Epigenetic changes are often observed in cancer, but demonstrating a direct role in driving malignancy has been difficult. Here, Stricker et al. reprogram malignant glioblastoma neural stem (GNS) cells to induced pluripotent stem cells. They find that differentiation into mesodermal lineages, but not neural lineages, erases the malignancy of the glioblastoma cell line. Imposing an epigenome associated with an alternative lineage can therefore suppress malignancy. However, in the neural lineage, resetting glioblastoma-associated epigenetic abnormalities is not sufficient to override the cancer genome.
Keywords: glioblastoma, epigenetics, DNA methylation, reprogramming, neural stem cell
Abstract
Epigenetic changes are frequently observed in cancer. However, their role in establishing or sustaining the malignant state has been difficult to determine due to the lack of experimental tools that enable resetting of epigenetic abnormalities. To address this, we applied induced pluripotent stem cell (iPSC) reprogramming techniques to invoke widespread epigenetic resetting of glioblastoma (GBM)-derived neural stem (GNS) cells. GBM iPSCs (GiPSCs) were subsequently redifferentiated to the neural lineage to assess the impact of cancer-specific epigenetic abnormalities on tumorigenicity. GiPSCs and their differentiating derivatives display widespread resetting of common GBM-associated changes, such as DNA hypermethylation of promoter regions of the cell motility regulator TES (testis-derived transcript), the tumor suppressor cyclin-dependent kinase inhibitor 1C (CDKN1C; p57KIP2), and many polycomb-repressive complex 2 (PRC2) target genes (e.g., SFRP2). Surprisingly, despite such global epigenetic reconfiguration, GiPSC-derived neural progenitors remained highly malignant upon xenotransplantation. Only when GiPSCs were directed to nonneural cell types did we observe sustained expression of reactivated tumor suppressors and reduced infiltrative behavior. These data suggest that imposing an epigenome associated with an alternative developmental lineage can suppress malignant behavior. However, in the context of the neural lineage, widespread resetting of GBM-associated epigenetic abnormalities is not sufficient to override the cancer genome.
The dominant model of cancer progression is the multistep accumulation of genetic changes that activate proto-oncogenes and silence tumor suppressors (Hanahan and Weinberg 2000). However, epigenetic mechanisms can also influence the activity of cancer-associated pathways through their effects on transcriptional regulation. Changes in histone modifications and noncoding RNAs have been reported during tumorigenesis (Jones and Baylin 2007). However, the canonical epigenetic defect in human cancer relates to DNA methylation. DNA hypermethylation at promoter regions has been functionally implicated in the stable silencing of PRC2 (Polycomb-repressive complex 2) targets and classical tumor suppressor genes (Jones and Baylin 2007; Widschwendter et al. 2007). Despite these advances, a lack of experimental approaches enabling simultaneous global restoration of a normal DNA methylome in cancer cells has limited our ability to assess whether methylation defects are critical drivers of the malignant state or, rather, secondary epiphenomena (Baylin and Bestor 2002).
Glioblastoma (GBM) is the most prevalent and aggressive form of human brain cancer. GBMs are driven` by an immature cancer stem cell population that displays many characteristics of normal neural stem (NS) cells (Ward and Dirks 2007). In vitro expansion of NS cells and GBM-derived stem cells as primary cell lines is possible by propagating them as neurospheres in suspension culture (Singh et al. 2003; Galli et al. 2004; Lee et al. 2006) or in adherent culture conditions (Pollard et al. 2009). Adherent GBM neural stem (GNS) cells retain stem cell characteristics and are tumor-initiating following xenotransplantation, recapitulating key features of the human disease such as extensive infiltration of surrounding brain regions (Pollard et al. 2009).
Patterns of DNA methylation in primary GBM specimens have recently been cataloged and include disruptions at many novel candidate tumor suppressors, such as the cell motility regulator TES (testis-derived transcript) as well as many polycomb repressor complex 2 (PRC2) target genes (Martinez et al. 2009). Silencing of tumor suppressors associated with “classic” genetically altered pathways (i.e., P53, RTK/PI3K, and CDK/RB) is not frequently observed.
Epigenetic reprogramming of genetically normal human somatic cells to a pluripotent stem cell state has recently been achieved through the expression of defined sets of transcription factors (Takahashi et al. 2007). This seminal work demonstrated that the epigenetic restrictions imposed by normal development are experimentally reversible using simple methods. More recently, it has been shown that transcription factor-mediated reprogramming can also be applied to human cancer cell lines (Carette et al. 2010; Miyoshi et al. 2010). However, several important issues remain unclear. First, can human cancer cells with highly aneuploid genomes be successfully reprogrammed? Second, if so, are cancer-specific epigenetic abnormalities erased? Third, does removal of these abnormal marks correlate with transcriptional changes and suppression of malignant behavior? Fourth, are these effects independent of the cell identity and developmental epigenome? In this study, we address these issues and demonstrate that transcription factor-mediated nuclear reprogramming can enable widespread resetting of cancer-specific DNA methylation marks in GNS cells. This enabled us to assess the relative contribution of the cancer epigenome to malignant cellular behavior.
Results
GNS cells can generate induced pluripotent stem cell (iPSC)-like colonies
We sought to identify GNS cell lines that might be readily reprogrammed in order to explore the functional consequences of resetting GBM-associated DNA methylation defects. Consistent with our previous studies, we confirmed that a panel of 14 GNS cell lines (derived from independent tumor specimens) express high levels of SOX2 and C-MYC but lack expression of the pluripotency-associated factors OCT4 and NANOG (Fig. 1A; Supplemental Fig. 1A–D). We therefore reasoned that some of these lines might be reprogrammable to pluripotency through delivery of only two transcription factors, OCT4 and KLF4, as has been reported for mouse NS cells (Kim et al. 2008). This strategy would enable us to avoid exogenous expression of C-MYC.
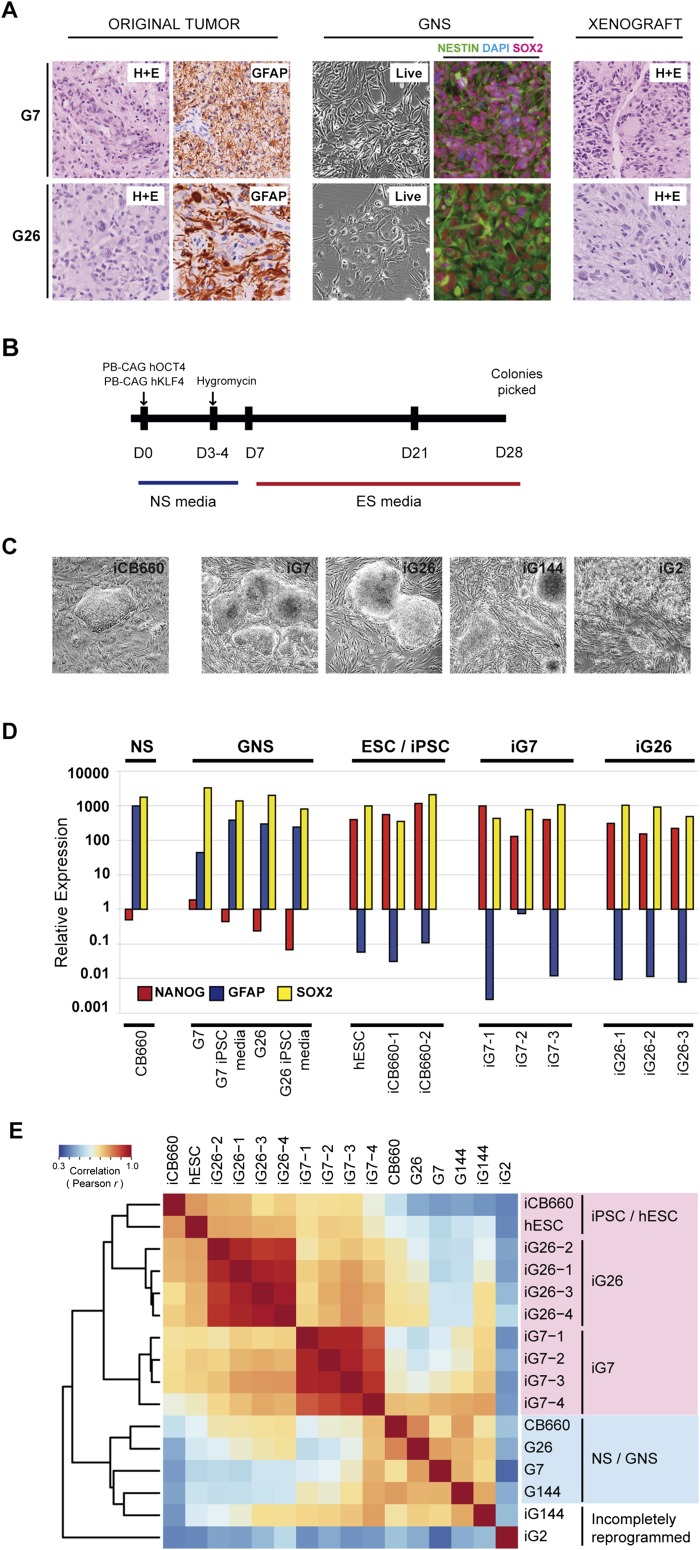
Figure 1.
GNS cells can be converted to an iPSC-like state. (A) GNS cell lines G7 and G26 were established from tumor samples obtained from two different patients. (Left panels) Original tumors show typical GBM histopathology (H&E) and GFAP immunoreactivity. G7 and G26 grow as adherent cell lines and are positive for the immature neural progenitor markers SOX2 and NESTIN. (Right panels) Upon xenotransplantation, they form tumors similar to the original patient tumor. (B) Strategy used for epigenetic reprogramming of GNS cells. Cells (2 × 106 to 6 × 106) were transfected with piggyBac vectors (KLF4 and OCT4 driven by a CAG promoter). Hygromycin selection was applied for at least 3 wk. Medium was changed to hESC condition after 1 wk. (C) Colonies resembling typical hESC colonies emerged after 4–7 wk for iG7, iG26, and control NS cells (iCB660). iG2 and iG144 were less well defined. Shown are typical examples after clonal colony picking and initial passaging (P3). (D) qRT–PCR for the critical pluripotency marker gene NANOG and the neural marker gene GFAP. Following reprogramming, these genes are activated and suppressed, respectively, and reach levels similar to control iPSCs or hESCs. SOX2 is expressed by ESCs, iPSCs, and NS cells. Levels were normalized to GAPDH. RNA was derived from iPSC and GiPSC culture passages 4–8. (E) Correlation analysis of hESCs, iPSCs, NS cells, GNS cells, and GiPSCs, based on expression levels of 90 markers present on the TLDA (Applied Biosystems) pluripotency panel. iG7 and iG26 are more similar to iPSCs and hESCs than to corresponding parental GNS cells or normal NS cells (CB660). iG2 and iG144 likely represent incompletely reprogrammed lines and failed to correlate with iPSCs/hESCs. RNA was derived from iPSC and GiPSC culture passages 4–8. See also Supplemental Figure 1.
We used the piggyBac transposon system to efficiently deliver excisable vectors PB-OCT4 and PB-KLF4 to GNS cell cultures and the genetically normal human NS cell line CB660 (Fig. 1B; Kaji et al. 2009). Remarkably, despite karyotypic abnormalities in GNS cells, seven of the 14 GNS cell lines tested gave rise to iPSC-like colonies at an efficiency similar to normal NS cells (Fig. 1C; Supplemental Table 1), and four of these could be continuously expanded (G2, G7, G26, and G144). iPSC clones derived from G7 and G26 showed consistent up-regulation of the pluripotency marker NANOG (>1000-fold) and down-regulation of the neural marker GFAP (>1000-fold) (Fig. 1D; Supplemental Fig. 1E).
To assess whether this indicated acquisition of an iPSC-like phenotype, we determined expression levels of pluripotency markers using the TaqMan low-density array (TLDA) human pluripotency microfluidic cards (Applied Biosystems). Cluster analysis confirmed that iG7 and iG26 expressed markers similar to human embryonic stem cells (hESCs) and control iPSCs (iCB660), whereas iG144 and iG2 appeared incompletely reprogrammed (Fig. 1E; Supplemental Fig. 1F). GNS cells that were directly replated into ESC culture medium on feeder cells (without transfection) never showed up-regulation of pluripotency markers (Fig. 1D). iG7 and iG26 colonies are immunopositive for the hESC surface markers Tra1-60, Tra1-81, SSEA4, Tra2-49, and Tra2-54 and show a strong nuclear NANOG signal at levels similar to control iPSCs (Fig. 2A). Thus, iG7 and iG26 represent GBM cells reprogrammed to an iPSC-like state (GBM iPSCs [GiPSCs]). Six clonal GiPSCs were analyzed in greater detail to explore the effects of reprogramming on the cancer epigenome (three independent lines from both G7 and G26; iG7-1, iG7-2, and iG7-3; iG26-1, iG26-2, and iG26-3).
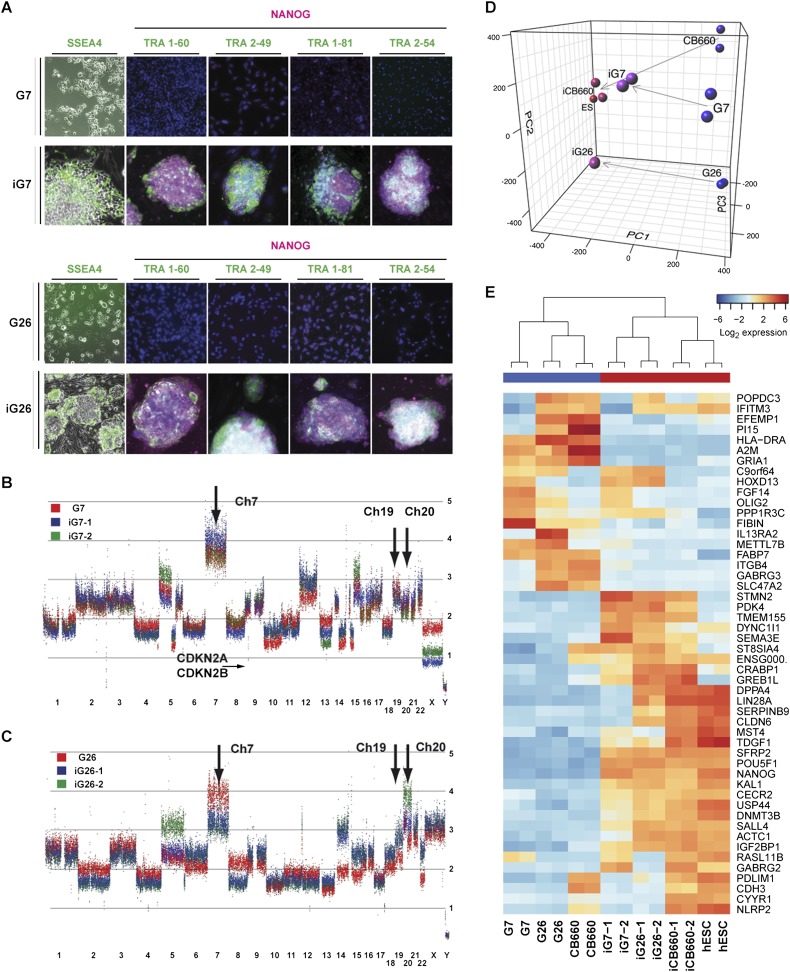
Figure 2.
Gene expression profiling and marker analysis confirms that iG7 and iG26 are reprogrammed to a hESC/iPSC state. (A) Immunocytochemistry for pluripotency marker NANOG and cell surface markers (SSEA4, TRA1-60, TRA2-49, TRA1-81, and TRA2-54). All tested iG7 and iG26 clonal cell lines (iG7-1, iG7-2, iG7-3; iG26-1, iG26-2, and iG26-3; P4–P10) were immunopositive for these pluripotency markers (iG7-1 and iG26-1 are shown), whereas parental GNS lines G7 and G26 were negative. SSEA4 immunostaining is shown in live cells. (B,C) Genomic analysis using Affymetrix SNP 6.0 microarrays for GNS cells (red) and their reprogrammed derivatives (GiPSCs, P8–P12; blue and green) identifies many hallmark genetic changes common to GBM, such as amplification of chromosomes 7, 19, and 20 (arrows) and losses of chromosomes 13, 14, and 15. G7 and iG7 also display a 400-kb deletion that includes CDKN2A and CDKN2B (small arrow). (D,E) PCA of global gene expression (D; see also Supplemental Fig. 2A) and hierarchical clustering (E) of the 50 most significantly differentially expressed genes for normal NS cells and GNS cells (CB660, G7, and G26), hESCs and two clonal GiPSCs (iG7-1, iG7-2; iG26-1, and iG26-2), and iPSCs (iCB660) confirms that iG7 and iG26 are extensively reprogrammed to an ESC-like state. Analyzed iPSCs and GiPSCs were between passages 6 and 11.
GiPSCs retain GBM genomes but display a transcriptional profile similar to ESCs
We next analyzed the genetic disruptions of the parental tumor cells using SNP arrays to confirm that they possess a genome typical of GBM. The spectrum of common genetic lesions underlying GBM is well established (Cancer Genome Atlas Research Network 2008). We identified in G7 and G26 many hallmark genetic alterations associated with GBM, including copy number increases of chromosomes 7, 19, and 20 and loss of chromosomes 13, 14, and 15 (Fig. 2B,C). G7 harbors a local 400-kb deletion at the CDKN2A (p16) locus, while G26 contains a mutation in the TP53 gene (R248Q) commonly observed in GBM (Supplemental Fig. 2B; data not shown). Gene expression profiling of G7 and G26 indicates that they are representative of different GBM subtypes (Verhaak et al. 2010), “proneural/classical” and “mesenchymal,” respectively (E Johnstone and P Bertone, pers. comm.; data not shown). Neither harbored IDH1 mutations that are characteristic of secondary GBMs or significant DNA hypermethylation at promoters commonly silenced in glioma-CpG island methylator phenotype (G-CIMP) tumors (Supplemental Figs. 2B, 3; Noushmehr et al. 2010). Together, these data support the original patient tumor diagnoses for G7 and G26 as primary GBM (Fig. 1A).
To determine the extent of reprogramming in GiPSCs, we carried out global transcriptome analyses. We assessed mRNA expression in iG7, iG26, and iCB660; the corresponding parental lines G7, G26, and CB660; and the hESC line Edi-2 as a comparative reference (Falk et al. 2012). Principal component analysis (PCA) of global expression and hierarchical clustering of differentially expressed genes indicates that all GiPSCs undergo dramatic transcriptional resetting and acquire a gene expression pattern closer to normal human iPSCs and ESCs than to NS cells (Fig. 2D,E; Supplemental Fig. 2A). Importantly, the patterns of structural chromosomal changes in parental GNS cells were largely retained through the reprogramming process in the GiPSCs (Fig. 2B,C), indicating that GiPSCs did not display significant genomic instability through the experimental procedure.
GNS cells display aberrant DNA methylation marks that are frequently observed in the human disease
To characterize cancer-specific DNA methylation abnormalities, we initially performed DNA methylation profiling using the Infinium Human Methylation27 BeadChip (Illumina). This array interrogates the DNA methylation state at CpG islands and promoters associated with ∼14,000 genes, providing a broad survey of affected loci. We compared methylation levels of GNS cells and NS cells to identify cancer-specific methylation variable positions (cMVPs) (Fig. 3A,B; Supplemental Fig. 2C). A total of 691 cMVPs were found between GNS cells (G26 and G7) and NS cells (252 hypomethylated and 439 hypermethylated) (Supplemental Table 2).
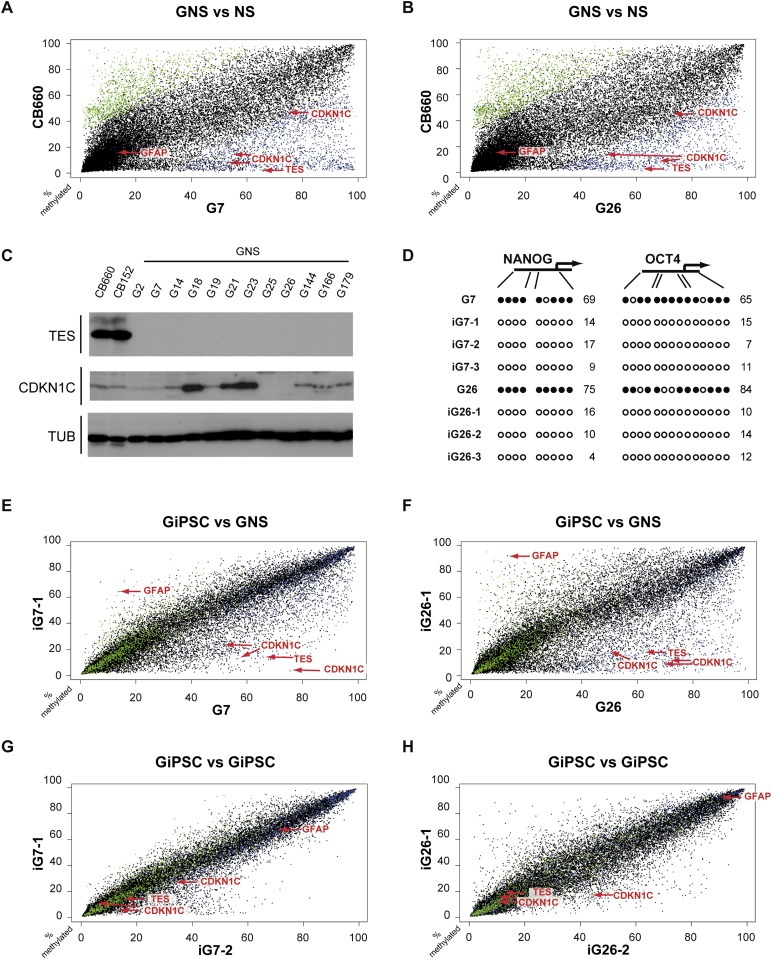
Figure 3.
GNS cells possess epigenetic anomalies common in GBM that can be reset following reprogramming of GNS cells. (A,B) Scatter plots depicting percentage of DNA methylation changes identified using Infinium Human Methylation27 BeadChip arrays (Illumina, Inc.). Each dot represents a distinct CpG site. cMVPs hypermethylated in both G7 and G26 appear in blue, and hypomethylated loci are in green. Normal NS cells (CB660) versus either G7 or G26 identify tumor-specific methylation changes on genes such as CDKN1C (p57KIP2) and TES, which were hypermethylated in both parental GNS lines (see also Supplemental Fig. 2). (C) Immunoblotting showing reduced expression of tumor suppressor genes TES (in all tested GNS lines) and CDKN1C (in several GNS lines, including G7 and G26) when compared with normal NS cells CB660 and CB152. (D) DNA methylation analysis of the OCT4 and NANOG promoters using pyrosequencing. An average methylation level of >50% is depicted as a black circle, while levels <50% are shown as white circles. GNS cells contain extensive DNA methylation at both promoters, and these are removed following reprogramming. Numbers are percentage of average DNA methylation at the OCT-4 and NANOG promoters. (E–H) Scatter plots depicting DNA methylation levels (percentage) analyzed with Infinium Human Methylation27 BeadChip arrays (Illumina, Inc.). Each dot represents a distinct CpG site. cMVPs hypermethylated in both G7 and G26 appear in blue, and hypomethylated cMVPs appear in green. (E,F) iG7-1(GiPSC) versus G7 (GNS) and iG26-1 (GiPSC) versus G26 (GNS) illustrate extensive changes in methylation patterns after reprogramming. (G,H) iG7-1 versus iG7-2 and iG26-1 versus iG26-2 illustrate similarities between individual GiPSC clonal lines. Analyzed GiPSC clones were between passages 6 and 11.
In primary GBMs, methylation changes are rare at tumor suppressors within the “classic” genetically disrupted CDK/RB, RTK, and P53 signaling pathways. Instead, hypermethylation is frequently detected at noncanonical genes/pathways, such as PRC2 targets (Widschwendter et al. 2007; Martinez et al. 2009). Consistent with these data, promoter hypermethylation of the tumor suppressor genes PTEN and TP53 was not detected in G7 or G26 or in other tested GNS cell lines. We noted a high proportion of PRC2 target genes in the set of cMVPs (22.1% [G7] and 12.1% [G26]; P = 0.0001). GNS cell-specific DNA hypermethylation also included the tumor suppressor genes cyclin-dependent kinase inhibitor 1C, (CDKN1C, encoding p57KIP2) and TES. Loss of CDKN1C expression is a frequent event in a large variety of tumors, including gliomas (Christensen et al. 2011), and is often accompanied by DNA hypermethylation (Kavanagh and Joseph 2011). TES is expressed in a wide range of adult human tissues, including brain (Tatarelli et al. 2000), and has been previously shown to inhibit tumorigenicity of human cancer cell lines (Tobias et al. 2001). DNA hypermethylation of the TES promoter is one of the most frequent epigenetic disruptions in primary GBM (∼60% of cases) (Mueller et al. 2007; Martinez et al. 2009; Christensen et al. 2011) and is also observed in other human cancers (Qiu et al. 2010; Weeks et al. 2010). We analyzed methylation profiles generated for 67 GBM samples by The Cancer Genome Atlas (TCGA) project. This revealed that TES hypermethylation significantly associates with patient survival (log rank P-value = 0.00232) and loss of TES expression (data not shown). Thus, patterns of DNA methylation in G7 and G26 faithfully mirror the human disease and include some of the most common epigenetic anomalies observed in primary GBM tumors.
To determine whether hypermethylation of TES and CDKN1C promoter regions correlated with transcriptional silencing in GNS cells (G7 and G26) we performed immunoblotting (Fig. 3C). While CDKN1C showed variable expression across GNS cell lines, TES was fully silenced in both GNS cell lines and was also undetectable in 10 additional cells lines assayed (Fig. 3C). Both genes were significantly down-regulated in G7 and G26 when compared with normal NS cells. These data, together with our recent expression profiling analysis (Engström et al. 2012), identify TES as one of the most consistently down-regulated genes in GNS cells. In keeping with their previously described functions, we found that exogenous expression of CDKN1C in G7 cells reduces cellular growth, while expression of TES reduces cellular motility (Supplemental Fig. 4).
GNS cell reprogramming involves widespread resetting of DNA methylation, including cancer-specific marks
We next assessed whether the reprogramming machinery would be able to reset any of the above-described cMVPs. A hallmark of successful transcription factor-based reprogramming is the removal of epigenetic restrictions, such as the promoter-specific DNA methylation of OCT4 and NANOG. As expected, the cancer cell lines G7 and G26, similarly to other somatic cells, showed pronounced DNA hypermethylation on CpG sites proximal to the OCT4 and NANOG transcriptional start sites (Fig. 3D). Coincident with transcriptional activation, these specific marks were erased during global reconfiguration of DNA methylation that takes place during the reprogramming process. These changes were largely consistent across profiles from clonal GiPSCs (Fig. 3E–H). In accordance with lineage reprogramming, the glial lineage marker GFAP was hypermethylated in both iPSCs and GiPSCs and also exhibited concomitant silencing of gene expression during reprogramming (Fig. 3E–H). Interestingly, the majority of the cMVPs identified in both G7 and G26 could be reset (450 of 691; 334 originally hypermethylated in GNS, and 116 originally hypomethylated in GNS), suggesting that these were not permanently imposed by the underlying genetic pathways. Hypermethylated CDKN1C and TES loci were demethylated during reprogramming, and this was validated by independent pyrosequencing (Supplemental Fig. 2D,E). These results confirm that aneuploid GNS cells can be reprogrammed using only two transcription factors and that associated epigenetic changes include erasure of abnormal GBM-associated patterns of DNA methylation.
GiPSCs generate immature multilineage teratomas in vivo
To test whether GiPSCs could engage in differentiation programs, iCB660, iG7, and iG26 cells were injected either subcutaneously or into the kidney capsules of NOD/SCID mice. In each case, compact and noninfiltrative tumors formed within 4–7 wk with macroscopic appearance as teratomas (iCB660, five of six; iG7, eight of eight; iG26, eight of nine) (Fig. 4A). As expected, teratomas did not form following injection of parental G7 and G26, and only glioma-like growths emerged (four of six and three of six).
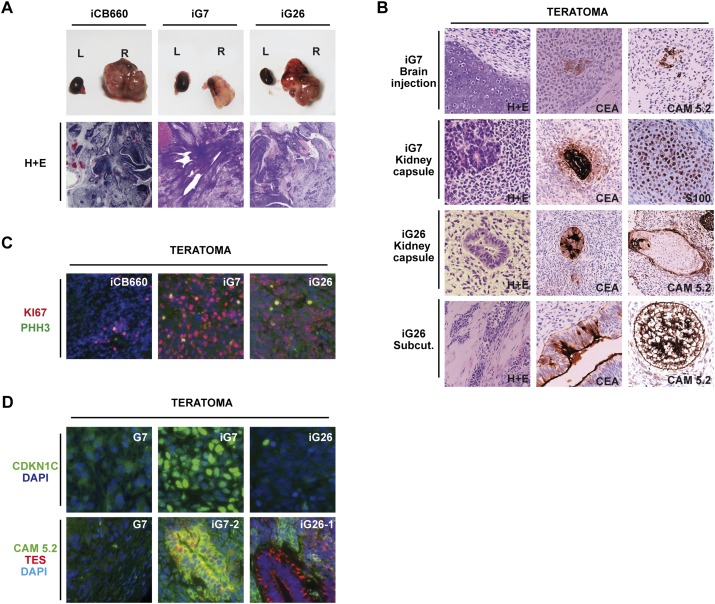
Figure 4.
GiPSCs form multilineage teratomas in vivo. (A) Appearance of teratoma-like tumors generated following transplantation of GiPSCs into the right kidney capsule (iG7 and iG26, P8–P11). (L) Left control kidney; (R) right control kidney. iG7 and iG26 tumors were similar in macroscopic appearance to normal iPSC-derived tumors (B) iG7 and iG26 gave rise to immature teratomas. Examples of immature neural-like rosettes (H&E; iG7, bottom panel; iG26, top panel) as well as more differentiated nonneural tissues such as mesenchymal cartilaginous differentiation (H&E; iG7, top panel) muscle (mesoderm; H&E; iG26, bottom panel), glandular structures (endoderm and CEA+) and nonneural ectoderm (hair follicle CAM5.2+). Similar results were obtained for tumors derived from both kidney capsule and subcutaneous injections. (Top panels) Brain transplantation of iG7 also gave rise to immature teratomas with regions of nonneural tissues. (C) Mitotic markers Ki67 and phospho-histone H3 (PPH3) are observed at higher frequency in GiPSC tumors than control iPSCs. (D) Teratomas contain CDKN1C- and TES-expressing tissues.
Teratoma formation is an intrinsic property of pluripotent cells and can be used to assess differentiation potential. Histological analysis (haematoxylin and eosin [H&E] staining) and immunohistochemistry confirmed that in contrast to parental GNS cells, all GiPSC-derived tumors displayed more complex patterns of differentiation, including regions representative of a variety of tissue types of all three germ layers (Fig. 4B; Supplemental Fig. 5A). These included glandular endodermal structures (CEA+), mesoderm (muscle and cartilage S100+), nonneural ectoderm (hair follicle and Cam5.2+), and neural rosettes (Nestin+) (Fig. 4B; Supplemental Fig. 5A). However, despite the presence of distinct tissue types, most of the tumor mass comprised densely packed cells with immature histological features that included many more mitotic figures or Ki67- or PHH3-immunopositive cells than control teratomas (Fig. 4C; Supplemental Fig. 5C,D).
OCT4 expression was only detected in a small subpopulation of teratoma cells at a frequency similar to control iPSCs, ruling out these tumors as malignant teratocarcinomas (Supplemental Fig. 5B). However, in both iG7 and iG26 teratomas, the majority of cells expressed the neural progenitor marker Nestin and Ki67 (Supplemental Fig. 5B). Thus, we found that GiPSCs are not “trapped” in the iPSC state and can engage in both neural and nonneural differentiation. Although GiPSCs can display multilineage differentiation, they are biased toward the neural lineage and remain highly proliferative.
Transcription of TES and CDKN1C can be activated following epigenetic resetting
In teratomas, TES and CDKN1C protein expression were typically detected within regions that also contained the nonneural epithelial marker Cam5.2 (Fig. 4D). These results suggest that despite widespread resetting of DNA methylation marks in the GiPSCs, re-expression of previously silenced tumor suppressors may be lineage-specific, and neural progenitors may continue to display unconstrained proliferation. To explore this further, we assessed tumor suppressor expression during in vitro differentiation to neural and nonneural lineages.
Either neural or nonneural in vitro differentiation protocols resulted in down-regulation of both mRNA and protein for the pluripotency markers NANOG and OCT4 with kinetics similar to control iPSCs (Fig. 5A; Supplemental Fig. 5E). To determine whether removal of abnormal DNA methylation at tumor suppressor genes correlated with re-expression of the mRNA and protein, we performed immunostaining, immunoblotting, and quantitative RT–PCR (qRT–PCR) for tumor suppressors TES and CDKN1C. We confirmed activation of gene expression for both genes within the differentiating population of GiPSCs and iPSCs (Fig. 5B,C). However, it was also evident that only a subpopulation of cells were immunopositive, suggesting that transcriptional regulation of tumor suppressors is strongly influenced by either developmental lineage or extent of differentiation.
Figure 5.
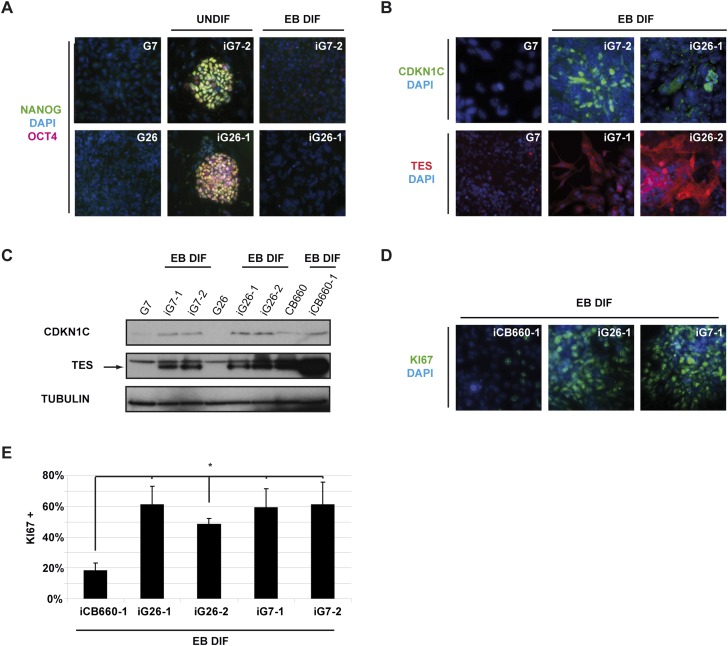
Demethylated tumor suppressor genes can be transcriptionally activated after reprogramming. (A) Immunocytochemistry for the pluripotency markers NANOG and OCT4 before and after in vitro differentiation to EBs. iG7 and iG26 lines are immunopositive for NANOG and OCT4, while the parental GNS lines G7 and G26 are not. NANOG and OCT4 immunoreactivity was down-regulated following differentiation. (B,C) Immunostaining and immunoblotting, respectively, show activation of previously silenced tumor suppressor genes TES and CDKN1C during EB differentiation. (D,E) Ki67 immunocytochemistry was used to score proliferating cells in EB differentiations (10-d + 7-d serum-containing medium) of GiPSC lines and revealed a significant and consistent increase in numbers of cycling cells between GiPSCs and normal iPSCs (iCB660) (P = 8–14). (*) P < 0.03, Student's t-test. See also Supplemental Figure 5.
Notably, differentiating GiPSC cultures formed more frequent and larger embryoid bodies (EBs) than normal iPSCs, with significantly more Ki67-immunopositive cells (more than twofold) (Fig. 5D,E). These data, together with the teratoma assays, suggested that despite epigenetic resetting, the progeny of GiPSCs were not readily able to exit cell cycle and terminally differentiate.
Neural progenitors can be isolated and expanded from differentiating GiPSC cultures
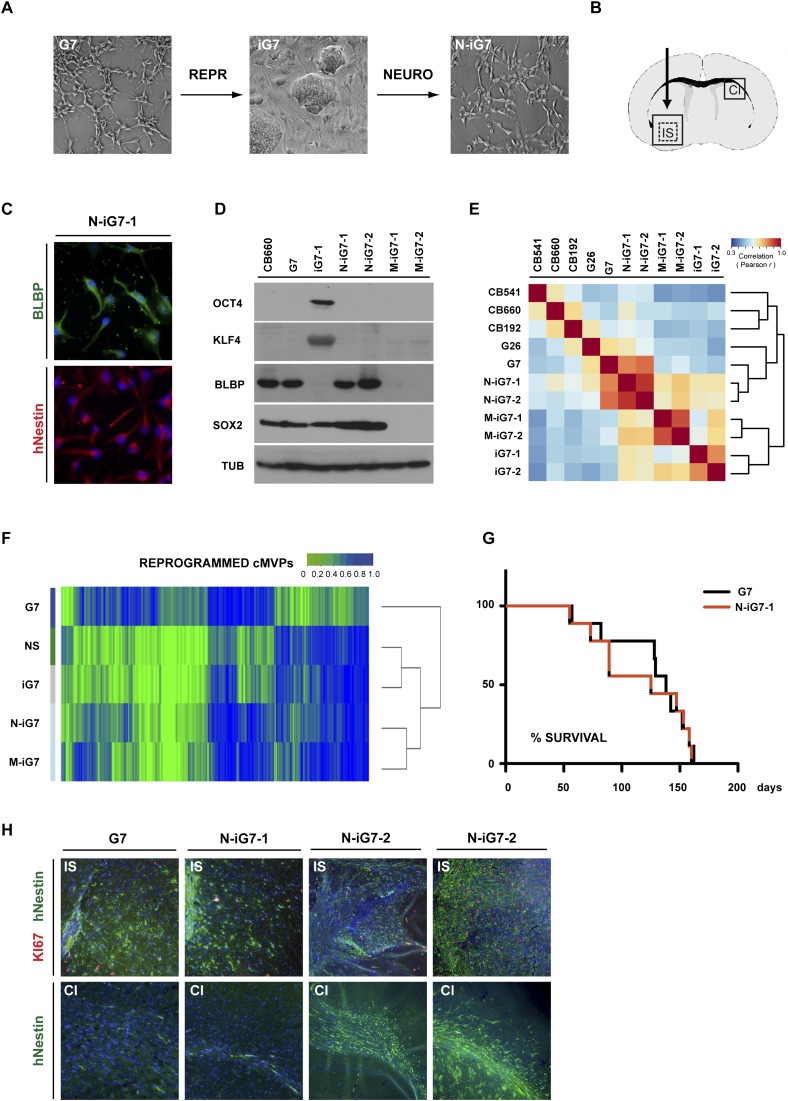
To rigorously assess the functional consequence of resetting cMVPs, we pursued in-depth functional characterization of one GNS cell line (G7) plus two independent GiPSC derivatives (iG7-1 and iG7-2) and four differentiated cultures derived from those (see below). We first differentiated GiPSCs in vitro to produce uniform cultures of NS cells (Pollard et al. 2009), as GNS cells share many characteristics of this cell type. Upon plating in serum-free neural induction medium, GiPSCs underwent rapid morphological changes and, within 7 d, expressed the early neuroepithelial marker PAX6 by qRT–PCR and immunostaining (Supplemental Fig. 5G,H). Neural differentiating cultures of iG7 could be captured and expanded as homogenous populations of neural progenitors (Fig. 6A). Within two to three passages, these cultures (designated N-iG7-1 and N-iG7-2) acquired the morphology typical of both normal NS cell cultures and parental G7 cells and uniformly expressed NS cell markers (e.g., NESTIN and BLBP) but not pluripotency-associated markers (e.g., NANOG and OCT4) (Fig. 6C,D). NS cell identity was confirmed following qRT–PCR analysis of 189 NS cell markers using custom TLDAs (Fig. 6E; Falk et al. 2012).
Figure 6.
GiPSC-derived neural progenitors remain malignant. Neural progenitors (N-iG7-1 and N-iG7-2) were generated by in vitro differentiation of GiPSCs (P12–P14) (A) and injected in the striatum of adult mice (B). Immunostaining (C), immunoblotting (D), and correlation analysis (E) of the expression of 189 NS cell markers using qRT–PCR on custom TLDAs. NS cells (CB660, CB541, and CB192), GNS cells (G7 and G26), GiPSCs (iG7-1 and iG7-2), and neural (N-iG7-1 and N-iG7-2) and nonneural progeny of GiPSCs (M-iG7-1 and M-iG72) show that neural differentiated GiPSC cultures express neural markers at levels comparable with G7. (F) Quantification and correlation of reprogrammed cMVPs. A majority (80% and 83%) of the normalized cMVPs persist in differentiating GiPSCs (N-iG7 and M-iG7). (G) Kaplan-Meier blot depicting the survival of a cohort of 18 adult mice that had 100,000 G7 or N-iG7 cells injected into the striatum. (H) Coronal sections of typical examples of forebrains from mice injected with 100,000 G7 or N-iG7-1 cells after 18 wk. (IS) Injection site; (CI) contralateral side. Immunohistochemistry for human Nestin (hNestin) and Ki67 indicates that N-iG7 cells are highly proliferative and disperse widely from the injection site. Similarly to G7, N-iG7 cells infiltrated the contralateral side of the brain in most cases.
To perform more comprehensive and higher-resolution methylation analysis in the G7 line and its derivatives, we profiled DNA methylation with Infinium Human Methylation 450K BeadChip arrays (Illumina). This enabled widespread assessment of the density and distribution of DNA methylation, including putative regulatory sites at CpG islands, shores, and shelves. Comparison of three normal NS cell lines (CB660, CB1130, and CB152) to G7 revealed 60,977 cMVPs, of which 77% (47,103) were hypermethylated and 23% (13,876) were hypomethylated. Permutation testing showed a significant enrichment (P = <0.0001) for cMVPs in regulatory regions, with >61% of cMVPs being located in CpG islands or CpG island shores.
Assessment of patterns of methylation in iG7 identified epigenetic resetting at >44% of all cMVPs genome-wide (55% of regulatory regions such as CpG islands, island shores, and shelves). These data identified hypermethylated loci in G7 that were removed in iG7 and confirmed our earlier observations for TES and CDKN1C. Multiple CpGs associated with the TES transcription start site and at a region downstream from the CDKN1C transcription start site were identified (Supplemental Fig. 6D). We also confirmed the association of cMVPs with PRC2 target genes, which was found to be highly significant by gene set enrichment analysis (P < 1.11 × 10−16). Remarkably, 85% (559) of PRC2 target genes were found to be associated with cMVPs. As expected, the vast majority (92%) of these cMVPs were hypermethylated in the cancer cells. Over half (53%) of PRC2 target genes are associated with cMVPs that become significantly demethylated during reprogramming (e.g., SFRP2, a negative regulator of Wnt signaling that is often a target of hypermethylation in cancer) (Supplemental Fig. 6C,D; Suzuki et al. 2004).
We next assessed whether epigenetic resetting of cMVPs was stable upon redifferentiation of GiPSCs to the NS cell type. Surprisingly, the majority (27,105, 83%) of normalized cMVPs persisted following differentiation of iG7 to NS cells, suggesting that cancer-related cMVPs are not immediately reacquired in the context of NS cell state as a result of the genetic defects in these cells (Fig. 6F). A similar proportion (80%) of PRC2 target cMVPs was stably maintained in the reset state, including SFRP2 (Supplemental Fig. 6C,D). While the transcription start site of TES remained demethylated during differentiation, a minority of cMVPs reacquired methylation during differentiation in vitro, including CDKN1C (Supplemental Fig. 6D). Together, these results indicate that epigenetic reconfiguration through iPSC reprogramming can be highly stable but suggest that certain loci may be specifically vulnerable to reacquisition of aberrant DNA methylation.
GiPSC-derived neural progenitors remain highly malignant despite epigenetic resetting
We next tested the tumor initiation potential of N-iG7 to determine the functional significance of the experimental resetting of the >25,000 cMVPs. To assess tumorigenicity, a cohort of mice was injected with 100,000 cells (either G7 or N-iG7-1; n = 18) (Fig. 6B). Surprisingly, in all cases, tumors emerged with kinetics similar to those of previously studied GNS cell lines (Pollard et al. 2009). No significant difference in overall survival between G7- and N-iG7-transplanted mice was observed (Fig. 6G). Tumor cells expressed the NS cell marker NESTIN and similar levels of the mitotic marker Ki67 as tumors originating from parental cell cultures (G7, 12.0% ± 0.028%; N-iG7-1, 12.7% ± 0.036%) (Fig. 6H). G7 and N-iG7 tumors were indistinguishable in their infiltrative behavior and typically crossed the midline to the contralateral side via the corpus callosum (G7, five of six; N-iG7-1, three of four; N-iG7-2, three of three) (Fig. 6H). These results suggest that the widespread resetting of cMVPs alone is not sufficient to alter malignant behavior of GBM tumor-initiating cells.
GiPSC-derived mesodermal progenitors are less malignant
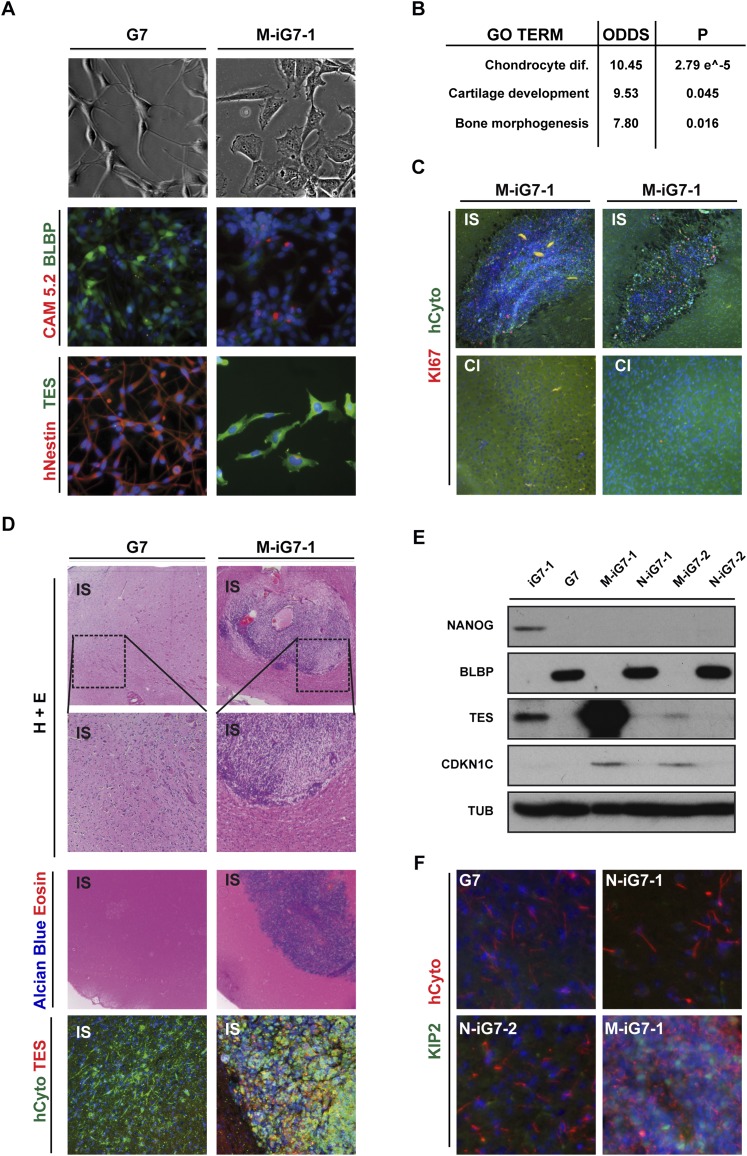
To test whether the malignant features of these cells are entirely defined by genetic alterations or, alternatively, might be suppressed by imposing more extensive epigenetic changes, we directed iG7 cells into a nonneural lineage in vitro. We derived homogeneous cultures of mesodermal progenitors from two independent iG7 clones (designated M-iG7-1 and M-iG7-2) through expansion of differentiating EBs in serum-containing medium (Supplemental Fig. 5F). These cells expressed T/Brachyury during the mesodermal differentiation process, later adopted a uniform morphology, and expressed varying levels of nonneural epithelial markers keratins 7 and 8 (Cam5.2) while silencing neural lineage genes (Fig. 7A). Gene ontology analysis indicated a mesodermal/cartilage progenitor identity for M-iG7 (Fig. 7B; Supplemental Table 4). The majority of normalized cMVPs persisted following mesodermal differentiation of iG7 to M-G7 (75% global and 83% PRC2 targets), suggesting that cMVPs are not immediately reacquired following differentiation (Fig. 6F; Supplemental Fig. 6C).
Figure 7.
GiPSC-derived mesodermal progenitors are no longer infiltrative. (A) Nonneural mesodermal cells (M-iG7-1 and M-iG7-2) were generated by in vitro differentiation of GiPSCs (P12–P14). Immunocytochemistry for the neural marker BLBP and Nestin and the nonneural epithelial marker keratins 7 and 8 (CAM5.2) confirms that in contrast to G7, M-iG7 cells show no expression of NS cell markers but partially express nonneuronal epithelial keratins. (B) Top three gene ontology terms (sorted by odds ratio) for those genes specifically induced in M-iG7 cells after differentiation. (C,D,F) Coronal sections of typical examples of forebrains from mice injected with 100,000 G7 and M-iG7-1 cells after 18 wk. (IS) Injection site; (CI) contralateral side. M-iG7-1 formed benign, noninfiltrative tumors that lacked pluripotency marker (see Supplemental Fig. 6E), expressed TES, and stained positive for alcian blue (indicative of cartilage). This contrasted with the highly infiltrative behavior of G7. See also Figure 6. Ki-67-positive cells were present in the benign mass but rare. (E) Immunoblotting for tumor suppressors, pluripotency marker NANOG, and radial glia/NS cell marker BLBP. CDKN1C (F) and TES (D) are only detectable in nonneural cultures (M-iG7-1 and M-iG7-2) or tumors.
The continued proliferation and homogeneity of M-iG7 offered the opportunity to assess the functional consequences of imposing an alternative developmental epigenome. Thus, we transplanted M-iG7 cells into the adult mouse brain to determine whether tumor development would be suppressed. All recipients survived until ∼18 wk, a time point at which mice injected with parental GNS cells had succumbed. Brains were harvested, and following histological analysis, we identified compact tumors that stained positive for alcian blue, indicative of cartilaginous tissue (Fig. 7C,D; Supplemental Fig. 6E). Interestingly, these tumors were benign and in all cases failed to infiltrate the surrounding brain (16 of 16) (Fig. 7C). Two mice in this cohort were left unprocessed and remained asymptomatic for >6 mo. In contrast to N-iG7, the mesodermal M-iG7 cultures and tumors were able to sustain expression of TES and CDKN1C mRNA and protein (Fig. 7D–F; Supplemental Fig. 6F).
To analyze key expression changes after reprogramming, we assayed a panel of 90 commonly misregulated or functionally relevant genes in GBM by qRT–PCR using custom-designed TaqMan microfluidic arrays (Engström et al. 2012). Thirty-six of these show expression differences (log2 > 2.5 or log2 < −2.5) between G7 and normal NS cells (CB660 and CB152) (Supplemental Fig. 7A,B). Eight of these 35 genes show a significant accumulation of cMVPs at the associated CpG island (HOXD10, PDGFRA, OLIG2, TERT, CCND2, FBLN2, IRX2, and TES). cMVPs at TES, CCND2, and IRX2 were reset in N-iG7 cells, but expression levels did not return to those observed in NS cells. cMVPs at TES, CCND2, and PDGFRA were reset in M-iG7 cells and were accompanied by a restoration of expression patterns (Supplemental Fig. 7A,B).
Taken together, our results indicate that in vitro manipulations inducing an alternative developmental lineage, with accompanying changes to the developmental epigenome and resetting of cancer-specific epigenetic abnormalities, are able to suppress malignant cellular behavior. Thus, the re-expression of demethylated tumor suppressor genes is profoundly influenced by the specific developmental lineage of the tumor cell.
Discussion
In this study, we demonstrated that iPSC reprogramming techniques can be successfully applied to highly aneuploid GBM cells. Exogenous expression of two reprogramming factors, OCT4 and KLF4, leads to conversion of malignant GNS cells to an iPSC-like state, and this is accompanied by erasure of a large proportion of GBM-associated cMVPs. Steering these reprogrammed cells along neural and mesodermal lineages has enabled us to assess for the first time the contribution of both cancer-specific epimutations and the developmental epigenome to the malignant properties of GBM-initiating cells.
We focused our analysis on DNA methylation, as this is the most commonly studied epigenetic mark in cancer biology, and DNA hypermethylation at promoter regions is the canonical example of an epimutation. Although demethylating agents, such as 5-azacytidine, have been widely used to study DNA methylation, these are toxic and lead to nonspecific global loss of DNA methylation, genomic instability, and loss of imprinting (Rizwana and Hahn 1998; Holm et al. 2005). This makes it challenging to interpret treatment-induced changes in cellular phenotypes. Thus, it has remained unclear whether global resetting of aberrant DNA methylation would be sufficient to restore normal cellular behavior and inhibit tumorigenicity of malignant cancer cells.
Our comparison of GNS cells with genetically normal NS cells identified >60,000 differentially methylated sites. Importantly, the identified sites of aberrant DNA methylation in G7 and G26 are disease-relevant and include some of the most common epigenetic anomalies associated with primary GBM tumors. Hypermethylation of PRC2 target genes and CDKN1C has been described for many human cancers (Schlesinger et al. 2007; Bennett et al. 2009; Fourkala et al. 2010; Avissar-Whiting et al. 2011). Hypermethylation of TES is among the most prevalent epigenetic alterations to have been reported in GBM (Martinez et al. 2009).
Conversion of GiPSCs into NS cells enabled us to assess the functional significance of the widespread resetting of aberrations in DNA methylation. Recent studies using immortalized mouse cells have speculated that removing DNA methylation from silenced tumor suppressors might restore normal cell cycle control (Ron-Bigger et al. 2010). However, NS cells generated from GiPSCs remained highly proliferative both in vitro and after orthotopic xenotransplantation. Thus, for the GBM cells analyzed here, reversing a large proportion of DNA methylation abnormalities did not override the activity of genetically altered pathways in driving unconstrained proliferation. Our findings would support a model in which epigenetic disruptions, such as changes to PRC2 targets, could have important functional roles during early tumor development (Feinberg et al. 2006), but their functional importance is diminished as genetic alterations accumulate.
GBMs are highly diverse in patterns of both genetic and epigenetic changes, and it will be important to extend this approach to a larger tumor series. Of interest will be to study other forms of GBM, particularly secondary GBM and pediatric cases, which display epigenetic features distinct from primary GBM (Schwartzentruber et al. 2012; Sturm et al. 2012). We investigated those genes that have been most commonly associated with aberrant DNA methylation in GBM tumors. However, it should be noted that iPSC reprogramming did not fully restore all epigenetic anomalies. The reprogramming process is also likely to have reset other types of epigenetic abnormalities that might have considerable influence on tumorigenicity, such as histone modifications or noncoding RNAs, and it will now be of interest to explore these further.
Differentiation of GiPSCs to mesodermal cells enabled us to test whether the malignant features of G7 are entirely defined by inherent genetic alterations or could be reduced by imparting more widespread developmental epigenetic changes. A defining feature of GBM cells is their capacity to infiltrate surrounding brain tissue. Tumors generated following orthotopic xenotransplantation of mesodermal GiPSC-derived cells showed reduced capacity to infiltrate surrounding brain tissue. One molecular explanation for this could be the sustained expression of CDKN1C and TES within the mesodermal progenitors. CDKN1C and TES are known regulators of cell cycle and cell motility, respectively (Pateras et al. 2006; Boeda et al. 2007), and we demonstrated that knockdown of TES in M-iG7 cells reduces their motility in vitro (Supplemental Fig. 4C). In NS-like cells derived from the GiPSCs, expression of CDKN1C and TES were not sustained despite removal of DNA hypermethylation.
In summary, our study demonstrates that highly aneuploid human cancer cells can be reprogrammed to an early embryonic state with concomitant removal of cancer-specific DNA methylation marks. By steering these cells along distinct developmental paths and testing functional outcomes in vivo, we showed that extensive resetting of cMVPs alone is not sufficient to suppress malignant cellular behavior and that there are lineage-specific requirements for sustained expression of tumor suppressor genes.
Materials and methods
Culture of NS and GNS cell lines
Fetal NS cell lines and GNS lines derived from human glioma samples have been described previously (Sun et al. 2008; Fael Al-Mayhani et al. 2009; Pollard et al. 2009;). Briefly, GNS and NS cells were cultured using serum-free basal medium supplemented with B27 and N2 culture supplements (Life Technologies). Growth factors EGF and FGF-2 (20 ng/mL) were added. Culture vessels were coated with Laminin (Sigma) at 10 μg/mL prior to use or during routine passage and was added directly to the culture medium at 1 μg/mL. GNS cells were routinely grown to confluence and dissociated using Accutase (Sigma).
iPSCs and hESCs were cultured in DMEM/F-12 supplemented with nonessential amino acids, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, and 20% knockout serum replacement (Life Technologies). To this, we added FGF-2 (10 ng/mL). Cells were plated on mitotically inactivated mouse embryonic fibroblasts. Cells were split every 7–14 d (1:3–1:10) or frozen in clumps using dissociation buffer (20% KSR, 1 mM CaCl2, 1 mg/mL collagenase IV, 0.25% trypsin in PBS) or freezing buffer (2 M DMSO, 1 M acetamide, 3 M propylene glycol in ESC medium).
Induced reprogramming and culture of hESCs and iPSCs
Fifteen GNS cell lines were tested for transcription factor-based reprogramming up to three times (G2, G7, G14, G18, G19, G21, G23, G25, G26, G30, G32, G144, G166, G179, and G144-D6) (Supplemental Table S1). The NS cell line CB660 was used in parallel as a normal control. Cells (2 × 106 to 6 × 106) were transfected using the Amaxa Nucleofector (Lonza) with 2 μg of PBhKlf4 and PBhOct4 (see the Supplemental Material) and 4 μg of the transposase PBase. Cells were seeded on a 10-cm dish precoated with gelatin (0.1%) and a layer of 2 × 104 mitotically inactivated mouse embryonic fibroblasts (hygromycin-resistant). Three days to 4 d later, 50 μg/mL hygromycin was added for selection of transfected cells. On day 7, NS cell culture medium was replaced with iPSC medium. Colonies emerged after 4 wk and were picked. PiggyBac cassettes were excised from GiPSCs differentiated progeny following delivery of Cre using adenovirus. Confirmation of excision was possible using the loss of dsRED expression and lack of OCT4 protein within differentiated cultures and tumors (Fig. 6C; Supplemental Fig. 6B).
RNA processing and microarray hybridization
Cells were dissociated using Accumax (Millipore), and feeder cells were depleted. RNA was extracted with Trizol (Invitrogen), followed by treatment with TURBO DNase (Ambion). RNA quality was assessed on the Agilent 2100 Bioanalyzer, and samples were processed for microarray hybridization according to the GeneChip whole-transcript sense target labeling assay (Affymetrix). Briefly, 2 μg of each sample was depleted of ribosomal RNA (RiboMinus, Invitrogen). Double-stranded cDNA was synthesized using random hexamers tagged with a 5′ T7 primer, and the products were amplified with T7 RNA polymerase to generate antisense cRNA. Reverse transcription was performed on the cRNA template using SuperScript III to yield ssDNA in the sense orientation, substituting dUTPs for dTTPs, and the cRNA was subsequently degraded via RNase H digestion. cDNA products were then nicked with uracil DNA glycosylase (UDG) and apurinic/apyrimidinic endonuclease 1 (APE 1) at sites of first-strand dUTP incorporation, followed by biotin labeling with terminal deoxynucleotidyl transferase (TdT). Affymetrix Exon Array 1.0 ST arrays were hybridized for 16 h at 45°C, washed, stained with streptavidin-phycoerythrin (SAPE) conjugate on a FS450 automated fluidics station, and imaged on a GCS3000 7G scanner (Affymetrix). Feature extraction was performed using Command Console 3.2.3, and hybridization quality was assessed with Expression Console 1.1.2 (Affymetrix). Gene expression analysis, functional category testing, and genome-wide methylation profiling methods are described in detail in the Supplemental Material. Exon array data are available in the ArrayExpress repository under accession E-MTAB-1273.
Quantitative real-time PCR
For qRT–PCR, RNA was extracted using RNeasy (Qiagen), including a DNase digestion step. cDNA was generated using SuperScript III (Invitrogen), and real-time PCR was carried out using TLDA microfluidic cards (Applied Biosystems). TaqMan fast universal PCR master mix was used with primers and probes as described in the Supplemental Material. TLDA data were analyzed using the Bioconductor package HTqPCR (Dvinge and Bertone 2009). PCA was carried out in R using the prcomp method in the R stats package to compute singular value decomposition, and results were visualized in OpenGL using RGL.
In vitro differentiation of iPSCs
For mesodermal differentiation, iPSC and GiPSC colonies were dissociated using Accumax and cultured for 7 d on nonadherent plates with serum-containing medium (GMEM; 10% fetal calf serum, 0.18 mM β-mercaptoethanol supplemented with sodium pyruvate and nonessential amino acids [Invitrogen]). EBs were mechanically dissociated and either plated on gelatin-coated vessels and cultured for an additional 7–10 d in the same medium supplemented with retinoid acid (5 nM) and BMP-4 (10 ng/mL) (EB differentiation) or grown in serum-containing medium until cultures showed an homogenous epithelial phenotype (M-iG7). For deriving neural cultures, iPSC and GiPSC colonies were plated down on Laminin-coated dishes in RHB-A medium until neuroepithelial rosettes were visible, then passaged once in RHB-A medium supplemented with EGF and FGF-2 (20 ng/mL) and either grown as neurospheres in RHB-A (supplemented with EGF and FGF [20 ng/mL]) or plated onto Laminin-coated dishes in the same medium condition (N-iG7). N-iG7 and M-iG7 cultures were subsequently passaged in RHB-A (supplemented with EGF and FGF) before experimental analysis.
Xenotransplantation
iPSCs, GiPSCs, and GNS cells were dissociated in clumps (Accumax), depleted of feeder cells, and diluted in PBS or 10% Matrigel in PBS, and 50,000 cells were transplanted for kidney capsule or subcutaneous injection into 6- to 9-wk-old NOD/SCID immunodeficient mice (NOD.CB17-Prkdcscid/NcrCrl, Charles River). Parental GNS cells failed to form tumors when transplanted in kidney capsule (G7, zero of four; G26, zero of four), but glioma-like tumors emerged using 250,000 cells. Additionally, 200,000 GiPSC (iG7-2) were injected using a stereotaxic frame into 6- to 8-wk-old NOD/SCID striatum following administration of general anesthesia as previously described.
Genomic analysis
GeneChip SNP 6.0 arrays (Affymetrix) were used to determine genetic alterations. Cells were dissociated, depleted of feeder cells, and processed using the DNA Wizard kit (Promega) to obtain genomic DNA. Data were analyzed using the Partek Genomics suite. Changes in DNA copy number were inferred using a hidden Markov model algorithm. Regions of gain were called using a threshold of 2.3 copies, and regions of loss were called with a cutoff at 1.7 copies. Copy number changes were called only when a minimum of 10 loci were above (or below) the threshold. SNP array data are available in the ArrayExpress repository under accession E-MTAB-1271.
Pyrosequencing
One microgram of DNA was bisulfite-treated using EZ DNA methylation kit (Zymo Research). Bisulfite-treated DNA was used for generating PCR-amplified templates for pyrosequencing using target-specific primers (Supplemental Material). Ten microliters of the biotinylated PCR products were sequenced according to the manufacturer's instructions. Pyrosequencing was carried on the PSQ HS 96 System and PyroMark MD System using Pyro Gold reagents (Biotage). Methylation was quantified using Pyro Q-CpG software that calculates the ratio of converted C's (T's) to unconverted C's at each CpG and expresses this as percentage DNA methylation. Average DNA methylation across the target region was determined for each sample and compared with the Batman and Infinium DNA methylation scores.
Genome-wide methylation profiling
One microgram of DNA was bisulfite-converted using the EZ DNA methylation kit (Zymo Research). The samples were then hybridized to Illumina Infinium Human 27K BeadArrays or the Illumina Infinium Human 450K BeadArrays according to the manufacturer's instructions. BeadStudio software (Illumina) was used to infer methylation scores from image intensities. Methylation data were quantile-normalized prior to calling MVPs. Using the distribution of differences between sample methylation scores, we defined a conservative threshold for calling differentially methylated regions based on the 95th percentile of the difference in methylation score. This resulted in a difference in methylation score of 31% between samples being considered as differentially methylated. PRC2 gene lists were defined from the gene set enrichment analysis database. Primary GBM 450K array methylation data were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga). Survival analysis was performed on 67 patients with a median survival of 202 d using the R package survival. Data for DNA methylation of GNS cells and reprogrammed derivatives are available in the ArrayExpress repository under accessions E-MTAB-1274 (450K arrays) and E-MTAB-1275 (27K arrays).
Immunocytochemistry
Cells in culture were fixed in 4% paraformaldehyde (PFA) for 10 min, washed three times in PBST, and transferred to blocking solution (3% goat serum, 1% BSA in PBST). Primary (listed in the Supplemental Material) and secondary antibodies were diluted in blocking solution, incubated overnight at 4°C and 2 h at room temperature, respectively, and separated by three washings. DAPI (0.5 ug/mL) was used to visualize nuclei.
Immunoblotting
Immunoblotting was performed using standard protocols, antibodies were diluted in 5% milk powder in PBST, and protein detection was carried out with HRP-coupled secondary antibodies and X-ray films.
Immunohistochemistry
Tumors were fixed between 12 and 24 h in 4% PFA embedded in paraffin and sectioned using a microtome (5–10 μm). Heat-induced epitope retrieval for 20 or 40 min in 10 mmol/L citrate buffer (pH 6) was used for CEA (M7072, Dako) or S100 (z0311, Dako) immunostaining. Primary antibodies were diluted 1:200 (S100), 1:25 (CEA), or 1:1 (CAM5.2, BD Biosciences). The Dako Envision HRP kit or biotin/avidin horseradish peroxidize procedure was used according to the manufacturers' protocols to visualize antibody binding. For fluorescent immunohistochemistry, sections were rehydrated using standard protocols (2× xylene for 15 min, 100% 3× ethanol for 10 min, 70% ethanol for 10 min). Epitope retrieval was performed in 10 mmol/L citrate buffer (pH 6) using a pressure cooker. Primary and secondary antibodies were diluted in 5% goat serum (diluted in PBST). For analysis of KI-67 immunoreactivity, at least three regions with highest, medium, and lowest KI-67 immunoreactivity in each section were analyzed. At least 3× 85 nuclei were counted for each class and cell line. Error bars depict the standard deviation. Standard histochemical protocols were used for haematoxylin, alcian blue, and eosin staining.
Acknowledgments
We thank William Mansfield for support with animal procedures, Margaret McLeish for histological preparations, and Tomi Bähr-Ivacevic (EMBL Genomics Core Facility, Heidelberg, Germany) for microarray processing. Ingrid Simonic (Addenbrooke's Hospital, Cambridge, UK) helped with SNP array analysis. Paulina Latos, Sascha Mendian, and Gillian Morrison provided guidance with nonneural iPSC differentiation protocols. Andrew Teschendorff helped in analysis and presentation of DNA methylation data. Christine Ender helped with virus production. Sophia Blake provided helpful comments on the manuscript. This work was supported by the Wellcome Trust, EMBL, and a project grant from Cancer Research UK (C25858/A9160). Y.T. and S.S. were supported by EMBO Long-Term Fellowships. A.S. is a Medical Research Council Professor. A.F. and S.B. were supported by the Wellcome Trust (grant 084071), and S.B. was also supported by a Royal Society Wolfson Research Merit Award. H.C. was supported by the Swedish Research Council and the Wenner-Gren foundation. S.P. was supported by a Wellcome Beit Memorial research fellowship, an Alex Bolt research fellowship, and the Brain Tumour Charity (grant 8/105).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.212662.112
References
- Avissar-Whiting M, Koestler DC, Houseman EA, Christensen BC, Kelsey KT, Marsit CJ 2011. Polycomb group genes are targets of aberrant DNA methylation in renal cell carcinoma. Epigenetics 6: 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S, Bestor TH 2002. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 1: 299–305 [DOI] [PubMed] [Google Scholar]
- Bennett LB, Schnabel JL, Kelchen JM, Taylor KH, Guo J, Arthur GL, Papageorgio CN, Shi H, Caldwell CW 2009. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer 48: 828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeda B, Briggs DC, Higgins T, Garvalov BK, Fadden AJ, McDonald NQ, Way M 2007. Tes, a specific Mena interacting partner, breaks the rules for EVH1 binding. Mol Cell 28: 1071–1082 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, Brummelkamp TR 2010. Generation of iPSCs from cultured human malignant cells. Blood 115: 4039–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, Wiemels JL, Nelson HH, Karagas MR, Wrensch MR, et al. 2011. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst 103: 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Bertone P 2009. HTqPCR: High-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics 25: 3325–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Tommei D, Stricker SH, Ender C, Pollard SM, Bertone P 2012. Digital transcriptome profiling of normal and glioblastoma-derived neural stem cells identifies genes associated with patient survival. Genome Med 4: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fael Al-Mayhani TM, Ball SL, Zhao JW, Fawcett J, Ichimura K, Collins PV, Watts C 2009. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods 176: 192–199 [DOI] [PubMed] [Google Scholar]
- Falk A, Koch P, Kesavan J, Takashima Y, Ladewig J, Alexander M, Wiskow O, Tailor J, Trotter M, Pollard S, et al. 2012. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS ONE 7: e29597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S 2006. The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Fourkala E-O, Hauser-Kronberger C, Apostolidou S, Burnell M, Jones A, Grall J, Reitsamer R, Fiegl H, Jacobs I, Menon U, et al. 2010. DNA methylation of polycomb group target genes in cores taken from breast cancer centre and periphery. Breast Cancer Res Treat 120: 345–355 [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, DiMeco F, Vescovi A 2004. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64: 7011–7021 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, Jaenisch R 2005. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8: 275–285 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K 2009. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458: 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh E, Joseph B 2011. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim Biophys Acta 1816: 50–56 [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. 2008. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454: 646–650 [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. 2006. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9: 391–403 [DOI] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M 2009. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics 4: 255–264 [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y, Mori M 2010. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci 107: 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller W, Nutt CL, Ehrich M, Riemenschneider MJ, von Deimling A, van den Boom D, Louis DN 2007. Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene 26: 583–593 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateras IS, Apostolopoulou K, Koutsami M, Evangelou K, Tsantoulis P, Liloglou T, Nikolaidis G, Sigala F, Kittas C, Field JK, et al. 2006. Downregulation of the KIP family members p27(KIP1) and p57(KIP2) by SKP2 and the role of methylation in p57(KIP2) inactivation in nonsmall cell lung cancer. Int J Cancer 119: 2546–2556 [DOI] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. 2009. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4: 568–580 [DOI] [PubMed] [Google Scholar]
- Qiu H, Zhu J, Yuan C, Yan S, Yang Q, Kong B 2010. Frequent hypermethylation and loss of heterozygosity of the testis derived transcript gene in ovarian cancer. Cancer Sci 101: 1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwana R, Hahn PJ 1998. CpG islands and double-minute chromosomes. Genomics 51: 207–215 [DOI] [PubMed] [Google Scholar]
- Ron-Bigger S, Bar-Nur O, Isaac S, Bocker M, Lyko F, Eden A 2010. Aberrant epigenetic silencing of tumor suppressor genes is reversed by direct reprogramming. Stem Cells 28: 1349–1354 [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D-AK, Tönjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res 63: 5821–5828 [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones DTW, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, et al. 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437 [DOI] [PubMed] [Google Scholar]
- Sun Y, Pollard S, Conti L, Toselli M, Biella G, Parkin G, Willatt L, Falk A, Cattaneo E, Smith A 2008. Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Mol Cell Neurosci 38: 245–258 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair K-W, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. 2004. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 36: 417–422 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Tatarelli C, Linnenbach A, Mimori K, Croce CM 2000. Characterization of the human TESTIN gene localized in the FRA7G region at 7q31.2. Genomics 68: 1–12 [DOI] [PubMed] [Google Scholar]
- Tobias ES, Hurlstone AF, MacKenzie E, McFarlane R, Black DM 2001. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene 20: 2844–2853 [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, Dirks PB 2007. Cancer stem cells: At the headwaters of tumor development. Annu Rev Pathol 2: 175–189 [DOI] [PubMed] [Google Scholar]
- Weeks RJ, Kees UR, Song S, Morison IM 2010. Silencing of TESTIN by dense biallelic promoter methylation is the most common molecular event in childhood acute lymphoblastic leukaemia. Mol Cancer 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158 [DOI] [PubMed] [Google Scholar]