SCFSlimb-mediated down-regulation of the condensin II subunit Cap-H2 is required to maintain proper organization and morphology of the interphase nucleus.
Abstract
Condensin complexes play vital roles in chromosome condensation during mitosis and meiosis. Condensin II uniquely localizes to chromatin throughout the cell cycle and, in addition to its mitotic duties, modulates chromosome organization and gene expression during interphase. Mitotic condensin activity is regulated by phosphorylation, but mechanisms that regulate condensin II during interphase are unclear. Here, we report that condensin II is inactivated when its subunit Cap-H2 is targeted for degradation by the SCFSlimb ubiquitin ligase complex and that disruption of this process dramatically changed interphase chromatin organization. Inhibition of SCFSlimb function reorganized interphase chromosomes into dense, compact domains and disrupted homologue pairing in both cultured Drosophila cells and in vivo, but these effects were rescued by condensin II inactivation. Furthermore, Cap-H2 stabilization distorted nuclear envelopes and dispersed Cid/CENP-A on interphase chromosomes. Therefore, SCFSlimb-mediated down-regulation of condensin II is required to maintain proper organization and morphology of the interphase nucleus.
Introduction
Eukaryotic genomes are spatially organized in a nonrandom manner (Kosak and Groudine, 2004; Misteli, 2007; Cremer and Cremer, 2010), and this 3D genomic structure is likely functionally important for control of gene expression (Laster and Kosak, 2010; Sanyal et al., 2011). Developments in chromosome conformation capture techniques suggest that interphase chromosomes exist as globule-like structures (chromosome territories) capable of long-range chromatin interactions (van Berkum et al., 2010; Sanyal et al., 2011). Studies probing genome-wide 3D structure and chromatin interactions revealed the organizational states of different cell types and developmental stages, making it possible to correlate gene expression patterns to 3D chromosome structures (Rajapakse et al., 2010; Rajapakse and Groudine, 2011). Although chromosomes adopt a variety of conformations that may facilitate gene expression, little is known about the mechanisms regulating chromosome conformation within interphase nuclei.
An example of chromosome organization with known biological function is homologue pairing in both somatic and meiotic cells (Wu and Morris, 1999; Duncan, 2002; Grant-Downton and Dickinson, 2004; McKee, 2004; Tsai and McKee, 2011). Pairing is critical for meiotic chromosome segregation and development of haploid gametes (Zickler, 2006), but pairing in somatic cells is less understood even though somatic pairing occurs in a variety of organisms. Homologue pairing in Drosophila melanogaster somatic cells can lead to transvection (Lewis, 1954; Henikoff and Dreesen, 1989; Wu and Morris, 1999; Duncan, 2002; Kennison and Southworth, 2002), which functions in trans-activation/inactivation of gene expression (Lewis, 1954). An extreme example of somatic homologous chromosome pairing is the Drosophila polyploid polytene chromosomes, where thousands of chromatin fibers align in a homology-dependent manner (Painter, 1933). Homologue pairing also functions in DNA damage repair (Rong and Golic, 2003). Despite these examples of chromosome organizational states and their functional relevance to gene regulation and genomic integrity, we lack a mechanistic understanding of how homologous chromosomes pair, unpair, and organize into territories. This information is especially wanting during interphase, when chromatin conformation likely has a major effect on transcription.
Condensins (I and II) are conserved protein complexes that condense chromatin and whose activities are especially evident in mitotic cells. Condensins I and II differ in composition: both have a heterodimer of Structural maintenance of chromosome subunits (Smc2 and Smc4) but contain different Chromosome-associated proteins (CAP-D2, -G, and -H for condensin I; CAP-D3, -G2, and -H2 for condensin II; Hirano and Hirano, 2004; Hirano, 2005). Their activities also differ: mitotic chromosomes are compacted laterally by condensin I and shortened axially by condensin II (Shintomi and Hirano, 2011). Interphase functions of condensins are diverse and less well studied (Hirano, 2005; Wood et al., 2010; Carter and Sjögren, 2012) but have been implicated in chromosome territory formation and homologue pairing in Drosophila (Hartl et al., 2008a,b; Bauer et al., 2012; Joyce et al., 2012).
Unlike condensin I, condensin II associates with chromatin throughout interphase and prevents homologous chromosome pairing in Drosophila. Pairing is facilitated by Su(Hw) and Topoisomerase II (Fritsch et al., 2006; Williams et al., 2007) and is antagonized by Cap-H2, which acts as an anti-pairing factor (Hartl et al., 2008a,b; Joyce et al., 2012). Other pairing factors have recently been identified (Joyce et al., 2012), but whether these function to directly modulate homologue pairing is unknown. Condensin II is also required during interphase to deposit and maintain the histone variant CENP-A at Xenopus laevis centromeres and for T cell development (Gosling et al., 2007; Bernad et al., 2011).
Our knowledge of the regulation of condensin II activity is mainly limited to mitosis, when the kinases Cdk1 and Plk1 act sequentially on condensin II, hyper-phosphorylating and activating the complex (Abe et al., 2011). In contrast, mechanisms regulating interphase condensin II are ill-defined. Condensin II is negatively regulated by MCPH1, a gene responsible for primary microcephaly, which competes with condensin II in binding chromatin and prevents premature chromosome condensation in G2 phase (Yamashita et al., 2011).
Here we show that the F-box protein Slimb (the fly homologue of human β-TrCP) localizes to chromatin and directly targets Cap-H2 for ubiquitination and degradation. Cap-H2 stabilization leads to chromosome unpairing and nuclear structural abnormalities. We also show that Slimb and Cap-H2 genetically interact in vivo to modulate chromosome pairing in ovarian nurse cell and salivary gland polytenes. To our knowledge, this is the first demonstration of condensin regulation by ubiquitination and Slimb association with chromatin to actively modulate interphase chromosome organization. These results demonstrate that condensin II activity must be closely regulated during interphase to prevent extensive changes to nuclear organization, which is achieved by the targeted degradation of Cap-H2.
Results
SCFSlimb ubiquitin ligase is required for chromatin reorganization
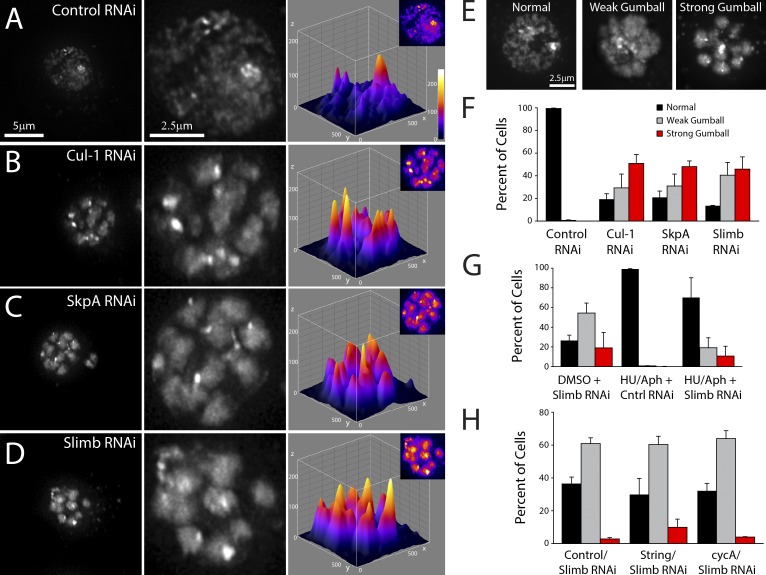
Previously, we observed that when Slimb was depleted in cultured Drosophila S2 cells, interphase chromatin became compacted into multiple, densely stained and approximately spherical globules (unpublished data). Therefore, we reasoned that an SCF complex acts as a negative regulator of interphase chromatin condensation. To test this, we reexamined micrographs collected for an unrelated RNAi screen of the SCF family (Rogers et al., 2009) to identify genes that alter chromatin morphology. Of 58 genes tested, only three (cul-1, skpA, and slimb) caused a clear compaction-like remodeling of interphase chromosomes (Fig. 1). Depletion of target proteins was confirmed by immunoblotting (Fig. S1 A). Surface plots of nuclear fluorescence intensity revealed that control cells typically display a single bright focus (likely the heterochromatic chromocenter) amid a relatively uniform and diffuse spherical nuclear pattern (Fig. 1 A). However, Cul-1, SkpA, or Slimb depletion caused dramatic chromatin reorganization into multiple globular structures (Fig. 1, B–D). Based on overall appearance, we refer to this as the “chromatin-gumball” phenotype, which manifests as either a weak or strong phenotype in the majority of the cul-1, skpA, or slimb RNAi-treated cells (Fig. 1, E and F). This phenotype is not unique to S2 cells, as Slimb depletion in S2R+ and Kc cells produced the same effect (Fig. S1, B and C). Thus, SCFSlimb regulates global chromatin remodeling.
Figure 1.
SCFSlimb RNAi promotes interphase chromatin compaction. (A–D) 7-d RNAi-treated S2 cells stained with Hoechst to visualize DNA. Depletion of Cul-1 (B), SkpA (C), or Slimb (D) but not control (A) promotes interphase chromatin compaction, generating a “gumball” phenotype. Cells are shown at low and high magnifications (left and middle). Shown on the right are 3D surface plots of the fluorescence intensities of the DNA (insets). (E) Representative images of DNA-stained RNAi-treated S2 cells displaying normal (wild-type), weak gumball, and strong gumball phenotypes. (F) Frequency histogram of the nuclear phenotypes in S2 cells after a 7-d depletion of the indicated proteins (n = 1,400–1,800 cells per treatment). (G) Chromatin of S-phase arrested cell compacts after slimb RNAi. S2 cells were treated daily with DMSO or S-phase arrested with hydroxyurea + aphidicolin for 6 d. Beginning on day 2, cells were also treated daily with control or slimb RNAi (see Fig. S2 A). Histogram shows the frequencies of nuclear phenotypes on day 6 (n = 1,100–1,600 cells per treatment). (H) S2 cells restricted to interphase form compact chromatin domains after slimb RNAi. Cells were treated daily with control, String, or cyclin A (cycA) dsRNA for 8 d. Stg RNAi promotes G2 arrest whereas cycA RNAi blocks mitotic entry. Beginning on day 4, cells were also treated daily with slimb RNAi (see Fig. S2 B). Histogram shows the frequencies of nuclear phenotypes on day 8 (n = 600–1,300 cells per treatment). Error bars indicate SEM.
Chromosome compaction occurs during interphase in the absence of Slimb
Chromosome condensation is a well-known behavior of mitotic chromosomes. It is less clear how extensively this occurs in interphase cells. SCFSlimb-depleted cells displaying chromatin-gumball phenotypes were negative for phospho-histone H3 staining (unpublished data), demonstrating that these cells were not mitotic. Moreover, slimb RNAi induces an accumulation of G1 cells (Rogers et al., 2009), again suggesting that chromatin-gumball formation occurs during interphase. However, this unique chromosome reshaping might be explained if chromosomes fail to decondense as cells exit mitosis in the absence of SCFSlimb. To test this, cells were arrested in different nonmitotic cell cycle phases, depleted of Slimb, and then analyzed for chromatin-gumball formation. First, S2 cells were arrested in S phase using the drugs hydroxyurea and aphidicolin and then depleted of Slimb while arrested (Fig. S2 A). Under these conditions, Slimb depletion again drove chromatin-gumballs formation (Fig. 1 G). Second, cells were depleted of either String/CDC25 phosphatase (to arrest cells in G2; Chen et al., 2007) or cyclin A (to block mitotic entry and induce endoreduplication; Mihaylov et al., 2002) and then co-depleted of Slimb (Fig. S2 B). These cells also formed chromatin gumballs, similar to the slimb RNAi-only treatment (Fig. 1 H). Collectively, these findings argue against the possibility that the phenotype results from a decondensation defect in cells exiting M phase. Instead, these data suggest that SCFSlimb functions during interphase to inhibit chromatin compaction into multiple dense domains.
SCFSlimb prevents abnormal dispersal of centromeric Cid protein and nuclear envelope defects
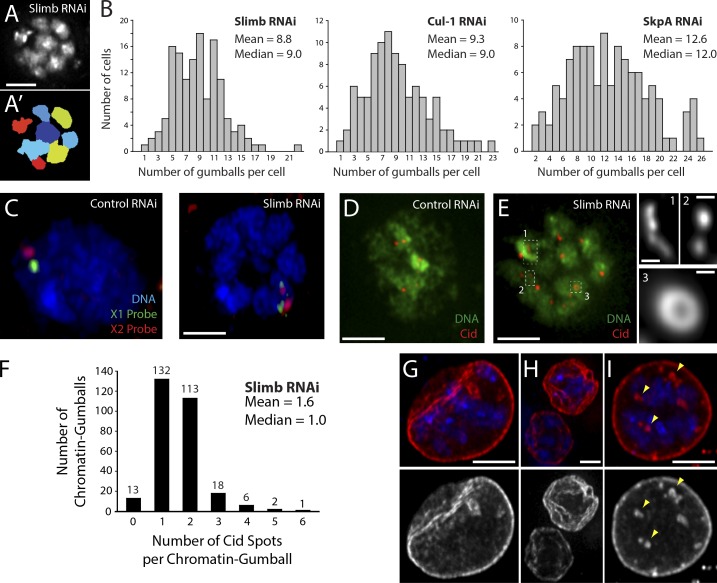
To characterize the chromatin domains induced by SCFSlimb depletion, we used software to identify, segment (Fig. 2, A and A′), and count the number of globular domains (“gumballs”; Fig. 2 B). Cells depleted of Cul-1, SkpA, or Slimb display a mean number of 9–13 globular gumballs. S2 cells possess a stable aneuploid genome with approximately two X chromosomes and two major autosomes, where each autosome is present in four copies (Zhang et al., 2010). If each discrete globular domain arose from a distinct major chromosome (not including the minute fourth chromosomes), then ∼10 gumballs per cell would be expected, which is consistent with our measurements. These findings suggest that SCFSlimb prevents interphase chromatin from undergoing extensive condensation into distinct chromosome globules.
Figure 2.
SCFSlimb depletion causes chromatin reorganization, abnormal dispersal of centromeric protein Cid, and nuclear envelope defects. (A) Representative image of a Slimb-depleted DNA-stained S2 cell displaying a strong chromatin-gumball phenotype. (A′) Segmented, pseudo-colored representation of the nucleus in A. (B) cul-1, skpA, or slimb RNAi cause interphase nuclei to reorganize their chromatin into 9–13 “gumball-like” compact globules. The number of globular segments per nucleus was measured from images as in A′ (n = 100–140 cells per histogram). (C) Control and Slimb-depleted Kc cells stained for DNA (blue) and two different euchromatic FISH probes specific for the X chromosome (green and red). FISH labeling shows paired homologous X chromosomes in controls but unpaired after slimb RNAi, decorating distinct chromatin domains. (D and E) Slimb depletion induces abnormal Cid dispersal. Shown are control and slimb RNAi-treated S2 cells immunostained for Cid (red). Hoechst-stained DNA, green. Boxes in E show abnormal Cid dispersal at higher magnification (labeled 1–3 on the right). (F) slimb RNAi increases the number of Cid spots per chromatin gumball. Cid spots were measured from 285 chromatin gumballs in 36 S2 cells. Numbers above the bars are the number of gumballs in each category. (G–I) slimb RNAi causes nuclear envelope defects. Slimb-depleted S2 cells immunostained for nuclear lamin (red; grayscale in bottom panels) show envelope invaginations (G), deformed, crumpled envelopes (H), and internalized nuclear microspheres (I, arrowheads). DNA, blue. Bars: (C–E and G–I) 2.5 µm; (E, insets) 0.25 µm.
If each chromatin gumball is a distinct chromosome, then each globular domain should contain one centromere. To test this, cells were immunostained for the centromere identifier protein Cid/CENP-A, and the number of Cid spots per globular domain was counted in Slimb-depleted cells (Fig. 2, D–F). Slimb-depleted cells had 1.6 ± 0.8 (mean ± SD) Cid spots per gumball (n = 285 gumballs). In addition, Cid spots in slimb RNAi cells were dispersed throughout the nucleus, sometimes appearing as closely juxtaposed pairs, short strings of spots, or bright doughnut shapes (Fig. 2 E). These observations are consistent with those of Joyce et al. (2012), showing a dispersal of pericentric heterochromatin FISH signal after slimb RNAi in Kc cells.
Because chromatin can be tethered to the inner nuclear membrane (Marshall, 2002) and SCFSlimb depletion promotes spatial reorganization of interphase chromatin, we examined if nuclear envelope morphology is affected. Treated cells were immunostained for nuclear lamin to visualize their nuclear envelopes. Strikingly, nuclei of Slimb-depleted cells frequently displayed severe morphological defects, such as long invaginations, a crumpled raisin-like appearance, or contained lamin-stained micronuclear spheres of variable number and size (Fig. 2, G–I). In some cases, individual cells displayed a combination of these phenotypes. Therefore, extensive interphase chromatin compaction is associated with defects in nuclear envelope morphology.
Slimb regulates chromosome structure through condensin II, not condensin I
Our results show that SCFSlimb depletion promotes interphase chromosome condensation, chromosome individualization, and formation of globular chromosome territories. Condensin complexes have been implicated in these processes (Chan et al., 2004; Hirano, 2005; Hartl et al., 2008b), which suggests that SCFSlimb and condensin are components of the same pathway regulating nuclear organization. To test if condensin activity is required to generate the slimb RNAi-induced phenotypes, we performed double RNAi against slimb and several condensin subunits and then assessed rescue of chromatin condensation. First, it was necessary to shorten the duration of condensin I RNAi treatment because significant cell death results from prolonged RNAi of condensin I subunits (Somma et al., 2003). When limited to 4 d, slimb RNAi alone produced weak chromatin-gumball phenotypes not observed in controls (Fig. S4, A, B, and G). Notably, the frequency of chromatin gumballs was not diminished when either condensin I subunit, Cap-D2 or Cap-G, was co-depleted with Slimb (no chromatin gumballs were observed in cells depleted of only Cap-D2 or Cap-G; Fig. S4, C–G). Thus, the condensin I complex is not required for chromatin reorganization caused by Slimb depletion.
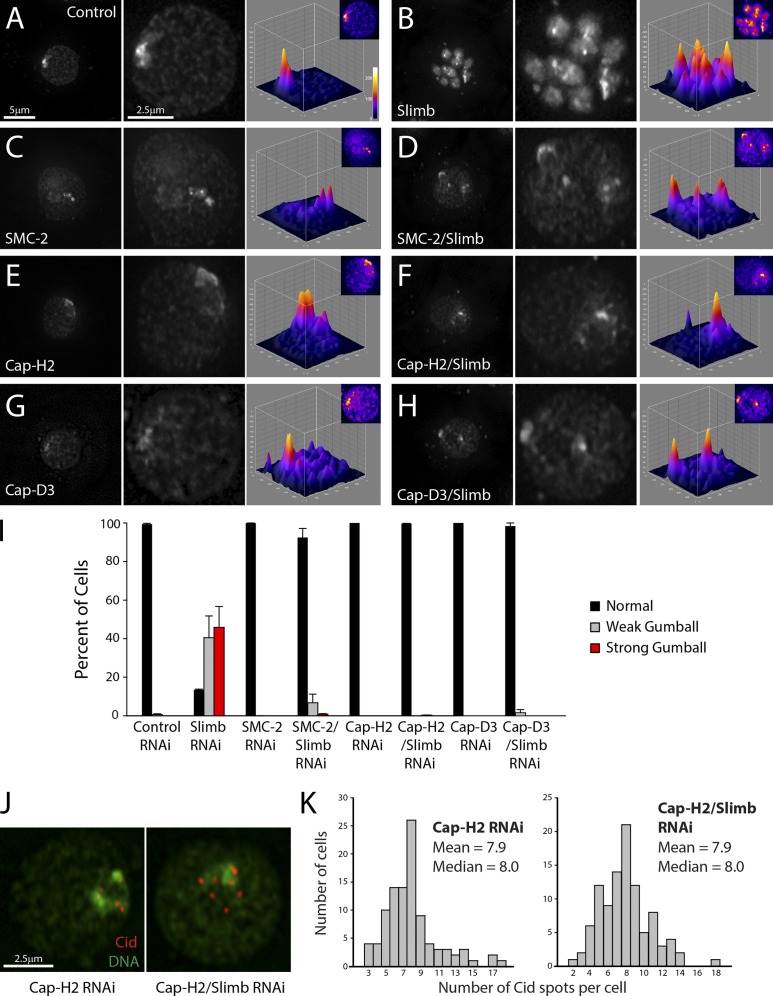
Similar experiments were performed with SMC-2 (shared by condensin I and II) and the condensin II subunits, Cap-D3 and Cap-H2. DNA morphology was assessed after 7-d double RNAi treatments to deplete Slimb and any one of the condensin subunits. As before, slimb RNAi produced cells containing both weak and strong chromatin gumballs (Fig. 3, B and I). However, smc-2/slimb, cap-H2/slimb, or cap-D3/slimb double RNAi strongly rescued this phenotype (no chromatin gumballs were observed when only condensin II subunits are depleted; Fig. 3, C–I). We conclude that condensin II is required to generate the chromatin-gumball phenotype induced by Slimb depletion.
Figure 3.
Depletion of condensin II subunits rescues slimb RNAi-induced nuclear phenotypes. (A–H) 7-d RNAi-treated S2 cells stained for DNA. Whereas slimb RNAi (B) promotes chromatin compaction, double RNAi of slimb and SMC-2 (D), cap-H2 (F), or cap-D3 (H) rescues this phenotype. Control (A), SMC-2 (C), cap-H2 (E), and cap-D3 (G) single RNAi-treated cells are shown at low and high magnifications (left and middle). Right, 3D surface plots of DNA fluorescence intensities. (I) Frequency histogram of nuclear phenotypes after day 7 RNAi (n = 1,200–1,500 cells per treatment). Error bars indicate SEM. (J) Day 7 double cap-H2/slimb RNAi rescues increase in Cid numbers. S2 cells immunostained for Cid (red). DNA, green. (K) Double cap-H2/slimb RNAi prevents an increase in Cid spots. The number of Cid spots per nucleus was counted from RNAi-treated interphase cells (100 cells per histogram).
To test whether condensin II is also responsible for the centromere abnormalities induced by Slimb depletion, RNAi-treated cells were immunostained for the centromere marker Cid. Cap-H2 and Slimb co-depletion rescued the defects in Cid morphology and the number of Cid spots per nucleus (Fig. 3, J and K; and Fig. S3 C).
Because Cap-H2 and other condensin II subunits serve as anti-pairing factors during interphase (Hartl et al., 2008a,b; Joyce et al., 2012), we tested whether Slimb also modulates this interphase function of condensin II. For this analysis, Kc cells were used instead of S2 cells to avoid the possibility that the segmental aneuploid S2 genome could confound the pairing measurements (Williams et al., 2007). Two FISH probes were used to label two different loci on each of the two X chromosomes. Homologue unpairing was monitored by counting the number of fluorescent spots within nuclei. If homologues pair, then only a single fluorescent spot is apparent for any given locus, whereas unpairing is manifested as two spots, one for each homologue. As shown by Joyce et al. (2012), euchromatic sequences on the X chromosome become unpaired in Slimb-depleted cells, unlike control and cap-H2 RNAi-treated cells (Figs. 2 C and S3, A and B). Slimb and Cap-H2 do functionally interact to regulate unpairing, because slimb/cap-H2 double RNAi completely rescued the homologue pairing defect (Fig. S3, A and B). Moreover, although Slimb depletion promotes homologue unpairing, each X chromosome probe (X1 and X2) clearly labeled distinct chromatin gumballs, again suggesting that each chromatin gumball is an individual chromosome (Fig. 2 C).
Finally, Cap-H2 mediates the nuclear envelope defects observed in Slimb-depleted cells (Fig. 2, G–I), as slimb/cap-H2 double RNAi rescued all nuclear envelope abnormalities (Fig. S4, H and I). Collectively, these findings suggest that, in the absence of Slimb, condensin II activity is responsible for reorganizing interphase chromatin, for the dispersal of centromeric DNA, and for nuclear envelope defects.
The condensin II subunit, Cap-H2, is a target for Slimb-mediated ubiquitination
The preceding experiments demonstrate a functional interaction between Slimb and condensin II, leading us to hypothesize that SCFSlimb depletion stabilizes condensin II activity. Because SCFSlimb catalyzes protein ubiquitination, a condensin II subunit could be a Slimb target for ubiquitin-mediated degradation. Consistent with this hypothesis, Drosophila Cap-H2 contains a potential Slimb-binding site near its carboxy terminus (Fig. 4 E); this sequence (DSGISS) fits the Slimb-binding consensus (DpSGXXp[S/T]; Rogers et al., 2009). No other Drosophila condensin subunit contains this motif.
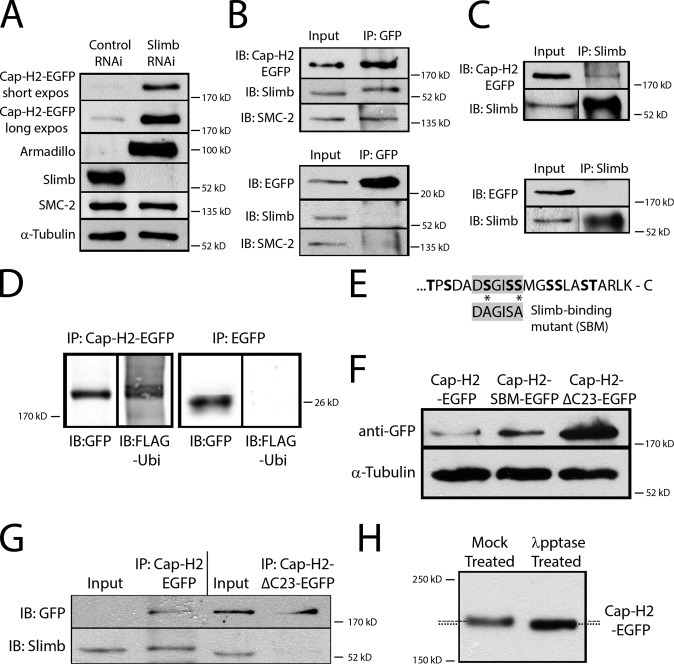
Figure 4.
Cap-H2 is degraded in a Slimb-dependent manner and stabilized by perturbing its interaction with Slimb. (A) An inducible Cap-H2-EGFP stable S2 cell line was control or slimb RNAi-treated for 7 d, and lysates were immunoblotted for the indicated proteins. Slimb depletion stabilizes Cap-H2-EGFP (and Armadillo) but not SMC-2. α-Tubulin, loading control. (B and C) Reciprocal immunoprecipitations and Western blots show that Slimb associates with Cap-H2. (B) Anti-GFP immunoprecipitates from cell lysates expressing Cap-H2-EGFP (top) or EGFP (bottom) probed with anti-GFP, Slimb, and SMC antibodies. (C) Anti-Slimb immunoprecipitates from cell lysates expressing Cap-H2-EGFP (top) or EGFP (bottom) probed for GFP and Slimb. (D) Cap-H2 is ubiquitinated. Anti-GFP immunoprecipitates from cell lysates transiently expressing 3×FLAG-ubiquitin and either Cap-H2-EGFP or EGFP probed for FLAG and GFP. (E) A Slimb-binding consensus (gray boxes) is encoded near the Cap-H2 carboxy terminus and flanked by multiple Ser/Thr residues (bold). A Cap-H2 Slimb-binding mutant (SBM) was engineered by substituting nonphosphorylatable Ala residues for two Ser residues (asterisks) within the Slimb-binding motif. Cap-H2-ΔC23-EGFP lacks these 23 terminal residues. (F) Anti-GFP immunoblots of lysates from cells transiently expressing Cap-H2-EGFP, Cap-H2-SBM-EGFP, or Cap-H2-ΔC23-EGFP. α-Tubulin, loading control. (G) Deletion of the Slimb-binding region disrupts Cap-H2 association with Slimb. Anti-GFP immunoprecipitates from cell lysates transiently expressing either inducible Cap-H2-EGFP or Cap-H2-ΔC23-EGFP probed for GFP and endogenous Slimb. (H) Cap-H2 is phosphorylated. Anti-GFP immunoprecipitates from day 7 slimb RNAi cells expressing inducible Cap-H2-EGFP were mock- or lambda phosphatase–treated. Compared with mock treatment (broken line), lambda phosphatase treatment alters the mobility of Cap-H2-EGFP to a faster migrating species (dotted line).
If Cap-H2 is a Slimb target, then Slimb depletion should stabilize Cap-H2. To test this, we generated an S2 stable line expressing inducible Cap-H2-EGFP, and then measured Cap-H2 levels in the lysates of RNAi-treated cells. Cap-H2-EGFP dramatically accumulated after slimb RNAi, as did our positive control Armadillo (a known Slimb substrate; Jiang and Struhl, 1998). In contrast, slimb RNAi did not affect the level of the condensin II subunit, SMC2 (Fig. 4 A). Thus, Slimb normally down-regulates Cap-H2.
We next examined if Slimb could associate with Cap-H2 using the stable line expressing Cap-H2-EGFP. When Cap-H2-EGFP was immunoprecipitated, endogenous Slimb and its binding protein SMC2 were also coimmunoprecipitated (Fig. 4 B). In the reciprocal experiment, Cap-H2-EGFP coimmunoprecipitated with endogenous Slimb (Fig. 4 C), confirming that Cap-H2 associates with Slimb.
Our hypothesis predicts that Cap-H2 is ubiquitinated in cells. To test this, immunoprecipitations were performed on lysates from cells coexpressing Cap-H2-EGFP and 3×FLAG-tagged ubiquitin. Cap-H2-EGFP was labeled with FLAG-ubiquitin, but the negative control EGFP was not (Fig. 4 D). Thus, Cap-H2 is a substrate for an endogenous ubiquitin ligase.
To test if Slimb ubiquitinates Cap-H2, we generated a Cap-H2 Slimb-binding mutant (Cap-H2-SBM-EGFP) by mutating two key residues in the binding consensus, changing DSGISS to DAGISA (S963A/S967A; Fig. 4 E). Because phosphorylation of the mutated serine residues is a prerequisite for Slimb recognition (Smelkinson and Kalderon, 2006), then Cap-H2-SBM should be stabilized if Slimb ubiquitinates Cap-H2. Surprisingly, expressed Cap-H2-SBM-EGFP was only marginally more stable than Cap-H2-EGFP (Fig. 4 F). However, maximal Slimb recognition often requires phosphorylation of multiple Ser/Thr residues that reside within and flank the Slimb-binding site (Smelkinson and Kalderon, 2006; Holland et al., 2010). Cap-H2 encodes nine residues within its last 23 amino acids that could be phosphorylated to generate a phosphodegron (Fig. 4 E). Therefore, we deleted the final 23 residues from Cap-H2 to make a new mutant (Cap-H2-ΔC23-EGFP), which lacks the Slimb-binding consensus site and the neighboring potential phosphorylation sites. Expression of the ΔC23 mutant produced a massive increase in Cap-H2 level, indicating that the complete Slimb-binding site includes residues immediately flanking the consensus motif (Fig. 4 F). Furthermore, endogenous Slimb does not coimmunoprecipitate with Cap-H2-ΔC23-EGFP from the lysate of mutant-expressing cells (Fig. 4 G). These results suggest that Slimb binds the carboxy-terminal Slimb-binding region in Cap-H2.
Finally, our model predicts that Cap-H2 phosphorylation promotes Slimb binding. The phosphorylation state of Cap-H2 in cells was evaluated using a gel-shift assay. Cap-H2-EGFP–expressing cells were RNAi-treated to deplete Slimb, after which Cap-H2-EGFP was immunoprecipitated from lysates and incubated with lambda phosphatase. When analyzed by SDS-PAGE, phosphatase treatment altered the mobility of Cap-H2-EGFP to a faster migrating species, which is consistent with the hypothesis that Cap-H2 is phosphorylated in cells (Fig. 4 H). Thus, Slimb depletion stabilizes phosphorylated Cap-H2. Collectively, our results support the model that Slimb binds and ubiquitinates phosphorylated Cap-H2.
Expression of stable Cap-H2 mutants induce chromosome reorganization, centromere dispersal, and nuclear envelope defects
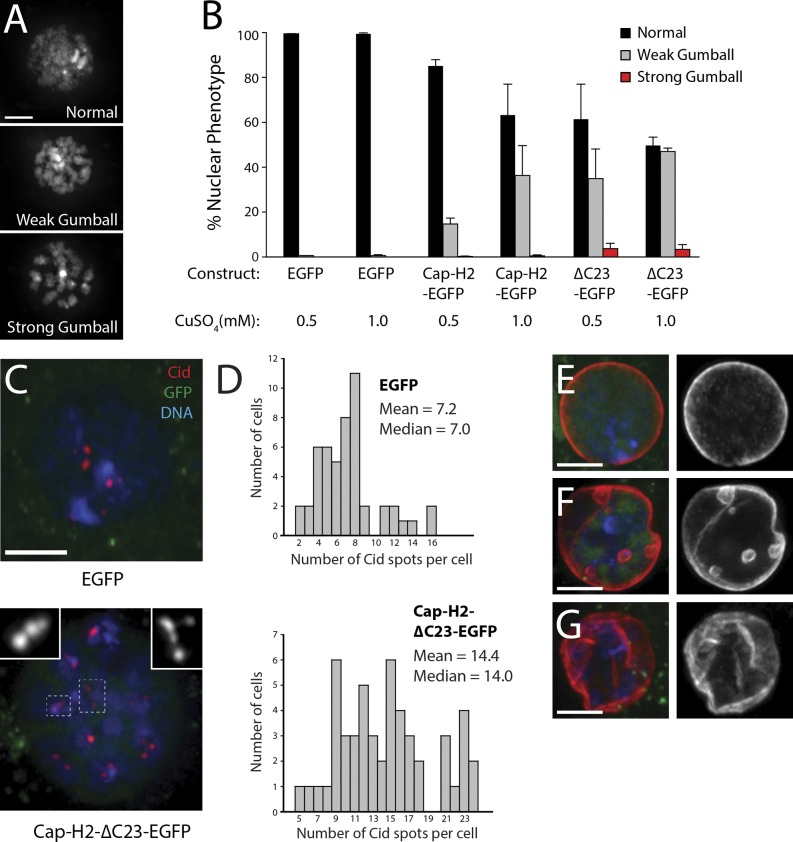
Our findings suggest that Cap-H2 destruction by Slimb inactivates condensin II, thereby preventing interphase chromatin reorganization. If correct, then expression of nondegradable Cap-H2 that cannot be targeted by Slimb should induce similar nuclear defects. We tested this prediction by overexpressing EGFP, Cap-H2-EGFP, or Cap-H2-ΔC23-EGFP in S2 cells and then analyzing their interphase chromatin organization (Fig. 5 A). As expected, transgenic Cap-H2 proteins were confined to nuclei (Fig. 5 C and not depicted). Although EGFP expression had no effect on interphase chromosome condensation, a high expression level of wild-type Cap-H2-EGFP induced a weak chromatin-gumball phenotype in ∼40% of cells (Fig. 5 B). Expression of Cap-H2-ΔC23-EGFP resulted in even greater frequencies of weak and strong chromatin gumballs (Fig. 5 B). Thus, Cap-H2 overexpression is sufficient to cause interphase chromatin compaction, and the effect is heightened by expression of nondegradable Cap-H2.
Figure 5.
Stable Cap-H2 promotes interphase chromatin compaction, Cid dispersal, and nuclear envelope defects. (A) DNA-stained S2 cells displaying representative nuclear phenotypes after overexpression of Cap-H2-ΔC23-EGFP. (B) Expression of stable Cap-H2 mutants induces chromatin compaction. Histogram of nuclear phenotype frequencies in cells expressing inducible EGFP, Cap-H2-EGFP, or Cap-H2-ΔC23-EGFP for 24 h. Expression levels were modulated by inducing with either 0.5 or 1 mM CuSO4 (n = 1,200–1,800 cells per treatment). Error bars indicate SEM. (C) Expression of stable Cap-H2 promotes abnormal Cid dispersal. S2 cells expressing EGFP (top; green) or Cap-H2-ΔC23-EGFP (bottom; green) and immunostained for Cid (red). Hoechst-stained DNA, blue. Boxed regions show abnormal Cid dispersal (insets, shown in higher magnification). (D) Stable Cap-H2 increases number of Cid spots in interphase cells. Numbers of Cid spots per nuclei were counted from interphase cells transiently expressing EGFP (top) or Cap-H2-ΔC23-EGFP (bottom; 51 cells per histogram). (E–G) Expression of stable Cap-H2 causes nuclear envelope defects. S2 cells expressing EGFP (E; green) or Cap-H2-ΔC23-EGFP (F and G; green) and immunostained for nuclear lamin (red; grayscale on the right). DNA, blue. Stable Cap-H2 expression induces invaginations and internalized nuclear microspheres (F) and crumpled envelopes (G). Bars, 2.5 µm.
Likewise, nondegradable Cap-H2 expression induced Cid dispersal. Cap-H2-ΔC23-EGFP–expressing cells contained short strings of multiple Cid spots on chromosome globules (Fig. 5 C) and had an increased number of Cid spots per nucleus (14.4 ± 5.1 [mean ± SD] vs. 7.2 ± 3.2 in control; Fig. 5 D), strikingly similar to that in Slimb-depleted cells (Fig. S3 C). Moreover, Cap-H2-ΔC23-EGFP–expressing cells displayed all of the nuclear envelope defects observed in Slimb-depleted cells (Fig. 5, F and G). Thus, up-regulation of condensin II activity is sufficient to induce centromere dispersal and nuclear envelope defects.
Previously, Slimb was found to localize to nuclei, but its association with chromatin was not determined (Rogers et al., 2009). To test this, nuclear fractions were isolated from S2 cells expressing EGFP and then further separated into nuclear-soluble and chromatin-bound fractions. Immunoblots confirmed that endogenous Slimb is nuclear and present in both the chromatin-bound and nuclear-soluble fractions (Fig. S4 J, left), revealing a novel association of Slimb with chromatin and suggesting that Slimb regulates Cap-H2 levels on chromatin. To test this prediction, subnuclear distributions of the Cap-H2-ΔC23-EGFP protein were determined. Cap-H2-ΔC23-EGFP levels massively accumulated in the chromatin-bound fraction, in contrast to Cap-H2-EGFP (Fig. S4 J, middle and right). Thus, eliminating the Slimb-binding site stabilizes Cap-H2 on chromatin, where its levels are normally low. These findings are consistent with the model that Slimb binds and ubiquitinates chromatin-bound Cap-H2, but do not exclude the possibility that non-chromatin-bound Cap-H2 is ubiquitinated.
Cap-H2 and Slimb genetically interact to regulate polytene chromosome pairing in vivo
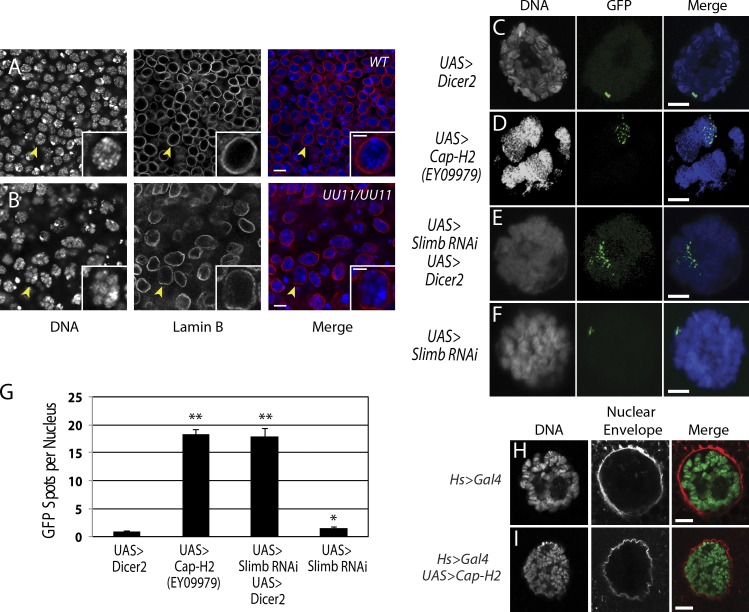
We next used Drosophila larvae to test the validity of our model in vivo by examining if interphase chromatin is reorganized in larvae with Slimb mutations. The proliferating diploid cells of wing imaginal disks from homozygous null Slimb mutant clones were stained for nuclear lamin and DNA. Whereas nuclear staining was roughly uniform in wild-type cells, chromatin was reorganized into compact, spherical structures surrounded by distorted nuclear envelopes in Slimb mutant cells (Fig. 6, A and B), similar to Slimb-depleted cultured cells. Thus, Slimb regulates chromatin organization in vivo.
Figure 6.
Slimb regulates chromatin organization in vivo, and its depletion causes unpairing of salivary gland polytene chromosomes. (A and B) Wild-type and FLP/FRT generated homozygous slimbUU11 null mutant clone of imaginal wing disk cells immunolabeled for lamin B (red). DNA, blue. Loss of Slimb induces chromatin gumballs. Insets show single nuclei (arrowheads) at higher magnification. (C–F) After heat shock, a transgenic line carrying Hs>LacI-GFP, LacO(250): Hs>Gal4 expresses LacI-GFP, which binds LacO arrays inserted on the second chromosome. Nuclei of salivary gland cells showing DNA (grayscale and blue) and organization of LacO arrays (green). UAS>Dicer2 expression does not disassemble polytenes, so LacI-GFP localizes to a single stripe (C). In contrast, Cap-H2 overexpression (D) or Slimb depletion (E) unpairs polytenes, causing LacI-GFP spots to disperse. Weak polytene unpairing is observed in cells possessing the UAS>slimb RNAi transgene without Dicer2, so some nuclei have two or three LacI-GFP spots (F). Error bars indicate SEM. (G) In vivo Cap-H2 overexpression or slimb RNAi causes polytene chromosomes to unpair. UAS>Dicer2 control salivary glands have one large LacI-GFP spot (n = 24 nuclei), whereas UAS>slimb-RNAi larvae display a small but significant increase in unpairing (*, P < 0.005; n = 24 nuclei). Polytene unpairing is sharply increased in both UAS>Cap-H2EY09979 and UAS>slimb-RNAi, UAS>Dicer2 lines (**, P < 10−12 and **, P < 10−16, respectively, compared with UAS>Dicer2 alone; n = 24 nuclei per treatment). (H and I) Salivary gland nuclei expressing Hs>Gal4 (H) or Hs>Gal4, UAS>Cap-H2EY09979 (I) stained for nuclear envelopes (wheat germ agglutinin, red). DNA, green. Bars: (A and B) 5 µm; (A and B, insets) 0.2 µm; (C–F, H, and I) 10 µm.
Previously, we’ve shown that the paired state of polytene chromosomes in larval salivary glands is antagonized by elevated condensin activity. Specifically, overexpressed Cap-H2 unpairs and disassembles polytene chromosomes; endogenous SMC2/4 and Cap-D3 are required for this effect (Hartl et al., 2008a). We propose that condensin II activity on chromosomes is normally suppressed by Slimb, thereby maintaining the paired status of the individual chromatids within the polytene structure. To test this, we performed a quantitative chromosome pairing assay using a fly line modified with transgenes that report the pairing state of chromosomes in vivo. A LacO array is inserted at a single site in the second chromosome, and this LacO sequence becomes labeled with LacI-GFP after heat shock. This line also contains a heat shock–inducible GAL4 transcription factor to drive expression of UAS-regulated transgenes (Hartl et al., 2008a). In this system, salivary gland polytene chromosomes are normally tightly paired and, after heat shock, contain a single fluorescent spot or stripe of LacI-GFP proteins bound to the many paired LacO array sequences. When pairing is disrupted, the aligned chromosomes bearing the LacO arrays separate, leading to multiple resolvable GFP spots; the extent of polytene unpairing is measured by counting the number of nuclear fluorescent spots.
Flies containing the pairing–reporter system were crossed with a second transgenic line that overexpresses wild-type Cap-H2 (UAS>Cap-H2EY09979). Chromosomes of salivary glands obtained from progeny overexpressing Cap-H2 were unpaired, as indicated by the significant increase in the number of GFP spots per nucleus (18 ± 4, mean ± SEM; Fig. 6, D and G). In contrast, progeny of a different cross that overexpressed Dicer2 (a ribonuclease that generates short interfering RNA from double-stranded RNA [dsRNA]) had one GFP spot per nucleus, demonstrating that chromosomes were still paired in this control (Fig. 6, C and G).
If Slimb suppresses the Cap-H2 level in vivo, then depleting Slimb should also cause unpairing of polytene chromosomes. The pairing–reporter line was crossed to another line containing two transgenes on different chromosomes: UAS>slimb-RNAi (which encodes the slimb hairpin used for RNAi) and UAS>Dicer2. Larvae of progeny receiving both slimb-RNAi and Dicer2 transgenes had completely (100%) unpaired salivary gland chromosomes, demonstrated by the significant increase in GFP spots per nucleus (18 ± 1.4; Fig. 6, E and G). Larvae that received only slimb RNAi without Dicer2 exhibited a mild but significant unpairing phenotype (1.5 ± 0.2 GFP spots per nucleus; Fig. 6, F and G). Thus, in salivary gland cells overexpressing Dicer2, slimb RNAi results in a phenotype identical to Cap-H2 overexpression (Fig. 6, D and G). We conclude that Slimb functions as a negative regulator of condensin II activity in vivo, probably through Slimb-mediated destruction of Cap-H2.
Cap-H2 overexpression in vivo also causes nuclear morphology defects. In 100% (n = 150) of the salivary polytene cells from control larvae, nuclei appeared round, and polytene banding patterns were not disrupted (Fig. 6 H). In contrast, all (100%) salivary cells from larvae overexpressing Cap-H2 (UAS>Cap-H2) had wrinkled nuclear envelopes (Fig. 6 I), and 32% exhibited intranuclear microspheres (Fig. S5). As in S2 cells, regulation of Cap-H2 levels in postmitotic polyploid cells maintains normal nuclear envelope morphology.
Cap-H2 and Slimb genetically interact to regulate chromosome pairing in polyploid nurse cells
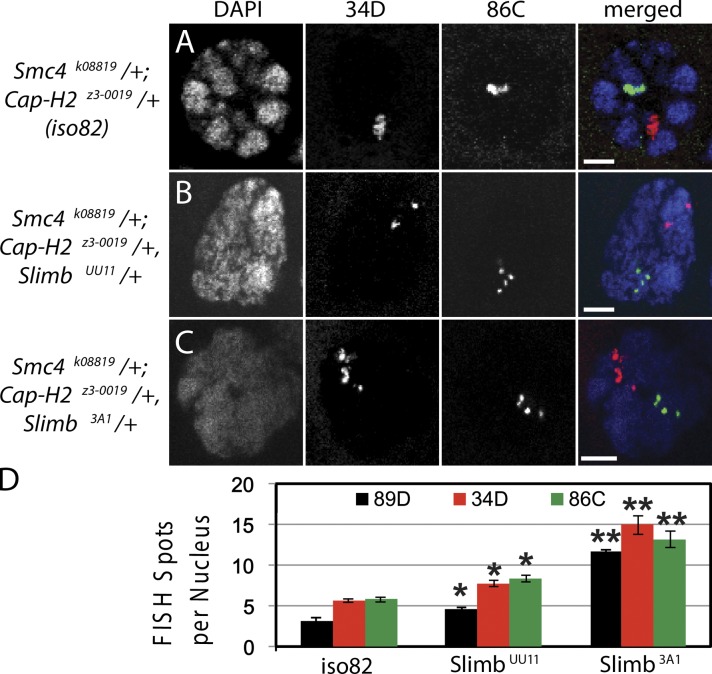
Unlike salivary gland cells, Drosophila nurse cells normally disassemble their polytene chromosomes to unpaired chromatids at a prescribed point during oogenesis, and condensin II subunits Cap-H2, Cap-D3, and SMC4 are required for this developmentally programmed event (Hartl et al., 2008a). Polytene unpairing is likely exerted through condensin II because the severity of unpairing can be modulated by crossing heterozygous, condensin II loss-of-function mutants to generate progeny with a varying number of functional alleles (Hartl et al., 2008a). For example, nurse cells from Smc4k08819/+; Cap-H2Z3-0019/+ double heterozygotes (with only a single allele encoding wild-type Cap-H2) have an intermediate unpairing defect, whereas Cap-H2Z3-0019 homozygous cells have severe defects (Hartl et al., 2008a). If Slimb targets Cap-H2 for destruction in nurse cells, then addition of a Slimb mutation should allow Cap-H2 protein to rise and rescue the intermediate Smc4k08819/+; Cap-H2Z3-0019/+ phenotype by increasing the extent of unpairing. To test this, DNA FISH probes to three different euchromatic loci (34D, 89D, and 86C) were used to measure the pairing status of the polytene chromosomes in mutant nurse cells; an increase in FISH spots indicates unpairing. In the Smc4k08819/+; Cap-H2Z3-0019/+ double heterozygous nurse cells, FISH probes confirmed an intermediate unpairing phenotype (Fig. 7, A and D). In contrast, when Slimb function is partially diminished by crossing the Smc4k08819/+; Cap-H2Z3-0019/+ line with either a null SlimbUU11/+ heterozygous mutant or a loss-of-function Slimb3A1/+ heterozygous mutant, unpairing is significantly increased in cells of the progeny (Fig. 7, B–D). Thus, partial inactivation of Slimb restores the ability of condensin II to promote polytene unpairing in nurse cells, which is consistent with Slimb acting as a negative regulator of condensin II.
Figure 7.
Slimb mutations suppress condensin II loss-of-function phenotypes in polyploid nurse cells. (A) Stage 10 nurse cells from double heterozygous Smc4k08819/+; Cap-H2z3-0019/+ flies in the iso82 genetic background were labeled with FISH probes to chromosomal positions 34D and 86C. Nurse cells maintain a pseudo-polytene structure, which is made evident by the DAPI-stained chromosomes and the clustered FISH spots indicating paired chromatids. Bars, 10 µm. (B and C) Two different Slimb alleles, UU11(B) and 3A1(C), carried on the same isogenic chromosome, were crossed with Smc4k08819/+; Cap-H2z3-0019/+, and triple heterozygous nurse cells were labeled with DAPI and FISH probes. (D) Stage 10 nurse cells triple labeled with FISH probes to chromosomal positions 34D, 86C, and 89D were quantified for number of spots per nucleus to determine the degree of polytene pairing. The number of FISH spots for each chromosomal position in Smc4k08819/+; Cap-H2z3-0019/+ was compared with Smc4k08819/+; Cap-H2z3-0019/+, SlimbUU11/+ (*, P < 10−4; n = 50 nuclei) and Smc4k08819/+; Cap-H2z3-0019/+, Slimb3A1/+ (**, P < 10−9; n = 50 nuclei). Error bars indicate SEM.
Discussion
In this study, we identify a mechanism that prevents interphase chromosomes from undergoing 3D spatial reorganization due to condensin II–mediated compaction. Condensin II activity must be limited during interphase to ensure proper homologue pairing and nuclear morphology. Nuclear morphology and organization must be actively maintained, and our data demonstrate that the SCFSlimb ubiquitin ligase is a requisite component of the maintenance mechanism that down-regulates Cap-H2 levels during interphase. SCFSlimb depletion in cultured cells leads to cytological compaction of individual interphase chromosomes, reminiscent of distinct chromosome territories, and, consequently, unpairs euchromatic sequences. Cap-H2 is a Slimb ubiquitination target that contains a Slimb-binding domain, associates with Slimb, and is regulated by ubiquitin-mediated proteolysis in a Slimb-dependent manner. In vivo interactions of SMC4 and Cap-H2 with Slimb suppress condensin II activity and prevent disruption of chromosome organization in Drosophila salivary gland and ovary cells. This is the first demonstration of condensin regulation by ubiquitin-mediated proteolysis in dividing and postmitotic cells.
Both Slimb and condensin II subunits localize to the nucleus throughout the cell cycle and bind chromatin. Cap-H2 on chromatin accumulates when Slimb association is disrupted. As with other Slimb substrates, Slimb recognition of Cap-H2 is probably dependent on phosphorylation, which likely occurs on the Cap-H2 carboxy terminus. Clearly, identification of the kinases that trigger Cap-H2 destruction is needed to fully characterize this pathway. Possibly, the activity and/or chromatin-targeting of this kinase are spatially and developmentally regulated to restrict Cap-H2 degradation to limited chromatin regions, thus locally controlling the 3D organizational state of chromosomes. Condensin II may also promote chromatin conformations that epigenetically influence gene expression patterns in a cell type–specific manner. For example, naive T lymphocyte interphase chromatin is transcriptionally quiescent and maintained in a relatively condensed state by condensin II. After T lymphocytes are exposed to an appropriate antigen, chromatin decondenses, and, simultaneously, transcription is up-regulated (Rawlings et al., 2011).
The pathway that we describe provides a mechanistic explanation for a recent study showing that Slimb depletion prevents homologue pairing in cultured Drosophila cells but is rescued by co-depletion of Cap-H2 (Joyce et al., 2012). Our findings suggest that SCFSlimb down-regulates Cap-H2 to directly modulate homologue pairing status in both cultured cells and polyploid cells in vivo. Slimb-modulated condensin II activity also controls interphase chromosome condensation and chromosome individualization effects consistent with vertebrate condensin II’s mitotic function as an axial compactor of chromosomes (Shintomi and Hirano, 2011; Green et al., 2012). Interestingly, of the four predicted Cap-H2 splice isoforms in flies, one of these, Cap-H2-RD, lacks a large N-terminal region as well as the C-terminal 64 amino acids encoding the Slimb-binding site present in the other three isoforms. A Slimb-resistant Cap-H2 may be necessary, for example during mitosis, when rapid and large increases of condensin II activity are needed, or during differentiation, when chromatin compaction facilitates gene regulation. The Slimb-binding domain may have evolved to allow high levels of somatic homologue pairing and polytene chromosome formation. Although the Slimb-binding domain of Cap-H2 is not found outside Drosophila, it is formally possible that other organisms use other ubiquitin ligases to down-regulate condensin II.
Slimb-depleted cells exhibit additional nuclear phenotypes, including an abnormal dispersal of the centromere histone, Cid/CENP-A. Deregulation of the Slimb-Cap-H2 mechanism is responsible for this phenotype. Consistent with these observations, condensin II has been shown to promote dispersal of pericentric heterochromatin in vivo and in cultured cells (Bauer et al., 2012; Joyce et al., 2012).
We also demonstrate new roles for both Slimb and Cap-H2 in maintaining the architecture of the nuclear envelope. Inappropriate activation of condensin II causes gross morphological changes in nuclear envelope structure. Hypercompaction of chromosomes tightly linked to the inner nuclear membrane could collapse the nuclear envelope and, in extreme cases, invaginate and internalize membrane patches forming intranuclear microspheres. Intriguingly, these deformed nuclear envelopes are similar in appearance to those in cells derived from Hutchinson-Gilford progeria syndrome patients (Scaffidi et al., 2005; Scaffidi and Misteli, 2006). Progeria and a variety of other syndromes, collectively known as laminopathies, are linked to mutations in nuclear lamins and are thought to alter gene expression, resulting in the disease state. It is tempting to speculate that the abnormal nuclear envelope shapes caused by mutations in lamin proteins are driven by condensin II. Chromatin is tethered to the nuclear periphery, through interactions with lamins or other envelope proteins, and may transduce forces to the nuclear envelope when interphase chromosomes undergo compaction. Cells with wild-type lamin function normally to withstand condensin-driven chromosome compaction. However, it is possible that in cells with compromised lamin structure, axial shortening of chromatin tethered to the envelope leads to distortions in nuclear envelope morphology. Future work is necessary to determine whether condensin II contributes to the abnormal nuclear morphology and etiology of these syndromes.
Materials and methods
Cell culture and double-stranded RNAi
Drosophila cell culture, in vitro dsRNA synthesis, and RNAi treatments were performed as described previously (Rogers et al., 2009). In brief, S2, Kc, and S2R+ cells were cultured in Sf900II media (Life Technologies). RNAi was performed in 6-well tissue culture plates. Cells (50–90% confluency) were treated with 10 µg of dsRNA in 1 ml of media and replenished with fresh media/dsRNA every other day for 4–7 d. Gene-specific primer sequences used to amplify DNA templates for RNA synthesis are shown in Table S1. Control dsRNA was synthesized from control DNA template amplified from a non-GFP sequence of the pEGFP-N1 vector (Takara Bio Inc.). Cell cycle arrest was induced by treating cells for at least 24 h with either 0.5 mM (final concentration) mimosine (for a G1-phase arrest), 1 µM hydroxyurea + 10 µM aphidicolin (S-phase arrest), 1.7 µM 20-hydroxyecdysone (G2-phase arrest), or 12 h of 30 µM colchicine (M-phase arrest), as described previously (Brownlee et al., 2011).
Immunofluorescence microscopy
For immunostaining, cultured cells were fixed and processed exactly as described previously (Rogers et al., 2009) by first spreading cells on concanavalin A–coated, glass-bottom dishes and then fixing with 10% formaldehyde in PBS at room temperature. Primary antibodies were diluted to concentrations ranging from 1 to 20 µg/ml; they included rabbit anti-Cid (produced in-laboratory), anti-lamin ADL84.12 (Developmental Studies Hybridoma Bank), and mouse and rabbit anti–phosphohistone H3 (EMD Millipore; Cell Signaling Technology). Secondary antibodies (conjugated with Cy2, Rhodamine Red-X, or Cy5; Jackson ImmunoResearch Laboratories, Inc.) were used at the manufacturer’s recommended dilutions. Hoechst 33342 (Life Technologies) was used at a final dilution of 3.2 µM. Cells were mounted in a solution of 0.1 M n-propyl galate, 90% (by volume) glycerol, and 10% PBS. Specimens were imaged with a DeltaVision Core system equipped with a microscope (IX71; Olympus), a 100× objective lens (NA 1.4), and a CoolSNAP HQ2 cooled-CCD camera (Photometrics). Images were acquired with SoftWoRx v1.2 software (Applied Precision). To characterize chromatin-gumball phenotypes, images were processed using the segmentation algorithm of Cell Profiler 2.0 (Broad Institute), and surface plots were generated using ImageJ (National Institutes of Health).
Each FISH probe was made from multiple BAC clones spanning ∼200–350 Kb: 15 µg of BAC DNA was digested with AluI, Rsa, MseI, MspI, HaeIII, and BfuCl overnight at 37°C, ethanol precipitated, and resuspended in ddH2O. DNA was denatured at 100°C for 1 min, and then 3′-end-labeled with unmodified aminoallyl dUTP and terminal deoxynucleotidyl transferase (Roche). After incubating for 2 h at 37°C, 5 mM EDTA was added to terminate the reaction. DNA was ethanol precipitated, resuspended in ddH2O, and then conjugated to fluorophores using ARES Alexa Fluor DNA labeling kits (A-21665, A21667, and A-21676; Invitrogen) for 2 h, according to the manufacturer’s instructions (Dernburg, 2000; Hartl et al., 2008a).
Ovaries were dissected in 1× PBS and fixed in 100 mM sodium cacodylate, 100 mM sucrose, 40 mM sodium acetate, 10 mM EGTA, and 3.7% formaldehyde for 4 min at 32°C. Ovaries were rinsed in 2× SSCT (0.3 M NaCl, 0.03 M sodium citrate, pH 7.0, and 0.1% Tween-20) and treated with 2 µg/ml RNase for 1 h. Germaria were teased apart in 2× SSCT and then washed sequentially with 20% formamide, 40% formamide, and 50% formamide in 2× SSCT (10 min per wash). Ovaries were then incubated at 37°C for 2 h in fresh 2× SSCT/50% formamide. 2 µl of each probe (34D, 89D, and 86C) labeled with different Alexa Fluor dyes was combined with 36 µl of hybridization solution (1.11% dextran sulfate and 55.5% formamide in 3.3× SSC [0.5 M NaCl and 0.05 M sodium citrate, pH 7.0]) and added to the tissue. Chromosomal DNA was then denatured for 2 min at 91°C and hybridized overnight at 37°C. Samples were washed three times with 2× SSCT/50% formamide (37°C, 20 min per wash), then sequentially washed with 2× SSCT/40% formamide and 2× SSCT/20% formamide, and three times with 2× SSCT at room temperature, 10 min per wash. Ovaries were rinsed in PBS/0.1% Tween-20 and stained with 0.1 µg/ml DAPI in PBS/0.005% Tween-20 for 10 min, followed by two 10-min washes with PBS/0.005% Tween-20. Nurse cell nuclei were stained with DAPI and then mounted in Vectashield (Vector Laboratories). Stage 10 egg chambers (and nurse cells within) were identified visually based on 50% oocyte volume, 50% nurse cell volume of the entire egg chamber. Stage 10 egg chambers were also recognized as having completed border cell migration and completed migration of oocyte-associate follicle cells away from the nurse cells where lateral pinching of the follicle cell epithelial layer was visible (at the oocyte–nurse cell border) and the germinal vesicle had completed its migration to the dorsal anterior region of the oocyte. Nurse cell nuclei and FISH signals were imaged with a confocal microscope (LSM 510 Meta; Carl Zeiss), a Plan-Apochromat 63×/1.4 NA objective lens, and acquisition software (LSM 510 Meta v4.0). Digitized 1 µm z-axis optical sections of samples were captured for analysis.
FISH probe preparation and hybridization on cultured cells were performed as described above for ovaries, except that cells were grown on coverslips and allowed to adhere to glass before fixation, as described previously (Williams et al., 2007; Joyce et al., 2012), and then imaged using a DeltaVision Core system identical to the system described in the first paragraph of this section. Images were acquired with SoftWoRx software (v1.2).
The number of FISH spots in each nucleus was counted manually in each z-slice for each FISH signal channel. Datasets were statistically analyzed by the two-tailed Student’s t test (unequal variance) to determine p-values.
The salivary gland polytene pairing assays were performed with transgenic lines carrying a 256-repeat array of the Lac-O sequence at chromosomal position 60F and carrying a heat shock–inducible transgene Hs>GFP-LacI, which encodes a fluorescent fusion protein that binds to the LacO arrays and marks the chromosomal insertion site of the LacO array (Vazquez et al., 2001). These lines also contained transgenes Hsp70>Gal4 and UAS>Cap-H2, as described previously (Hartl et al., 2008a). Expression of GFP-LacI and Cap-H2 was controlled with heat shock at 37°C for 1 h and allowed to recover for 2–4 h at 25°C, then salivary glands were dissected as follows. Third instar larvae were dissected in PBS/0.1% Triton X-100 (PBT), and glands were fixed for 10 min in PBS/4% formaldehyde. Glands were rinsed three times with PBT, stained for 10 min with 0.1 µg/ml DAPI in PBS/0.005% Triton X-100, and then washed twice with PBT for 10 min. The Carl Zeiss confocal system described earlier in this section was used to obtain 1-µm z-axis optical sections of samples. All samples were imaged with identical settings (exposure time, illumination intensity, gain, etc.). The number of GFP spots per nucleus was counted manually from digital images displayed with LSM Image Browser software (Carl Zeiss). At least three different biological replicates were imaged for the GFP spot quantification, and a minimum of three glands per replicate were analyzed. Data were statistically analyzed with the two-tailed Student’s t test (equal variance).
To visualize salivary gland nuclear envelopes, glands were dissected in PBS and fixed in 10% formaldehyde in PBS for 5 min, incubated for 20 min in PBS containing 1 mg/ml wheat germ agglutinin conjugated to Alexa Fluor 488 (Virtanen and Wartiovaara, 1976), and then stained with 10 ng/ml DAPI in PBS for 10 min and washed twice with PBS (10 min per wash). Glands were mounted in Vectashield and imaged with the Carl Zeiss confocal system.
The slimbUU11 null allele was recombined onto an FRT82 chromosome. These lines were crossed to a UbxFLP line also containing the FRT82 chromosome with a Minute mutation. Clones inheriting two Minute mutant chromosomes were outcompeted by the slimbUU11/slimbUU11 homozygous mutant clonal cells. Larval wing disc mutant clones nuclei were stained with lamin Dm0 mouse monoclonal antibodies (ADL84.12; Developmental Studies Hybridoma Bank). Disc nuclei images shown are single confocal sections collected acquired with a confocal microscope (TCS; Leica), a Plan-Apochromat 63×/1.4 NA objective lens, and Application Suite FA software (Leica), and then assembled using Photoshop CS4 (Adobe), as described previously (Windler and Bilder, 2010). At least five samples were analyzed for each experiment.
Immunoblotting
S2 cell extracts were produced by lysing cells in PBS, 0.1% Triton X-100. The Bradford protein assay (Bio-Rad Laboratories) was used to measure lysate protein concentrations. Laemmli sample buffer was then added and samples were boiled for 5 min. The efficiency of RNAi was determined by Western blotting of treated cell lysates; equal total protein was loaded for each sample, and the integrated densities of chemiluminescent bands (measured with ImageJ) were normalized relative to the integrated densities of the loading control. Either endogenous α-tubulin or GAPDH were used as a loading control.
Antibodies
E. coli–expressed full-length GST- or MPB-Cid proteins were purified on either glutathione-Sepharose or amylose resin. Rabbit polyclonal antisera were raised against GST-tagged purified Cid protein (provided by S. Rogers, University of North Carolina, Chapel Hill), and the corresponding MBP fusion was used for antibody affinity purification by precoupling to Affigel 10/15 resin (Bio-Rad Laboratories). Antibodies were affinity purified from antisera using resin with coupled peptide. Additional antibodies used for Western blots include polyclonal anti-Slimb (Brownlee et al., 2011), anti–α-tubulin DM1a (Sigma-Aldrich), anti-FLAG (Sigma-Aldrich), anti-GAPDH (Imgenex), monoclonal anti-Arm (provided by M. Peifer, University of North Carolina, Chapel Hill, Chapel Hill, NC), anti-Cap-D2 and SMC-2 (provided by M. Heck, University of Edinburgh, Edinburgh, Scotland, UK), anti–cyclin A (provided by H. Richardson, University of Adelaide, Adelaide, Australia), anti–Cullin-1 (provided by R. Duronio, University of North Carolina, Chapel Hill), anti-SkpA (Rogers et al., 2009), and monoclonal anti-GFP JL8 (Takara Bio Inc.). HRP-conjugated secondary antibodies (Sigma-Aldrich and Jackson ImmunoResearch Laboratories, Inc.) were prepared according to the manufacturer’s instructions and used at 1:1,500 dilutions.
Constructs and transfection
cDNA encoding Cap-H2-EGFP was subcloned into the inducible metallothionein promoter pMT vector. Site-directed mutagenesis was performed using QuikChange II (Agilent Technologies). Expression of all constructs was induced by addition of 0.5–2 mM CuSO4 to the media. Transient transfections were performed using the Nucleofector II (Lonza) according to manufacturer’s instructions. Stable S2 cell lines were selected by cotransfection with pCoHygro (Life Technologies) plasmid and treated for 3–4 wk with Hygromycin B (Life Technologies).
Immunoprecipitation
Polyclonal anti-Slimb antibody was bound to equilibrated Protein A–coupled Sepharose (Sigma-Aldrich) by gently rocking overnight at 4°C in 0.2 M sodium borate. For GFP immunoprecipitations, GFP-binding protein (GBP; Rothbauer et al., 2008) was fused to the Fc domain of human IgG (pIg-Tail; R&D Systems), tagged with His6 in pET28a (EMD Millipore), expressed in E. coli, and purified on Talon resin (Takara Bio Inc.) according to manufacturer’s instructions. GBP was bound to Protein A–coupled Sepharose, cross-linked to the resin using dimethyl pimelimidate, and rocked for 1 h at 22°C; the coupling reaction was then quenched in 0.2 M ethanolamine, pH 8.0, and rocked for 2 h at 22°C. Antibody or GBP-coated beads were washed three times with 1.5 ml of cell lysis buffer (CLB; 100 mM Tris, pH 7.2, 125 mM NaCl, 1 mM DTT, 0.1% Triton X-100, and 0.1 mM PMSF). Transfected S2 cells were induced to express recombinant Cap-H2 with 1–2 mM CuSO4. After 24 h, transfected cells were lysed in CLB, clarified by centrifugation, and then diluted to 2–5 mg/ml in CLB. Antibody-coated beads were mixed with lysate for 40 min at 4°C, washed three times with CLB, and then boiled in Laemmli sample buffer. Lambda phosphatase (New England Biolabs, Inc.) treatments were performed for 1 h at 37°C. In vivo ubiquitination assays were performed by coexpressing Plk4-GFP (Rogers et al., 2009) or Cap-H2-GFP constructs with 3×FLAG–tagged Drosophila ubiquitin (CG32744), driven under the inducible metallothionein promoter pMT vector, immunoprecipitated using anti-GFP JL8 antibody, and analyzed by anti-FLAG immunoblotting.
Chromatin fractionation experiments
S2 cells were transfected by Amaxa nucleofection (Lonza), according to manufacturer’s instructions, with 2 µg pMT-EGFP, pMT-Cap-H2-EGFP, or pMT-Cap-H2ΔC23-EGFP. Transcription of the transgene (under control of the metallothionein promoter of pMT) was induced with 1 mM CuSO4. After 24 h, the transfected cells were harvested and then fractionated into whole cell lysate (L), cytoplasmic (S2), nuclear-soluble (S3), and chromatin (P3) fractions as described previously (Wysocka et al., 2001). In brief, ∼107 cells were collected, washed with cold PBS, and resuspended at 4 × 107 cells/ml in buffer A: 10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, and protease inhibitor cocktail (Roche). Cells were lysed by the addition of 0.1% Triton X-100 and then incubated on ice for 8 min (fraction L). Nuclei were collected by centrifugation (5 min, 1,300 g, 4°C), the initial supernatant was further cleared by high-speed centrifugation (5 min, 20,000 g, 4°C), and the final supernatant was collected (fraction S2). The pelleted nuclei were washed once in buffer A, and nuclear membranes were lysed for 30 min in 3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and protease inhibitor cocktail (buffer B). The insoluble chromatin (fraction P3) and soluble (fraction S3) fractions were separated by centrifugation (5 min, 1,700 g, 4°C). The insoluble chromatin pellet was washed once with buffer B and resuspended in SDS loading buffer. The protein concentration of each fraction was determined by a Bradford’s assay (Bio-Rad Laboratories), and 20 µg of protein from each fraction was immunoblotted with anti-GFP, anti-Slimb, and anti–lamin B (ADL84.12-c; Developmental Studies Hybridoma Bank).
Online supplemental material
Fig. S1 shows RNAi efficiency and that slimb RNAi promotes chromatin-gumball formation in cultured S2R+ and Kc cells. Fig. S2 outlines the experimental strategies used to arrest cells in interphase. Fig. S3 shows that chromosome pairing in interphase Kc cells is blocked by slimb RNAi but is rescued by slimb/cap-H2 double RNAi and that Cid localization is disrupted in the nuclei of slimb RNAi-treated cells. Fig. S4 shows that slimb/condensin I double RNAi does not rescue chromatin-gumball formation, that slimb/cap-H2 double RNAi rescues nuclear envelope defects, and that Slimb associates with chromatin and controls Cap-H2 levels on chromatin. Fig. S5 shows that Cap-H2 overexpression induces nuclear envelope defects in larval salivary gland cells. Table S1 lists the primer sequences used to generate dsRNA in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207183/DC1.
Supplementary Material
Acknowledgments
We thank M. Peifer, R. Duriono, H. Richardson, S. Rogers, and M. Heck for reagents, P. Krieg for comments on the manuscript, and E. Joyce and C.-T. Wu for sharing results before publication.
G.C. Rogers is grateful for support from the National Cancer Institute (NCI; grant P30 CA23074) and GI SPORE (NCI/National Institutes of Health grant P50 CA95060). This work was supported by NIH grant GM069462 and by support from the Arizona Center on Aging to G. Bosco as well as grants from NIH (GM068675 and GM090150), the American Cancer Society (RSG-07-040-01), and the Burroughs Wellcome Fund to D. Bilder.
Footnotes
Abbreviations used in this paper:
- dsRNA
- double-stranded RNA
- GBP
- GFP-binding protein
References
- Abe S., Nagasaka K., Hirayama Y., Kozuka-Hata H., Oyama M., Aoyagi Y., Obuse C., Hirota T. 2011. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 25:863–874 10.1101/gad.2016411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C.R., Hartl T.A., Bosco G. 2012. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 8:e1002873 10.1371/journal.pgen.1002873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., Sánchez P., Rivera T., Rodríguez-Corsino M., Boyarchuk E., Vassias I., Ray-Gallet D., Arnaoutov A., Dasso M., Almouzni G., Losada A. 2011. Xenopus HJURP and condensin II are required for CENP-A assembly. J. Cell Biol. 192:569–582 10.1083/jcb.201005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C.W., Klebba J.E., Buster D.W., Rogers G.C. 2011. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195:231–243 10.1083/jcb.201107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S.D., Sjögren C. 2012. The SMC complexes, DNA and chromosome topology: right or knot? Crit. Rev. Biochem. Mol. Biol. 47:1–16 10.3109/10409238.2011.614593 [DOI] [PubMed] [Google Scholar]
- Chan R.C., Severson A.F., Meyer B.J. 2004. Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167:613–625 10.1083/jcb.200408061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Archambault V., Kar A., Lio’ P., D’Avino P.P., Sinka R., Lilley K., Laue E.D., Deak P., Capalbo L., Glover D.M. 2007. Multiple protein phosphatases are required for mitosis in Drosophila. Curr. Biol. 17:293–303 10.1016/j.cub.2007.01.068 [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M. 2010. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2:a003889 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F. 2000. In situ hybridization to somatic chromosomes. Drosophila Protocols. Sullivan W., Ashburner M., Hawley R.S., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York: 25–55 [DOI] [PubMed] [Google Scholar]
- Duncan I.W. 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36:521–556 10.1146/annurev.genet.36.060402.100441 [DOI] [PubMed] [Google Scholar]
- Fritsch C., Ploeger G., Arndt-Jovin D.J. 2006. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 14:451–464 10.1007/s10577-006-1068-z [DOI] [PubMed] [Google Scholar]
- Gosling K.M., Makaroff L.E., Theodoratos A., Kim Y.H., Whittle B., Rui L., Wu H., Hong N.A., Kennedy G.C., Fritz J.A., et al. 2007. A mutation in a chromosome condensin II subunit, kleisin beta, specifically disrupts T cell development. Proc. Natl. Acad. Sci. USA. 104:12445–12450 10.1073/pnas.0704870104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R.T., Dickinson H.G. 2004. Plants, pairing and phenotypes—two’s company? Trends Genet. 20:188–195 10.1016/j.tig.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Green L.C., Kalitsis P., Chang T.M., Cipetic M., Kim J.H., Marshall O., Turnbull L., Whitchurch C.B., Vagnarelli P., Samejima K., et al. 2012. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J. Cell Sci. 125:1591–1604 10.1242/jcs.097790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T.A., Smith H.F., Bosco G. 2008a. Chromosome alignment and transvection are antagonized by condensin II. Science. 322:1384–1387 10.1126/science.1164216 [DOI] [PubMed] [Google Scholar]
- Hartl T.A., Sweeney S.J., Knepler P.J., Bosco G. 2008b. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 4:e1000228 10.1371/journal.pgen.1000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Dreesen T.D. 1989. Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc. Natl. Acad. Sci. USA. 86:6704–6708 10.1073/pnas.86.17.6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15:R265–R275 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Hirano M., Hirano T. 2004. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 23:2664–2673 10.1038/sj.emboj.7600264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., Cleveland D.W. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Struhl G. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 391:493–496 10.1038/35154 [DOI] [PubMed] [Google Scholar]
- Joyce E.F., Williams B.R., Xie T., Wu C.T. 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8:e1002667 10.1371/journal.pgen.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J.A., Southworth J.W. 2002. Transvection in Drosophila. Adv. Genet. 46:399–420 10.1016/S0065-2660(02)46014-2 [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Groudine M. 2004. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 18:1371–1384 10.1101/gad.1209304 [DOI] [PubMed] [Google Scholar]
- Laster K., Kosak S.T. 2010. Genomic Pangea: coordinate gene regulation and cell-specific chromosomal topologies. Curr. Opin. Cell Biol. 22:314–319 10.1016/j.ceb.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Lewis E.B. 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88:225–239 10.1086/281833 [DOI] [Google Scholar]
- Marshall W.F. 2002. Order and disorder in the nucleus. Curr. Biol. 12:R185–R192 10.1016/S0960-9822(02)00724-8 [DOI] [PubMed] [Google Scholar]
- McKee B.D. 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta. 1677:165–180 10.1016/j.bbaexp.2003.11.017 [DOI] [PubMed] [Google Scholar]
- Mihaylov I.S., Kondo T., Jones L., Ryzhikov S., Tanaka J., Zheng J., Higa L.A., Minamino N., Cooley L., Zhang H. 2002. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 22:1868–1880 10.1128/MCB.22.6.1868-1880.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Painter T.S. 1933. A new method for the study of chromosome rearrangements and the plotting of chromosome maps. Science. 78:585–586 10.1126/science.78.2034.585 [DOI] [PubMed] [Google Scholar]
- Rajapakse I., Groudine M. 2011. On emerging nuclear order. J. Cell Biol. 192:711–721 10.1083/jcb.201010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Scalzo D., Tapscott S.J., Kosak S.T., Groudine M. 2010. Networking the nucleus. Mol. Syst. Biol. 6:395 10.1038/msb.2010.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings J.S., Gatzka M., Thomas P.G., Ihle J.N. 2011. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 30:263–276 10.1038/emboj.2010.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., Rogers S.L. 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S., Golic K.G. 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics. 165:1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., Leonhardt H. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 7:282–289 [DOI] [PubMed] [Google Scholar]
- Sanyal A., Baù D., Martí-Renom M.A., Dekker J. 2011. Chromatin globules: a common motif of higher order chromosome structure? Curr. Opin. Cell Biol. 23:325–331 10.1016/j.ceb.2011.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T. 2006. Good news in the nuclear envelope: loss of lamin A might be a gain. J. Clin. Invest. 116:632–634 10.1172/JCI27820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., Gordon L., Misteli T. 2005. The cell nucleus and aging: tantalizing clues and hopeful promises. PLoS Biol. 3:e395 10.1371/journal.pbio.0030395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Hirano T. 2011. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 25:1464–1469 10.1101/gad.2060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smelkinson M.G., Kalderon D. 2006. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr. Biol. 16:110–116 10.1016/j.cub.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Somma M.P., Fasulo B., Siriaco G., Cenci G. 2003. Chromosome condensation defects in barren RNA-interfered Drosophila cells. Genetics. 165:1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.H., McKee B.D. 2011. Homologous pairing and the role of pairing centers in meiosis. J. Cell Sci. 124:1955–1963 10.1242/jcs.006387 [DOI] [PubMed] [Google Scholar]
- van Berkum N.L., Lieberman-Aiden E., Williams L., Imakaev M., Gnirke A., Mirny L.A., Dekker J., Lander E.S. 2010. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 39:1869 10.3791/1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J., Belmont A.S., Sedat J.W. 2001. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11:1227–1239 10.1016/S0960-9822(01)00390-6 [DOI] [PubMed] [Google Scholar]
- Virtanen I., Wartiovaara J. 1976. Lectin receptor sites on rat liver cell nuclear membranes. J. Cell Sci. 22:335–344 [DOI] [PubMed] [Google Scholar]
- Williams B.R., Bateman J.R., Novikov N.D., Wu C.T. 2007. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics. 177:31–46 10.1534/genetics.107.076356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windler S.L., Bilder D. 2010. Endocytic internalization routes required for delta/notch signaling. Curr. Biol. 20:538–543 10.1016/j.cub.2010.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A.J., Severson A.F., Meyer B.J. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11:391–404 10.1038/nrg2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.T., Morris J.R. 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9:237–246 10.1016/S0959-437X(99)80035-5 [DOI] [PubMed] [Google Scholar]
- Wysocka J., Reilly P.T., Herr W. 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21:3820–3829 10.1128/MCB.21.11.3820-3829.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita D., Shintomi K., Ono T., Gavvovidis I., Schindler D., Neitzel H., Trimborn M., Hirano T. 2011. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J. Cell Biol. 194:841–854 10.1083/jcb.201106141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Malone J.H., Powell S.K., Periwal V., Spana E., Macalpine D.M., Oliver B. 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8:e1000320 10.1371/journal.pbio.1000320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. 2006. From early homologue recognition to synaptonemal complex formation. Chromosoma. 115:158–174 10.1007/s00412-006-0048-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.