-Borate-EDTA gels (Bio-Rad), stained with SYBR gold stain (Invitrogen), and quantified with ImageJ software (National Institutes of Health). Triplicate samples in the linear range were run to determine isoform ratios. Real-time PCR primers were designed with the software Primer3 (SourceForge) and verified by electrophoresis to generate single, correctly sized products. PCR reactions were prepared with SYBR Green master mix (Applied Biosystems), run on a MasterCycler ep realplex (Eppendorf), and quantified by the ΔΔCt method. Primer sets are listed in Supplemental Table S5.
Tandem affinity immunopurification
Flag-HA-tagged PQBP1 and the Y65C and ΔAG mutants were purified from HeLa cell extracts by tandem affinity purification and visualized by silver stain as recently described (Adelmant et al. 2012). Specific details are provided in the Supplemental Material.
Sample preparation and mass spectrometry analysis
Sample preparation and liquid chromatography/tandem mass spectrometry analysis were done with slight modifications to a recently described method (Rozenblatt-Rosen et al. 2012). Specific details are provided in the Supplemental Material.
Western blotting
Cellular proteins were extracted with 1× RIPA buffer (1× PBS, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitors). Ten micrograms to 30 μg of protein was loaded onto each lane of a NuPAGE 4%–12% Bis-Tris gel. Proteins were transferred onto PVDF membranes and probed with the following primary antibodies diluted in 1× PBST with 5% nonfat milk: α-PQBP1 (Santa Cruz Biotechnology), α-Flag (Sigma), α-HA (Roche), α-actin (Chemicon), and α-SF3B1 (MBL).
CLIP
HeLa cells were cross-linked at 4000 mJ/cm2 with a UV Stratalinker (Stratgene) and suspended in 1× RIPA buffer supplemented with protease inhibitors, 10 mM DTT, and 0.4 U/μL RNaseOUT (Invitrogen). Lysates were treated briefly with RQ1 RNase-free DNase (Promega) and cleared by centrifugation. Dynal beads (Invitrogen) that were precoupled with 10 μg of antibody or nonrelevant IgG were incubated with the supernatants and rocked for 4 h at 4°C. Beads were then washed three times with the lysis buffer plus 300 mM NaCl and twice with 1× PNK buffer (50 mM Tris-Cl at pH 7.4, 10 mM MgCl2, 0.5% NP-40). RNA–protein complexes were eluted and treated with Protease K (Ambion), and RNAs were extracted by acid phenol/chloroform. Reverse transcription was performed with a 1:1 mix of oligo-dT and random hexamers, and cDNAs were analyzed by real-time PCR.
Immunofluorescence
Neurons were fixed with 4% paraformaldehyde and 2% sucrose/PBS for 8 min. Cells were then washed three times with 1× PBS and treated with 1× GDB (gelatin dilution buffer) buffer (0.3% Triton, 0.1% gelatin) for 15 min. Cells were then incubated with antibodies, stained with DAPI, and imaged on a Nikon Ti inverted fluorescence microscope. For costaining, we swapped the fluorophores on secondary antibodies to confirm there was no bleed-through in the fluorescence channels.
Annexin V staining
Neurons were incubated with FITC-conjugated Annexin V (BioLegend) diluted 1:50 in medium for half an hour at 37°C. Neurons were then directly imaged alive on a Nikon Ti inverted fluorescence microscope.
RNA-seq sample preparation
Libraries were prepared from total RNA following the instructions of the TruSeq RNA sample preparation kit (Illumina). Samples were then run on Illumina HiSeq 2000. Detailed computational methods for AS target profiling are provided in the Supplemental Material. The reported sequencing data have been deposited in NCBI's Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO series accession number GSE44402.
GO functional analysis
GO term enrichments for proteins associated with PQBP1 in HeLa cells were determined with the DAVID (Database for Annotation, Visualization, and Integrated Discovery) Bioinformatics Database (Dennis et al. 2003; Huang et al. 2009). Functional GO enrichments of PQBP1 AS targets in neurons were determined with GoMiner software (Zeeberg et al. 2003).
Acknowledgments
We thank Hsi-Wen Liao, Athar Malik, David Lipton, and Seth Margolis for help with neuron culture and RNAi; Alan Mardinly for help with Sholl analysis; Jennifer Waters for help with imaging; and Daniel Ducat and Yvonne Chen for thoughtful critiques. We are indebted to Michael Greenberg's laboratory for the generous sharing of mouse embryonic cortical neurons. Microscope imaging was done in the Nikon Imaging Center at Harvard Medical School. Sequencing was performed by the biopolymer facility at Harvard Medical School. This work was supported by grants GM36373 and GM057476 to P.A.S. from the National Institutes of Health.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.212308.112.
References
- Adelmant G, Calkins AS, Garg BK, Card JD, Askenazi M, Miron A, Sobhian B, Zhang Y, Nakatani Y, Silver PA, et al. 2012. DNA ends alter the molecular composition and localization of Ku multicomponent complexes. Mol Cell Proteomics 11: 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgul C, Moulding DA, Edwards SW 2004. Alternative splicing of Bcl-2-related genes: Functional consequences and potential therapeutic applications. Cell Mol Life Sci 61: 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM 2008. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40: 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarova CF, Hims MM, Bolz H, Abu-Safieh L, Patel RJ, Papaioannou MG, Inglehearn CF, Keen TJ, Willis C, Moore AT, et al. 2002. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet 11: 87–92 [DOI] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. 2012. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75: 437–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3. [PubMed] [Google Scholar]
- Doherty P, Moolenaar CE, Ashton SV, Michalides RJ, Walsh FS 1992. The VASE exon downregulates the neurite growth-promoting activity of NCAM 140. Nature 356: 791–793 [DOI] [PubMed] [Google Scholar]
- Dredge BK, Polydorides AD, Darnell RB 2001. The splice of life: Alternative splicing and neurological disease. Nat Rev Neurosci 2: 43–50 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M Jr, Mody I, Black DL 2011. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet 43: 706–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ, Black DL 2001. Alternative RNA splicing in the nervous system. Prog Neurobiol 65: 289–308 [DOI] [PubMed] [Google Scholar]
- Hernández-Deviez DJ, Casanova JE, Wilson JM 2002. Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci 5: 623–624 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gécz J, Sefiani A, Hoffmann K, Moser B, Haas S, et al. 2003. Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet 35: 313–315 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Saeki M, Kato S 1999. Association of two nuclear proteins, Npw38 and NpwBP, via the interaction between the WW domain and a novel proline-rich motif containing glycine and arginine. J Biol Chem 274: 36513–36519 [DOI] [PubMed] [Google Scholar]
- Kunde SA, Musante L, Grimme A, Fischer U, Muller E, Wanker EE, Kalscheuer VM 2011. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum Mol Genet 20: 4916–4931 [DOI] [PubMed] [Google Scholar]
- Lahrtz F, Horstkorte R, Cremer H, Schachner M, Montag D 1997. VASE-encoded peptide modifies NCAM- and L1-mediated neurite outgrowth. J Neurosci Res 50: 62–68 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB 2006. Splicing regulation in neurologic disease. Neuron 52: 93–101 [DOI] [PubMed] [Google Scholar]
- Liu L, Haines S, Shew R, Akeson RA 1993. Axon growth is enhanced by NCAM lacking the VASE exon when expressed in either the growth substrate or the growing axon. J Neurosci Res 35: 327–345 [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G 2007. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Lührmann R 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J 23: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE 2006. SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J 20: 1680–1682 [DOI] [PubMed] [Google Scholar]
- McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, et al. 2001. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 10: 1555–1562 [DOI] [PubMed] [Google Scholar]
- Moore CD, Thacker EE, Larimore J, Gaston D, Underwood A, Kearns B, Patterson SI, Jackson T, Chapleau C, Pozzo-Miller L, et al. 2007. The neuronal Arf GAP centaurin α1 modulates dendritic differentiation. J Cell Sci 120: 2683–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Kunde S-A, Sulistio TO, Fischer U, Grimme A, Frints SGM, Schwartz CE, Martínez F, Romano C, Ropers H-H, et al. 2010. Common pathological mutations in PQBP1 induce nonsense-mediated mRNA decay and enhance exclusion of the mutant exon. Hum Mutat 31: 90–98 [DOI] [PubMed] [Google Scholar]
- Okazawa H, Rich T, Chang A, Lin X, Waragai M, Kajikawa M, Enokido Y, Komuro A, Kato S, Shibata M, et al. 2002. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron 34: 701–713 [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN 2007. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 30: 399–423 [DOI] [PubMed] [Google Scholar]
- Qi Y, Hoshino M, Wada Y-I, Marubuchi S, Yoshimura N, Kanazawa I, Shinomiya K-I, Okazawa H 2005. PQBP-1 is expressed predominantly in the central nervous system during development. Eur J Neurosci 22: 1277–1286 [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, et al. 2012. Interpreting cancer genomes using systematic host network perturbations by tumor virus proteins. Nature 487: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffell JL, Walsh FS, Doherty P 1994. Expression of NCAM containing VASE in neurons can account for a developmental loss in their neurite outgrowth response to NCAM in a cellular substratum. J Cell Biol 125: 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87: 387–406 [PMC free article] [PubMed] [Google Scholar]
- Small SJ, Akeson R 1990. Expression of the unique NCAM VASE exon is independently regulated in distinct tissues during development. J Cell Biol 111: 2089–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson RE, Bennett CW, Abidi F, Kleefstra T, Porteous M, Simensen RJ, Lubs HA, Hamel BCJ, Schwartz CE 2005. Renpenning Syndrome comes into focus. Am J Med Genet 134: 415–421 [DOI] [PubMed] [Google Scholar]
- Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ, et al. 2010. Y65C missense mutation in the WW domain of the Golabi–Ito–Hall syndrome protein PQBP1 affects its binding activity and deregulates pre-mRNA splicing. J Biol Chem 285: 19391–19401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu J-S, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. 2005. Nova regulates brain-specific splicing to shape the synapse. Nat Genet 37: 844–852 [DOI] [PubMed] [Google Scholar]
- Varani G, Nagai K 1998. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct 27: 407–445 [DOI] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, et al. 2001. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 8: 375–381 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Walsh FS, Doherty P 1997. Neural cell adhesion molecules of the immunoglobulin superfamily: Role in axon growth and guidance. Annu Rev Cell Dev Biol 13: 425–456 [DOI] [PubMed] [Google Scholar]
- Wang G-S, Cooper TA 2007. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waragai M, Lammers CH, Takeuchi S, Imafuku I, Udagawa Y, Kanazawa I, Kawabata M, Mouradian MM, Okazawa H 1999. PQBP-1, a novel polyglutamine tractbinding protein, inhibits transcription activation by Brn-2 and affects cell survival. Hum Mol Genet 8: 977–987 [DOI] [PubMed] [Google Scholar]
- Waragai M, Junn E, Kajikawa M, Takeuchi S, Kanazawa I, Shibata M, Mouradian MM, Okazawa H 2000. PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5-15kD/dim1p via the carboxylterminal domain. Biochem Biophys Res Commun 273: 592–595 [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, et al. 2003. GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol 4: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]