Abstract
Aneuploidy, an aberrant number of chromosomes, has been recognized as a feature of human malignancies for over a century, but compelling evidence for causality was largely lacking until mouse models for chromosome number instability were used. These in vivo studies have not only uncovered important new insights into the extremely complex aneuploidy–cancer relationship but also into the molecular mechanisms underlying proper and aberrant chromosome segregation. A series of diverse mouse models for the mitotic checkpoint protein BubR1 has provided evidence for a provocative novel link between aneuploidization and the development of age-related pathologies.
Introduction
Aneuploidy refers to a state in which the number of chromosomes in a cell is not an exact multiple of the haploid set. Chromosomal instability (CIN), on the other hand, defines a condition in which cells are unable to accurately segregate whole chromosomes (whole CIN [W-CIN]) or prone to structural chromosome rearrangements (structural CIN [S-CIN]), including translocations, deletions, and duplications of large parts of chromosomes (Ricke et al., 2008). CIN genes are commonly classified as genes that increase the rate of numerical and/or structural chromosome alterations when mutated (Michor et al., 2005). In the early 1900s, Theodor Boveri hypothesized that aneuploidy was a causal feature of human cancers. This long-standing hypothesis was difficult to test until the development of targeted approaches to genetically manipulate mice and the discovery of genes and mechanisms that act to prevent chromosome number instability. Although the relationship between aneuploidy and tumorigenesis is characterized by ever increasing complexity, aneuploidy-prone mouse models revealed that the effect of W-CIN on tumorigenesis is highly dependent on the gene that is defective, including its other cellular functions, the extent or nature of the gene defect, the affected tissue or cell type, and the context of other cancer gene mutations (Ricke et al., 2008). Studies designed to explore the role of BubR1 in cancer uncovered a surprising link between abundance of this mitotic regulator and the rate of aging (Baker et al., 2004, 2013). This provided a molecular entry point for studies on age-related aneuploidization and its potential role in tissue/organ degeneration. The impact of aneuploidization on physiological homeostasis seems negative, but accumulating evidence suggests that select tissues are subject to orchestrated aneuploidization as part of normal tissue development (Rehen et al., 2001; Duncan et al., 2012b). Here, we highlight the recent advances in understanding the physiological impact of aneuploidy and CIN using mouse models as well as the new mechanistic insights these studies provided into proper and aberrant chromosome segregation.
Mechanistic insights into CIN gene function and malfunction
Early attempts to understand the aneuploidy–cancer relationship were hampered by a lack of information about the molecular genetic basis of mitosis, which is believed to involve hundreds of genes (Stirling et al., 2011). Although much of what is currently known about the molecules and mechanisms that drive chromosome segregation originates from in vitro studies, mouse models have been invaluable tools for obtaining mechanistic information for various reasons. First, gene-targeted and transgenic mice offer a clean genetic system in which all cells are afflicted in the absence of confounding preexisting genetic aberrations. Second, gene expression can be up- or down-regulated in a graded fashion, which has helped uncover the multifaceted nature of several CIN genes. Third, knockin mutations targeting specific domains of certain mitotic regulators have been instrumental for delineating their modular functions. Fourth, CIN genes can be analyzed in a wide variety of cell types residing in their natural tissue context, allowing for the identification of any mechanistic diversity in the execution of mitosis between distinct cell types.
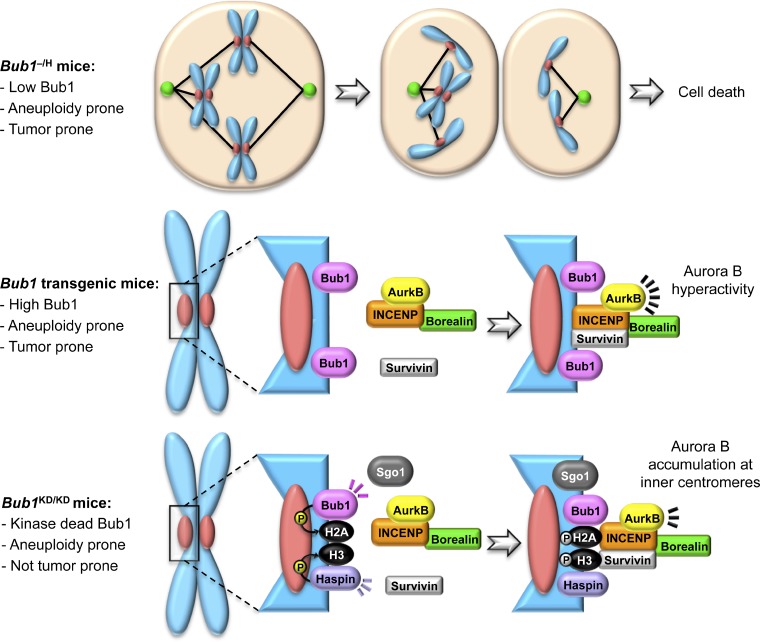
The novel mechanistic insights gained from mouse modeling are perhaps best exemplified by studies of the mitotic checkpoint gene Bub1 (Fig. 1), for which seven different targeted mutations (Jeganathan et al., 2007; Perera et al., 2007; Leland et al., 2009; Schliekelman et al., 2009; Ricke et al., 2012) and several transgenic strains have been created (Cowley et al., 2005; Ricke et al., 2011). For example, conditional knockout alleles for Bub1 uniquely demonstrated that premature centromeric separation is a consequence of mitotic checkpoint weakening (Perera et al., 2007). Complete ablation of Bub1 kinase activity unveiled how Bub1-mediated histone H2A phosphorylation promotes Aurora B inner centromeric localization (Ricke et al., 2012), whereas Bub1 overexpression uncovered that Bub1 carefully controls the level of Aurora B activity to prevent chromosome missegregation and aneuploidization (Ricke et al., 2011). Hypomorphic alleles of Bub1 further underscored that the level at which this mitotic regulator is expressed is pivotal for chromosomal stability, with Bub1 insufficiency resulting in aberrant kinetochore assembly and checkpoint activity (Jeganathan et al., 2007).
Figure 1.
New mechanistic insights from various Bub1 mouse models. Bidirectional deviations from normal Bub1 levels and inactivation of Bub1 enzymatic activity universally cause aneuploid cells to accumulate in mice. Analysis of the underlying mechanisms of chromosome missegregation in each mouse model has provided important new insights into the multifaceted nature of this mitotic regulator. For instance, in addition to confirming that Bub1 plays a critical role in kinetochore assembly and mitotic checkpoint activity, studies of Bub1 hypomorphic mice (top) revealed that Bub1 acts as a crucial trigger to induce cell death after chromosome missegregation (Jeganathan et al., 2007). On the other hand, transgenic mice that overexpress Bub1 (middle) revealed the novel concept of Aurora B hyperactivity and linked it to chromosome missegregation (Ricke et al., 2011). Unlike Bub1 hypomorphic or transgenic mice, mice lacking Bub1 kinase activity (bottom) harbor significant aneuploidy without a predisposition to cancer. In this model, it was revealed that accumulation of Aurora B at inner centromeric regions is mediated by Bub1-mediated histone H2A phosphorylation at T121 in a shugoshin-independent manner (Ricke et al., 2012). P, phosphorylation.
Studies of various Mad2 mutant mice indicate that differential effects imparted by bidirectional deviation of protein expression may be a more common feature of mitotic regulators. Both up- and down-regulation of Mad2 predispose to aneuploidy, with Mad2 haploinsufficiency weakening mitotic checkpoint signaling and Mad2 overexpression hyperstabilizing microtubule–kinetochore attachments (Michel et al., 2001; Kabeche and Compton, 2012). On the other hand, bidirectional deviation of BubR1 results in divergent effects on aneuploidy, with BubR1 overexpression providing protection against aneuploidy and BubR1 insufficiency perturbing accurate chromosome segregation (Baker et al., 2004, 2013). That Mad2 and BubR1 overexpression have opposite effects on chromosome segregation is intriguing given that both function in a complex to inhibit anaphase-promoting complex (APC)/cyclosomeCdc20 (Kulukian et al., 2009). The divergence may simply reflect potential differences in level of overexpression or fundamental differences in protein function.
A key advantage of using mouse models to decipher the mechanisms by which CIN genes operate is that gene malfunction can be directly correlated to effects on health and disease. This has been particularly important to advance our insight into the intricate aneuploidy–cancer relationship.
Aneuploidy and cancer
Although aneuploidy has been long recognized to be a defining feature of cancer cell genomes, inferring the significance of chromosomal aberrancies in tumorigenesis has remained a challenge. Whereas some researchers argue that aneuploidy is a primary force driving tumorigenesis (Duesberg et al., 1998), others contend that aneuploidy is simply a side effect of malignant transformation (Zimonjic et al., 2001). The confusion is in part caused by the heterogeneous nature of tumors, the broad landscape of genetic mutations a cancer cell harbors to thwart protective pathways (Wood et al., 2007), and the observation that few solid tumors undergo identical CIN events (Mitelman, 2000). Here, we first recognize the tremendous complexity of the cancer–aneuploidy issue, then discuss the various lines of evidence from mouse models that aneuploidy drives cancer, and finally provide an alternative look at the aneuploidy–cancer connection.
Multilevel complexity of the aneuploidy–cancer relationship.
In addition to an incomplete understanding about the molecular genetic basis of mitosis, there are at least seven more layers of complexity regarding the actions of aneuploidy and CIN in human cancer.
(1) Recurrent chromosome gains/losses are rare in human cancers. Although specific chromosome translocations often classify hematologic malignancies (Mitelman, 2000), recurrent gains/losses of specific chromosomes are extremely rare in any human cancer type, complicating the interpretation of whether whole chromosome reshuffling is crucial or irrelevant. Recent studies suggest that chromosome reshuffling in human tumors is not entirely arbitrary (Ozery-Flato et al., 2011; Duijf et al., 2013), but the role of co-occurrence of losses or gains of specific chromosomes in tumor evolution remains entirely unclear.
(2) Inconsistency in aneuploidy measurement and interpretation. A database of published karyotypic abnormalities found in neoplastic diseases, now containing ∼60,000 cases (Höglund et al., 2002), is available to study the prevalence and frequency of karyotypes among tumor types. However, key challenges remain in analyzing the available information, including the lack of uniformity in data collection, the overreliance on metaphase spread karyotyping (which biases toward proliferating cells), and the limited knowledge about the degree of intratumor karyotypic heterogeneity (McGranahan et al., 2012).
(3) Challenges in measuring CIN. CIN has been proposed to facilitate tumor adaptation (Gutenberg et al., 2010; Lee et al., 2011) and is a predictor for poor prognosis and treatment refractory tumors (Carter et al., 2006; Bakhoum et al., 2011; Birkbak et al., 2011). Despite these clinical implications, few methods measure the dynamic nature of CIN in tumors. One exception is FISH, which infers CIN from intratumor variation of chromosome copy number. A surrogate assessment for CIN is its molecular gene signature, as the total transcriptional activity of a tumor can be reflective of unbalanced chromosome load, and chromosomally unstable tumors often aberrantly express chromosome integrity regulators (Upender et al., 2004; Carter et al., 2006; Gao et al., 2007; Pavelka et al., 2010).
(4) The integral link between aneuploidy and W-CIN. Several studies provide evidence for a vicious cycle in which chromosome number imbalances undermine faithful chromosome segregation, causing further aneuploidization. The most compelling evidence is that certain aneuploid yeast strains are prone to additional karyotypic changes (St Charles et al., 2010; Sheltzer et al., 2011; Zhu et al., 2012). Consistent with this notion, some cells from humans with autosomal trisomies gain or lose other chromosomes at elevated rates compared with cells from diploid individuals (Amiel et al., 2006; Reish et al., 2006, 2011).
(5) Temporal importance of CIN in tumors. Theoretically, CIN can emerge and act throughout the entire tumor process. However, the aneuploidy status of mature tumors provides little information about the timing and impact of numerical chromosome changes during tumor evolution. For instance, CIN occurring early during tumorigenesis may be masked by late-stage genetic alterations promoting karyotypic stability.
(6) An apparent inseparable nature of W-CIN and S-CIN. Several lines of evidence suggest that impaired mitotic fidelity creates DNA damage that adversely impacts genome integrity. Structural chromosomal damage may occur when lagging chromosomes are trapped in the cytokinesis furrow (Janssen et al., 2011). Alternatively, micronuclei formation caused by lagging chromosomes may drive loss of structural integrity through breakage–fusion–bridge cycles or the more extreme process of chromosome pulverization (Guerrero et al., 2010; Crasta et al., 2012). The latter process may explain the phenomenon of “chromothripsis,” during which chromosomes undergo extensive rearrangements (Hastings et al., 2009; Liu et al., 2011; Stephens et al., 2011).
(7) Aneuploidy induces complex cellular responses impacting cell fate. Compelling evidence from cultured cells suggests that aneuploidization is associated with engagement of certain cellular stress pathways, including those responding to genotoxic, proteotoxic, metabolic, or proliferative stress (Torres et al., 2007; Williams et al., 2008; Li et al., 2009, 2010; Thompson and Compton, 2010; Sheltzer et al., 2012). The ability of aneuploid tumor cells to counteract these potentially negative effects on cell growth and survival could be tumor type dependent.
Evidence for causality.
Three independent lines of evidence from mouse models support the hypothesis that there is a causal relationship between aneuploidy and tumorigenesis. First, if aneuploidy were a causal feature of tumorigenesis, one would expect that increasing aneuploidization in mice would increase tumor predisposition. Indeed, most of the several dozen chromosomally unstable mouse models are tumor prone (Pfau and Amon, 2012). This includes mice with aberrancies in mitotic checkpoint signaling (Michel et al., 2001; Iwanaga et al., 2007; Jeganathan et al., 2007; Weaver et al., 2007; Li et al., 2009; Schliekelman et al., 2009), centrosome duplication (van Ree et al., 2010), spindle assembly (Aguirre-Portolés et al., 2012; Zhang et al., 2012), microtubule–kinetochore attachment (Sotillo et al., 2007; Weaver et al., 2007; Diaz-Rodríguez et al., 2008), or attachment error correction (Fernández-Miranda et al., 2011; Ricke et al., 2011), suggesting that tumor propensity is independent of the mechanism driving the aneuploidy. As in vitro studies have linked aberrant chromosome segregation to structural chromosomal abnormalities (Guerrero et al., 2010; Janssen et al., 2011; Crasta et al., 2012), it will now be important to carefully analyze the available W-CIN models for evidence of S-CIN predisposition.
Second, if aneuploidy was a driving force in tumorigenesis, one might predict that protection against aneuploidization would attenuate tumor formation. One mouse model that suppresses chromosome missegregation is a transgenic mouse strain that overexpresses the mitotic checkpoint protein BubR1. Indeed, spontaneous, carcinogen, and genetically induced tumorigenesis are all reduced in these mice (Baker et al., 2013). BubR1 is unique in that its overexpression protects against aneuploidy, as overexpression of other mitotic regulators, such as Bub1, Mad2, UbcH10, and Hec1, increases aneuploidization (Sotillo et al., 2007; Diaz-Rodríguez et al., 2008; van Ree et al., 2010; Ricke et al., 2011). That BubR1 overabundance protects against aneuploidization is remarkable considering that overexpression of Mad2, a mitotic regulator that similarly acts to prevent precocious APC/cyclosome activation, induces aneuploidy through hyperstabilization of microtubules to kinetochores (Sotillo et al., 2007; Kabeche and Compton, 2012).
Third, genetic alterations that promote chromosome missegregation have long been proposed to drive tumorigenesis through loss of whole chromosomes containing key tumor suppressor genes. Specifically, it has been shown that whole chromosome missegregation, caused by Bub1 hypomorphism, promotes loss of heterozygosity to potentiate tumorigenesis in two different tumor suppressor backgrounds, p53 and APC (Baker et al., 2009; Baker and van Deursen, 2010). Whether this is a universal feature of CIN and what contexts drive these key events remain unclear, as Bub1 hypomorphism initiates the loss of key tumor suppressors in restricted genetic contexts. Moreover, aneuploidy driven by haploinsufficiency of either Bub1 or Bub3 was unable to promote p53 loss of heterozygosity (Kalitsis et al., 2005; Baker et al., 2009). Therefore, understanding which CIN genes cooperate to promote this type of event will require further clarity.
Although many W-CIN mouse models are tumor prone, why some mouse strains with CIN are susceptible to tumorigenesis and others are not remains a key unanswered question. Curiously, the incidence of tumorigenesis does not correlate with aneuploidy levels, although technical limitations preclude a systematic, animal-wide analysis of aneuploidy. Additionally, aneuploidy in proliferating cells, measured using karyotyping, may not be representative of nondividing cells, measured using FISH, particularly if aneuploidy prevents proliferation in those tissues (Torres et al., 2007; Williams et al., 2008). Tumor spectrum is also independent of the error driving aneuploidy. For example, mice with chromosome instability caused by DNA replication defects develop tumors with a similar spectrum as canonical W-CIN mice (Chuang et al., 2010; Kawabata et al., 2011). Similarly, mice with aneuploidy as a result of a variety of mitotic defects develop similar tumor types (Jeganathan et al., 2007; Schliekelman et al., 2009; van Ree et al., 2010; Fernández-Miranda et al., 2011; Ricke et al., 2011; Zhang et al., 2012). This implies that the stochastic nature of aneuploidization is sufficient to drive the tumor process rather than any specific activities.
A hierarchical view of the aneuploidy–cancer connection.
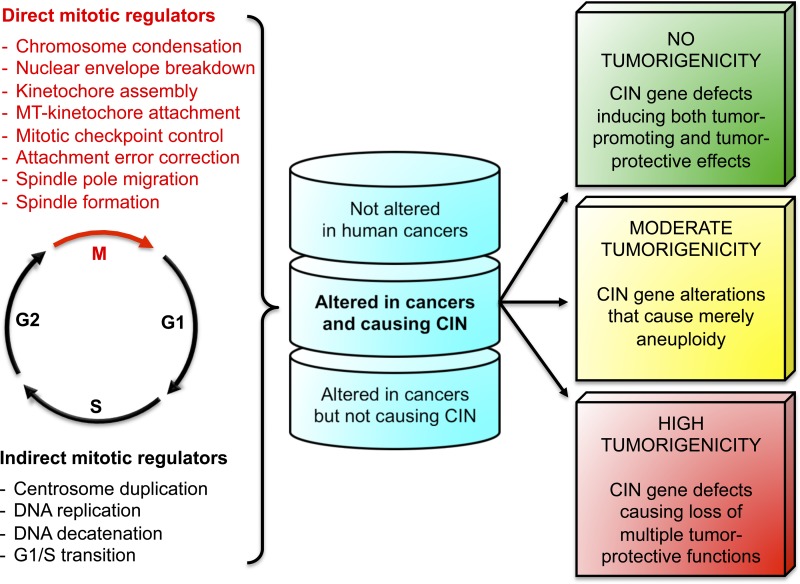
Studies on the molecular genetic basis of chromosome segregation have largely centered on understanding the workings of various mitotic processes, including chromosome condensation, kinetochore assembly, spindle formation, spindle pole migration, microtubule–kinetochore attachment, nuclear envelope breakdown, mitotic checkpoint activation, and attachment error correction (Fig. 2). Here, we classify components acting in these processes as direct mitotic regulators. However, increasing evidence suggests that various cellular processes occurring outside of mitosis can be key determinants of segregation accuracy. We define components implicated in these nonmitotic processes as indirect mitotic regulators (Fig. 2). For example, cells with supernumerary centrosomes demonstrate an increased frequency of lagging chromosomes during anaphase, resulting from aberrant microtubule–kinetochore attachment before centrosome clustering and anaphase onset (Ganem et al., 2009; Silkworth et al., 2009). Moreover, incomplete DNA duplication combined with precocious mitotic entry has been proposed to drive anaphase bridges or lagging chromosomes (Chan et al., 2009; Kawabata et al., 2011; Remeseiro et al., 2012). Although incomplete DNA replication results in the linkage of sister chromatids, improper resolution of other forms of topological linkages, such as DNA catenation and cohesin ring assembly, may impair chromosome segregation fidelity (Wang et al., 2010; Xu et al., 2010; Solomon et al., 2011; Remeseiro et al., 2012). Finally, proteins that regulate transcriptional or posttranslational expression of mitotic regulators may indirectly impact mitotic fidelity. One example is Mad2, whose expression is controlled by retinoblastoma (Rb) and E2F, the p53–p21 pathway, and the E3 ubiquitin ligase SCFβTrCP (Warren et al., 2002; Hernando et al., 2004; Guardavaccaro et al., 2008; Manning and Dyson, 2011; Schvartzman et al., 2011).
Figure 2.
A hierarchy-based view of the aneuploidy–cancer relationship. Hundreds of genes are thought to directly or indirectly influence chromosomal stability. Whereas the functions of direct mitotic regulators are limited to mitosis, cellular processes acting outside of mitosis that influence segregation accuracy are defined as indirect mitotic regulators. Genetic modification of CIN genes in mice suggests that certain gene alterations are more cancer relevant than others, leading us to propose a hierarchal CIN gene model. This model not only takes into consideration aneuploidy rates but also other cancer-critical functions CIN genes might possess and the frequency with which a particular CIN gene alteration occurs in human cancers. We recognize three classes of CIN gene alterations within both the direct and indirect mitotic regulators: CIN genes that are not or rarely found altered in human cancers, CIN gene alterations found in human cancers but not causing aneuploidy, and genes that are altered and act causally to promote chromosome missegregation. Animal modeling studies predict three potential cancer-related outcomes for the latter class of CIN gene alterations: (1) no tumorigenicity (or inhibiting tumor development), (2) moderate tumorigenicity, and (3) high tumorigenicity. The latter CIN gene defects presumably are multifaceted and act through numerous tumor-protective functions, whereas those with only moderate tumorigenicity perhaps act solely in the generation of aneuploid cells. Alternatively, CIN genes with counteracting tumor-promoting and tumor-protecting functions may have no tumorigenicity predisposition. One example for this final type is BubR1, through which deregulation induces both aneuploidization and cellular senescence.
From a basic science perspective, it is important to understand how each of the several hundred direct/indirect mitotic regulators contribute to the accuracy of chromosome segregation. From a cancer biology perspective, it is imperative to identify the CIN genes that are altered in human tumors and to determine which of these gene alterations drive neoplastic transformation (Fig. 2). Unfortunately, our knowledge about both the identity and the effects of CIN genes altered in human malignancies is very limited. Gene expression profiles that predict CIN status and treatment outcome could assist these efforts. One such profile consists of 70 aberrantly expressed genes, referred to as CIN70, many of which are implicated in DNA replication or chromosome segregation (Carter et al., 2006). An alternative signature of 11 overexpressed genes associated with tumor aggressiveness and poor prognosis is enriched for mitotic factors, including CcnB1, Bub1, and Hec1/Ndc80 (Glinsky et al., 2005). Animal modeling will be instrumental in discriminating between alterations in CIN gene expression that represent true oncogenic events compared with alterations simply caused by the increased proliferative index that tumors have.
Based on currently available data from mouse modeling, we envision that CIN gene alterations that are found in human cancers and induce aneuploidization will fall into one of three classes (Fig. 2). First, the particular CIN gene defect found in human cancers counteracts tumorigenesis, such as observed for BubR1 overexpression (Baker et al., 2013). Another example is BubR1 hypomorphism, which besides aneuploidy promotes senescence, a widely recognized anticancer mechanism (Baker et al., 2008; Rodier and Campisi, 2011). Second, the CIN gene aberrancy has little or no impact on tumorigenesis, as is the case for securin knockout mice (Wang et al., 2001; Chesnokova et al., 2005). This could be caused by low aneuploidization rates or abrogation of the tumor-promoting effect of aneuploidy through other cellular functions of the affected CIN gene. Third, the altered CIN gene aggressively drives tumorigenesis.
We propose that more potent CIN genes are those that possess multiple tumor-suppressive activities that are simultaneously perturbed. One example of a CIN gene alteration in this class would be Bub1 hypomorphism, which has two prominent features that could influence tumor predisposition in addition to aneuploidization. First, it assists in eliminating aneuploid cells after aberrant mitoses (Jeganathan et al., 2007), thus coupling chromosome missegregation with increased survival. Second, it increases micronuclei formation (Jeganathan et al., 2007), which has been implicated in structural aberrancies (Guerrero et al., 2010; Crasta et al., 2012). Other examples might be established cancer-critical gene mutations frequently found in human cancers, including Rb loss and oncogenic Ras (Kamata and Pritchard, 2011; Coschi and Dick, 2012). In certain cells, loss of Rb has been shown to potentiate aneuploidy, perhaps through deregulated E2F activities, which target S- and M-phase genes, including Mad2 (Zheng et al., 2002; Hernando et al., 2004; Baker et al., 2009; Conklin et al., 2012). In vitro oncogenic Ras directly perturbs the accuracy of mitosis before transformation (Denko et al., 1994; Woo and Poon, 2004; Baker et al., 2013), potentially by deregulating prometaphase (Sarthy et al., 2007; Luo et al., 2009). Thus, these CIN genes could represent the top of the aneuploidy hierarchy by driving tumorigenesis through multiple mechanisms.
Aneuploidy during development and aging
Besides cancer, aneuploidization has recently been linked to two other physiological processes, development and aging. Indeed, in select tissues, such as brain and liver, aneuploidization seems to be an integral part of normal organ development (Rehen et al., 2001; Kingsbury et al., 2005; Duncan et al., 2012b), raising the intriguing concept that aneuploidy in some settings may not be detrimental and perhaps even be beneficial. The emerging connection between aneuploidy and aging is particularly fascinating, as aging is known to be the main risk factor for chronic diseases and declining health. In this section, we first review the novel concept of orchestrated aneuploidization during development and then the provocative link between aneuploidization and the development of age-related pathologies.
Orchestrated aneuploidization.
Studies designed to understand the development and function of the mammalian central nervous system revealed aneuploidy in a significant proportion of normal human and mouse brain cells, including mitotic cells and postmitotic neurons (Rehen et al., 2001, 2005; Yang et al., 2003; Kingsbury et al., 2005). The biological relevance of aneuploidization in the developing and mature brain remains speculative. One theory is that aneuploidy promotes cellular diversity in the brain, thus perhaps contributing to the plasticity necessary for complex functions such as learning and memory (Kingsbury et al., 2006; Faggioli et al., 2012). Hepatocytes are also subject to orchestrated aneuploidization. They first become polyploid and then undergo reductive division, a process characterized by massive chromosome loss and the creation of near-diploid aneuploid cells (Duncan et al., 2009, 2010, 2012b; Faggioli et al., 2011; Gentric et al., 2012). It has been suggested that this process may grant the tissue a selective advantage to guard against varied and unknown assaults (Duncan et al., 2012a).
A key open question is whether orchestrated aneuploidy as part of a developmental process applies to tissues other than liver and brain. It will also be important to further explore the molecular mechanisms and functional implications of orchestrated aneuploidy, as studies into the adjustment of neurons and hepatocytes to chromosome imbalances may provide novel insights into the cellular responses to aneuploidy. One possibility is that each specific cell type buffers against the adverse effects of aneuploidy by regulating the expression of detrimental aneuploidy-induced targets. Alternatively, chromosome-specific events may allow the accumulation of certain gene products that provide cells with an advantage for a particular phenotype.
Aneuploidy and accelerated aging.
Age-related aneuploidization has been well documented for oocytes and is considered to be the main cause of female reproductive infertility (Nagaoka et al., 2012). Men are known to be subject to age-related loss of the Y chromosome in several tissues, but the physiological impact of this phenomenon has remained unclear (Jacobs et al., 1963; Pierre and Hoagland, 1972). Initial evidence for a connection between regulators of chromosome segregation and somatic aging was provided by a study designed to investigate the aneuploidy–cancer relationship through a series of mice with graded reduction in BubR1 (Baker et al., 2004). Mutant mice carrying two hypomorphic BubR1 alleles and expressing ∼10% of normal BubR1 levels were prone to aneuploidy as anticipated but, surprisingly, instead of tumors, developed a series of progeroid and age-related pathologies including short lifespan, sarcopenia, growth retardation, cataracts, fat loss, impaired wound healing, and reduced dermal thickness (Fig. 3; Baker et al., 2004).
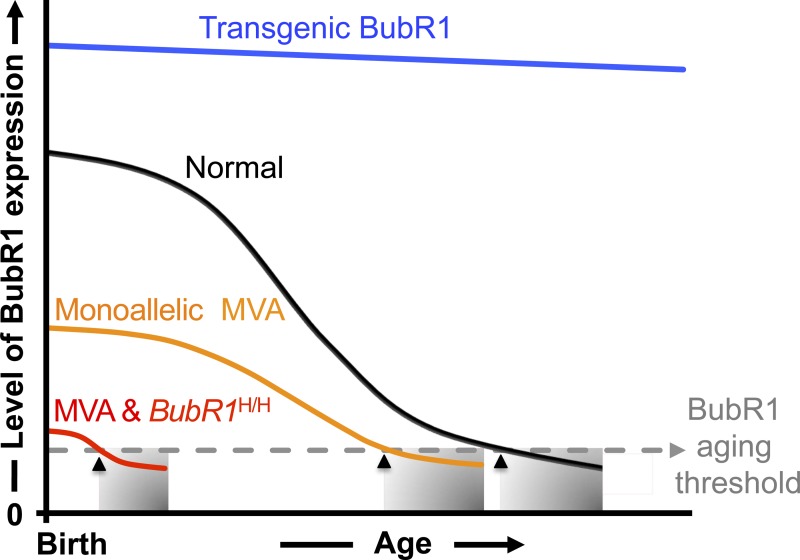
Figure 3.
A minimum threshold level of BubR1 delays age-related pathologies. Several lines of evidence provide a link between BubR1 level and time of age-related pathology onset. During natural aging (black line), BubR1 levels decline in various tissues (Baker et al., 2004, 2013). Mutant mice with very low levels of BubR1 at birth prematurely develop age-related phenotypes with a shortened lifespan (red line). Similarly, many patients with mosaic variegated aneuploidy (MVA) harbor mutations within BubR1 that diminish steady-state BubR1 protein levels (Suijkerbuijk et al., 2010). Even the presence of a single MVA allele negatively impacts health- and lifespan in mice (Wijshake et al., 2012), implying that carriers of an MVA mutation may reach the critical BubR1 threshold level earlier than normal individuals (orange line). Another line of evidence that the amount of BubR1 impacts health has been revealed through artificially elevating BubR1 to extend both health- and lifespan (blue line; Baker et al., 2004, 2013). Together, these results suggest that a threshold level of BubR1 (gray dashed line) is required to prevent onset (black arrows) of tissue deterioration (shaded regions).
Studies on individuals with a rare human recessive autosomal disorder called mosaic variegated aneuploidy (MVA) syndrome have subsequently reinforced the link between BubR1 insufficiency and progeroid disease (Hanks et al., 2004; Matsuura et al., 2006). MVA is a pediatric syndrome implicated in the literature as a hereditary cancer syndrome based on increased risk for childhood cancers such as rhabdomyosarcoma, Wilms’ tumor, and leukemia (Limwongse et al., 1999; Hanks et al., 2004, 2006; Matsuura et al., 2006; García-Castillo et al., 2008). However, MVA is a poorly characterized heterogeneous disease that can also be classified as a progeroid syndrome based on features such as short lifespan, growth retardation, facial dysmorphisms, and cataract formation. The majority of MVA patients have mutations in BUBR1, either biallelic mutations with one allele harboring a missense mutation and the other a nonsense mutation or monoallelic mutations combined with allelic variants producing low amounts of wild-type BubR1 (Hanks et al., 2004; Matsuura et al., 2006). Overall, BubR1 protein levels are typically very low in patients with BUBR1 mutations, largely because mutant BubR1 proteins produced by these alleles tend to be unstable (Suijkerbuijk et al., 2010).
Mice that are doubly haploinsufficient for the mitotic checkpoint genes Bub3 and Rae1 (Babu et al., 2003) represent a second aneuploidy-prone mouse strain with an accelerated aging phenotype, although the rate of premature aging is less profound than in BubR1 hypomorphic mice (Baker et al., 2006). However, the myriad of other aneuploidy mouse models have not been reported to exhibit early traits of early aging. At the surface, this argues against the idea that aneuploidy is sufficient to accelerate aspects of the aging process, but this may be premature for several reasons. First, most aneuploidy models were generated for the purpose of studying cancer predisposition, with mice typically being sacrificed between 14 and 18 mo to thoroughly screen for tumors. Thus, most of these studies would have missed accelerated aging phenotypes that develop later in life but nonetheless prematurely. Second, age-related deterioration may not be overt in most aneuploid models or may be restricted to select tissues. Such was the case for mice harboring one engineered allele that mimics a BubR1 nonsense mutation found in MVA patients with biallelic BubR1 mutations. In depth analyses of this model revealed shortened lifespan and accelerated onset of sarcopenia, cataracts, and fat loss (Wijshake et al., 2012). Third, not all MVA patients have mutations in BubR1, implying that other genes are linked to this progeroid syndrome. One such candidate is Cep57, a gene encoding a centrosomal protein, which is mutated in a subset of MVA patients (Snape et al., 2011). Therefore, a thorough evaluation of other aneuploidy-prone models is needed to determine the impact on aneuploidy on a broad range of tissues.
A likely possibility is that the aneuploidy and aging relationship is as complex as aneuploidy and cancer, such that there exists a hierarchy of CIN genes that also contribute to aging. Perhaps, aneuploidy-associated genes that are strongly linked with early aging, such as BubR1, have multiple functions in preventing tissue deterioration. For example, BubR1 could counteract both aneuploidization and cellular stresses that engage senescence response pathways (Naylor et al., 2013). Consistent with this idea, the principal biomarker for senescent cells, p16Ink4a, is expressed at elevated levels in BubR1 progeroid mice (Baker et al., 2008). Clearing of these p16-positive cells, genetically or pharmacologically, delays progeroid features (Baker et al., 2008, 2011), providing a crucial link between senescence and aging. Clearly, a thorough evaluation of other aneuploidy-prone models is needed to determine the impact of aneuploidy on a broad range of tissues. For this, a system level approach may be useful to screen for phenotypic alterations in a variety of tissues (Guan et al., 2012).
Aneuploidy and natural aging.
The link between BubR1 and early aging raises the question as to whether BubR1 is implicated in natural aging. One observation consistent with such a role is that BubR1 levels decline in various tissues with chronological aging, at least in mice (Baker et al., 2004, 2008). The underlying mechanisms are poorly understood and may occur at both transcriptional and posttranslational levels. BubR1 expression could simply decline as a result of reduced cell proliferation with aging, but a study on transgenic mice that constitutively overexpress BubR1 and are not subject to an age-related drop in BubR1 seem to argue against this (Baker et al., 2013). BubR1 transgenic mice live longer than normal mice and have an increased healthspan (the period during which an organism is free from serious or chronic disease, including cancer) characterized by attenuated muscle and renal atrophy, glomerulosclerosis, and increased cardiac function.
These studies further uncovered that aneuploidization is a hallmark of aging (Baker et al., 2013), raising the possibility that age-related aneuploidy contributes to tissue dysfunction. Consistent with this idea, reduced senescence and tissue deterioration in BubR1 transgenic mice tightly correlated with attenuated age-related aneuploidy (Baker et al., 2013). How BubR1 overexpression counteracts chromosome missegregation remains under investigation, with early evidence suggesting that defects in mitotic checkpoint control and microtubule–kinetochore attachment are ameliorated (Baker et al., 2013). This would imply that both these mitotic processes are subject to age-related decline and at least partially responsible for age-related aneuploidy. Interestingly, the degree of aneuploidization with aging tissue is dependent on proliferative index, as highly proliferative tissues and stem cells show relatively low rates, and largely postmitotic tissues demonstrate higher rates (Baker et al., 2013). One potential explanation is that tissues and cell types with an increased proliferation index are inherently more protected against chromosome segregation than cells that occasionally proliferate. Alternatively, euploid cells may outcompete aneuploid cells in highly proliferating tissues because of the antiproliferative influence of aneuploidization (Williams et al., 2008).
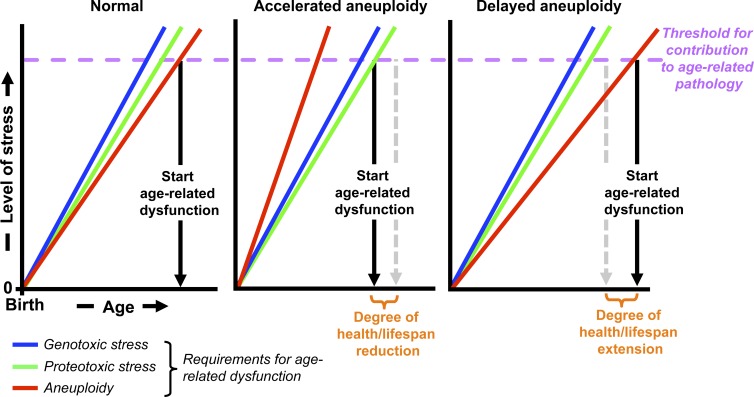
In Fig. 4, we have presented a hypothetical model for how aneuploidization might modulate health- and lifespan based on the available data from wild-type mice and the various models of accelerated and attenuated aneuploidy. It is important to note that aneuploidy is not the only age-related stress and that the effects of varying aneuploidy rates on tissue and organ deterioration have to be considered in the context of a variety of other aging-related stresses. It will be important to further test this provocative model in future experiments.
Figure 4.
Hypothetical models for how aneuploidy might impact health- and lifespan. Working model for how aneuploidy rates might modulate age-related tissue dysfunction. The model takes into consideration the widely held view that tissue aging is driven, at least in part, by diverse cellular stresses causing progressive accumulation of various types of macromolecular damage/stress (Finkel et al., 2007). For simplicity, only three kinds of stresses have been included in the model: aneuploidy, proteotoxic, and genotoxic stress. Accelerated aneuploidy caused by CIN in and of itself is not sufficient to initiate age-related deterioration but may expedite aging by reducing the time needed to reach the cumulative stress threshold. On the other hand, delayed aneuploidization, such as through increased BubR1, may attenuate the aneuploidy stress component and reduce the combined stress sum, thereby extending healthy lifespan. We note that accelerated aneuploidy may influence the threshold of other stresses to trigger pathology because aneuploidy itself can activate genotoxic and proteotoxic stress (Williams et al., 2008; Sheltzer et al., 2011; Tang et al., 2011). Thus, the rate of aneuploidy may alter the slope of other cellular stresses (not depicted).
Conclusions and future studies
The burst of animal modeling that started over a decade ago to critically test Boveri’s theory has provided compelling evidence that CIN provides selective pressure to initiate and propagate malignant transformation. However, the biological consequences of aneuploidy are clearly not limited to tumorigenesis, as aneuploidy correlates also with age-related tissue degeneration and rather paradoxically with benign gain-of-function processes such as in neural and liver cells. Thus, one important unifying theme emerging from the animal studies is the heterogeneity of phenotypes for both cancer and aging among the animals with different CIN gene defects. Perhaps the proposed hierarchical view of CIN genes that takes into consideration functions of these genes outside of mitosis may facilitate future studies aimed at deciphering the basis for this heterogeneity.
One clear future goal is to identify those CIN genes that are most critical for human cancers and to understand why their deregulation is particularly potent. This includes studies aimed at understanding how these alterations derail the chromosome segregation process and which additional tumor-protective molecular activities these genes may have. Improved future insights into the shielding mechanisms that allow nonneoplastic aneuploidy-prone tissues to tolerate aneuploidy’s adverse effects may actually help clarify how aneuploidy acts to negatively impact cells under certain circumstances. Importantly, the potential pervasiveness of orchestrated or age-related mosaic aneuploidy in adult tissues may have repercussions on whether aneuploidy can be exploited as a therapeutic target for cancer treatments. Another important goal will be to decipher the molecular mechanisms that drive new phenomena such as orchestrated and age-related aneuploidization.
In this review, we have highlighted that the establishment of novel aneuploidy-prone mutant strains is an important approach enabling insights into the multifaceted physiological impacts of aneuploidy. This is in part necessary because of the modular nature of mitotic regulators, which function in multiple aspects of mitotic fidelity. Although mutants expressing varying levels of these regulators are valuable, more precise targeting of specific protein activities may yield further insights into functions previously masked and highlight new roles. Thus, although studies using chromosomally unstable mice have exposed layers of complexities in the biological outcomes induced by aneuploidy, these studies also signal that innovative and fresh perspectives are required to shed new light on the potential physiological role of aneuploidy. Finally, the only known gene alteration that counteracts aneuploidization and tumorigenesis in the absence of any overt adverse effects is BubR1 overexpression. Understanding how BubR1 exerts its beneficial effects at a modular level may provide important entry points for the design of small molecule-based therapies mimicking the effects of high BubR1.
Acknowledgments
We apologize for omitting citation of numerous papers as a result of space limitations. The authors thank Drs. Paul Galardy, Darren Baker, Hyun-Ja Nam, and Liviu Malureanu for helpful comments on this manuscript.
J.M. van Deursen is supported by the National Institutes of Health (grants CA96985, CA126828, and AG041122) and the Paul Glenn and Noaber Foundations.
Footnotes
Abbreviations used in this paper:
- APC
- anaphase-promoting complex
- CIN
- chromosomal instability
- MVA
- mosaic variegated aneuploidy
- Rb
- retinoblastoma
- S-CIN
- structural CIN
- W-CIN
- whole CIN
References
- Aguirre-Portolés C., Bird A.W., Hyman A., Cañamero M., Pérez de Castro I., Malumbres M. 2012. Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 72:1518–1528 10.1158/0008-5472.CAN-11-1971 [DOI] [PubMed] [Google Scholar]
- Amiel A., Goldzak G., Gaber E., Fejgin M.D. 2006. Molecular cytogenetic characteristics of Down syndrome newborns. J. Hum. Genet. 51:541–547 10.1007/s10038-006-0395-4 [DOI] [PubMed] [Google Scholar]
- Babu J.R., Jeganathan K.B., Baker D.J., Wu X., Kang-Decker N., van Deursen J.M. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341–353 10.1083/jcb.200211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., van Deursen J.M. 2010. Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle. 9:1711–1716 10.4161/cc.9.9.11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Malureanu L., Perez-Terzic C., Terzic A., van Deursen J.M. 2006. Early aging–associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172:529–540 10.1083/jcb.200507081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Perez-Terzic C., Jin F., Pitel K.S., Niederländer N.J., Jeganathan K., Yamada S., Reyes S., Rowe L., Hiddinga H.J., et al. 2008. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat. Cell Biol. 10:825–836 10.1038/ncb1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Jin F., Jeganathan K.B., van Deursen J.M. 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 16:475–486 10.1016/j.ccr.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Wijshake T., Tchkonia T., LeBrasseur N.K., Childs B.G., van de Sluis B., Kirkland J.L., van Deursen J.M. 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 479:232–236 10.1038/nature10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D.J., Dawlaty M.M., Wijshake T., Jeganathan K.B., Malureanu L., van Ree J.H., Crespo-Diaz R., Reyes S., Seaburg L., Shapiro V., et al. 2013. Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat. Cell Biol. 15:96–102 10.1038/ncb2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum S.F., Danilova O.V., Kaur P., Levy N.B., Compton D.A. 2011. Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin. Cancer Res. 17:7704–7711 10.1158/1078-0432.CCR-11-2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak N.J., Eklund A.C., Li Q., McClelland S.E., Endesfelder D., Tan P., Tan I.B., Richardson A.L., Szallasi Z., Swanton C. 2011. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71:3447–3452 10.1158/0008-5472.CAN-10-3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38:1043–1048 10.1038/ng1861 [DOI] [PubMed] [Google Scholar]
- Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11:753–760 10.1038/ncb1882 [DOI] [PubMed] [Google Scholar]
- Chesnokova V., Kovacs K., Castro A.V., Zonis S., Melmed S. 2005. Pituitary hypoplasia in Pttg-/- mice is protective for Rb+/- pituitary tumorigenesis. Mol. Endocrinol. 19:2371–2379 10.1210/me.2005-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.H., Wallace M.D., Abratte C., Southard T., Schimenti J.C. 2010. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet. 6:e1001110 10.1371/journal.pgen.1001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Baker J., Sage J. 2012. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun. 3:1244 10.1038/ncomms2254 [DOI] [PubMed] [Google Scholar]
- Coschi C.H., Dick F.A. 2012. Chromosome instability and deregulated proliferation: an unavoidable duo. Cell. Mol. Life Sci. 69:2009–2024 10.1007/s00018-011-0910-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D.O., Muse G.W., Van Dyke T. 2005. A dominant interfering Bub1 mutant is insufficient to induce or alter thymic tumorigenesis in vivo, even in a sensitized genetic background. Mol. Cell. Biol. 25:7796–7802 10.1128/MCB.25.17.7796-7802.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K., Ganem N.J., Dagher R., Lantermann A.B., Ivanova E.V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482:53–58 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko N.C., Giaccia A.J., Stringer J.R., Stambrook P.J. 1994. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc. Natl. Acad. Sci. USA. 91:5124–5128 10.1073/pnas.91.11.5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodríguez E., Sotillo R., Schvartzman J.M., Benezra R. 2008. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc. Natl. Acad. Sci. USA. 105:16719–16724 10.1073/pnas.0803504105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P., Rausch C., Rasnick D., Hehlmann R. 1998. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl. Acad. Sci. USA. 95:13692–13697 10.1073/pnas.95.23.13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijf P.H., Schultz N., Benezra R. 2013. Cancer cells preferentially lose small chromosomes. Int. J. Cancer. 132:2316–2326 10.1002/ijc.27924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Hickey R.D., Paulk N.K., Culberson A.J., Olson S.B., Finegold M.J., Grompe M. 2009. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 5:e1000385 10.1371/journal.pgen.1000385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. 2010. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 467:707–710 10.1038/nature09414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Hanlon Newell A.E., Bi W., Finegold M.J., Olson S.B., Beaudet A.L., Grompe M. 2012a. Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Invest. 122:3307–3315 10.1172/JCI64026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Hanlon Newell A.E., Smith L., Wilson E.M., Olson S.B., Thayer M.J., Strom S.C., Grompe M. 2012b. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 142:25–28 10.1053/j.gastro.2011.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F., Vezzoni P., Montagna C. 2011. Single-cell analysis of ploidy and centrosomes underscores the peculiarity of normal hepatocytes. PLoS ONE. 6:e26080 10.1371/journal.pone.0026080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F., Wang T., Vijg J., Montagna C. 2012. Chromosome-specific accumulation of aneuploidy in the aging mouse brain. Hum. Mol. Genet. 21:5246–5253 10.1093/hmg/dds375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Miranda G., Trakala M., Martín J., Escobar B., González A., Ghyselinck N.B., Ortega S., Cañamero M., Pérez de Castro I., Malumbres M. 2011. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development. 138:2661–2672 10.1242/dev.066381 [DOI] [PubMed] [Google Scholar]
- Finkel T., Serrano M., Blasco M.A. 2007. The common biology of cancer and ageing. Nature. 448:767–774 10.1038/nature05985 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Furge K., Koeman J., Dykema K., Su Y., Cutler M.L., Werts A., Haak P., Vande Woude G.F. 2007. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc. Natl. Acad. Sci. USA. 104:8995–9000 10.1073/pnas.0700631104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Castillo H., Vásquez-Velásquez A.I., Rivera H., Barros-Núñez P. 2008. Clinical and genetic heterogeneity in patients with mosaic variegated aneuploidy: delineation of clinical subtypes. Am. J. Med. Genet. A. 146A:1687–1695 10.1002/ajmg.a.32315 [DOI] [PubMed] [Google Scholar]
- Gentric G., Desdouets C., Celton-Morizur S. 2012. Hepatocytes polyploidization and cell cycle control in liver physiopathology. Int. J. Hepatol. 2012:282430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky G.V., Berezovska O., Glinskii A.B. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115:1503–1521 10.1172/JCI23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Gorenshteyn D., Burmeister M., Wong A.K., Schimenti J.C., Handel M.A., Bult C.J., Hibbs M.A., Troyanskaya O.G. 2012. Tissue-specific functional networks for prioritizing phenotype and disease genes. PLOS Comput. Biol. 8:e1002694 10.1371/journal.pcbi.1002694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D., Frescas D., Dorrello N.V., Peschiaroli A., Multani A.S., Cardozo T., Lasorella A., Iavarone A., Chang S., Hernando E., Pagano M. 2008. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 452:365–369 10.1038/nature06641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero A.A., Gamero M.C., Trachana V., Fütterer A., Pacios-Bras C., Díaz-Concha N.P., Cigudosa J.C., Martínez-A C., van Wely K.H. 2010. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc. Natl. Acad. Sci. USA. 107:4159–4164 10.1073/pnas.0912143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenberg A., Gerdes J.S., Jung K., Sander B., Gunawan B., Bock H.C., Liersch T., Brück W., Rohde V., Füzesi L. 2010. High chromosomal instability in brain metastases of colorectal carcinoma. Cancer Genet. Cytogenet. 198:47–51 10.1016/j.cancergencyto.2009.12.006 [DOI] [PubMed] [Google Scholar]
- Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Méhes K., Nash R., Robin N., et al. 2004. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 36:1159–1161 10.1038/ng1449 [DOI] [PubMed] [Google Scholar]
- Hanks S., Coleman K., Summersgill B., Messahel B., Williamson D., Pritchard-Jones K., Strefford J., Swansbury J., Plaja A., Shipley J., Rahman N. 2006. Comparative genomic hybridization and BUB1B mutation analyses in childhood cancers associated with mosaic variegated aneuploidy syndrome. Cancer Lett. 239:234–238 10.1016/j.canlet.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10:551–564 10.1038/nrg2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E., Nahlé Z., Juan G., Diaz-Rodriguez E., Alaminos M., Hemann M., Michel L., Mittal V., Gerald W., Benezra R., et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 430:797–802 10.1038/nature02820 [DOI] [PubMed] [Google Scholar]
- Höglund M., Gisselsson D., Säll T., Mitelman F. 2002. Coping with complexity. multivariate analysis of tumor karyotypes. Cancer Genet. Cytogenet. 135:103–109 10.1016/S0165-4608(01)00645-8 [DOI] [PubMed] [Google Scholar]
- Iwanaga Y., Chi Y.H., Miyazato A., Sheleg S., Haller K., Peloponese J.M., Jr, Li Y., Ward J.M., Benezra R., Jeang K.T. 2007. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 67:160–166 10.1158/0008-5472.CAN-06-3326 [DOI] [PubMed] [Google Scholar]
- Jacobs P.A., Brunton M., Court Brown W.M., Doll R., Goldstein H. 1963. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 197:1080–1081 10.1038/1971080a0 [DOI] [PubMed] [Google Scholar]
- Janssen A., van der Burg M., Szuhai K., Kops G.J., Medema R.H. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 333:1895–1898 10.1126/science.1210214 [DOI] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L., Compton D.A. 2012. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr. Biol. 22:638–644 10.1016/j.cub.2012.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Fowler K.J., Griffiths B., Earle E., Chow C.W., Jamsen K., Choo K.H. 2005. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer. 44:29–36 10.1002/gcc.20215 [DOI] [PubMed] [Google Scholar]
- Kamata T., Pritchard C. 2011. Mechanisms of aneuploidy induction by RAS and RAF oncogenes. Am. J. Cancer Res. 1:955–971 [PMC free article] [PubMed] [Google Scholar]
- Kawabata T., Luebben S.W., Yamaguchi S., Ilves I., Matise I., Buske T., Botchan M.R., Shima N. 2011. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell. 41:543–553 10.1016/j.molcel.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury M.A., Friedman B., McConnell M.J., Rehen S.K., Yang A.H., Kaushal D., Chun J. 2005. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc. Natl. Acad. Sci. USA. 102:6143–6147 10.1073/pnas.0408171102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury M.A., Yung Y.C., Peterson S.E., Westra J.W., Chun J. 2006. Aneuploidy in the normal and diseased brain. Cell. Mol. Life Sci. 63:2626–2641 10.1007/s00018-006-6169-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.J., Endesfelder D., Rowan A.J., Walther A., Birkbak N.J., Futreal P.A., Downward J., Szallasi Z., Tomlinson I.P., Howell M., et al. 2011. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71:1858–1870 10.1158/0008-5472.CAN-10-3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland S., Nagarajan P., Polyzos A., Thomas S., Samaan G., Donnell R., Marchetti F., Venkatachalam S. 2009. Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc. Natl. Acad. Sci. USA. 106:12776–12781 10.1073/pnas.0903075106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fang X., Wei Z., York J.P., Zhang P. 2009. Loss of spindle assembly checkpoint–mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 185:983–994 10.1083/jcb.200904020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fang X., Baker D.J., Guo L., Gao X., Wei Z., Han S., van Deursen J.M., Zhang P. 2010. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc. Natl. Acad. Sci. USA. 107:14188–14193 10.1073/pnas.1005960107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limwongse C., Schwartz S., Bocian M., Robin N.H. 1999. Child with mosaic variegated aneuploidy and embryonal rhabdomyosarcoma. Am. J. Med. Genet. 82:20–24 [DOI] [PubMed] [Google Scholar]
- Liu P., Erez A., Nagamani S.C., Dhar S.U., Kołodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A., et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 146:889–903 10.1016/j.cell.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Emanuele M.J., Li D., Creighton C.J., Schlabach M.R., Westbrook T.F., Wong K.K., Elledge S.J. 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 137:835–848 10.1016/j.cell.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., Dyson N.J. 2011. pRB, a tumor suppressor with a stabilizing presence. Trends Cell Biol. 21:433–441 10.1016/j.tcb.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura S., Matsumoto Y., Morishima K., Izumi H., Matsumoto H., Ito E., Tsutsui K., Kobayashi J., Tauchi H., Kajiwara Y., et al. 2006. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. A. 140:358–367 [DOI] [PubMed] [Google Scholar]
- McGranahan N., Burrell R.A., Endesfelder D., Novelli M.R., Swanton C. 2012. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 13:528–538 10.1038/embor.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V., Benezra R. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 409:355–359 10.1038/35053094 [DOI] [PubMed] [Google Scholar]
- Michor F., Iwasa Y., Vogelstein B., Lengauer C., Nowak M.A. 2005. Can chromosomal instability initiate tumorigenesis? Semin. Cancer Biol. 15:43–49 10.1016/j.semcancer.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Mitelman F. 2000. Recurrent chromosome aberrations in cancer. Mutat. Res. 462:247–253 10.1016/S1383-5742(00)00006-5 [DOI] [PubMed] [Google Scholar]
- Nagaoka S.I., Hassold T.J., Hunt P.A. 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13:493–504 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor R.M., Baker D.J., van Deursen J.M. 2013. Senescent cells: a novel therapeutic target for aging and age-related diseases. Clin. Pharmacol. Ther. 93:105–116 10.1038/clpt.2012.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozery-Flato M., Linhart C., Trakhtenbrot L., Izraeli S., Shamir R. 2011. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol. 12:R61 10.1186/gb-2011-12-6-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W.D., Saraf A., Florens L., Sanderson B.W., Hattem G.L., Li R. 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 468:321–325 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 13:566–579 10.1016/j.devcel.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Pfau S.J., Amon A. 2012. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 13:515–527 10.1038/embor.2012.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre R.V., Hoagland H.C. 1972. Age-associated aneuploidy: loss of Y chromosome from human bone marrow cells with aging. Cancer. 30:889–894 [DOI] [PubMed] [Google Scholar]
- Rehen S.K., McConnell M.J., Kaushal D., Kingsbury M.A., Yang A.H., Chun J. 2001. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl. Acad. Sci. USA. 98:13361–13366 10.1073/pnas.231487398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen S.K., Yung Y.C., McCreight M.P., Kaushal D., Yang A.H., Almeida B.S., Kingsbury M.A., Cabral K.M., McConnell M.J., Anliker B., et al. 2005. Constitutional aneuploidy in the normal human brain. J. Neurosci. 25:2176–2180 10.1523/JNEUROSCI.4560-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reish O., Brosh N., Gobazov R., Rosenblat M., Libman V., Mashevich M. 2006. Sporadic aneuploidy in PHA-stimulated lymphocytes of Turner’s syndrome patients. Chromosome Res. 14:527–534 10.1007/s10577-006-1050-9 [DOI] [PubMed] [Google Scholar]
- Reish O., Regev M., Kanesky A., Girafi S., Mashevich M. 2011. Sporadic aneuploidy in PHA-stimulated lymphocytes of trisomies 21, 18, and 13. Cytogenet. Genome Res. 133:184–189 10.1159/000323504 [DOI] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Carretero M., Martínez P., Drosopoulos W.C., Cañamero M., Schildkraut C.L., Blasco M.A., Losada A. 2012. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31:2076–2089 10.1038/emboj.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., van Ree J.H., van Deursen J.M. 2008. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 24:457–466 10.1016/j.tig.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., Jeganathan K.B., van Deursen J.M. 2011. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J. Cell Biol. 193:1049–1064 10.1083/jcb.201012035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., Jeganathan K.B., Malureanu L., Harrison A.M., van Deursen J.M. 2012. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J. Cell Biol. 199:931–949 10.1083/jcb.201205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F., Campisi J. 2011. Four faces of cellular senescence. J. Cell Biol. 192:547–556 10.1083/jcb.201009094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy A.V., Morgan-Lappe S.E., Zakula D., Vernetti L., Schurdak M., Packer J.C., Anderson M.G., Shirasawa S., Sasazuki T., Fesik S.W. 2007. Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol. Cancer Ther. 6:269–276 10.1158/1535-7163.MCT-06-0560 [DOI] [PubMed] [Google Scholar]
- Schliekelman M., Cowley D.O., O’Quinn R., Oliver T.G., Lu L., Salmon E.D., Van Dyke T. 2009. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 69:45–54 10.1158/0008-5472.CAN-07-6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J.M., Duijf P.H., Sotillo R., Coker C., Benezra R. 2011. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 19:701–714 10.1016/j.ccr.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer J.M., Blank H.M., Pfau S.J., Tange Y., George B.M., Humpton T.J., Brito I.L., Hiraoka Y., Niwa O., Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science. 333:1026–1030 10.1126/science.1206412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer J.M., Torres E.M., Dunham M.J., Amon A. 2012. Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. USA. 109:12644–12649 10.1073/pnas.1209227109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape K., Hanks S., Ruark E., Barros-Núñez P., Elliott A., Murray A., Lane A.H., Shannon N., Callier P., Chitayat D., et al. 2011. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat. Genet. 43:527–529 10.1038/ng.822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon D.A., Kim T., Diaz-Martinez L.A., Fair J., Elkahloun A.G., Harris B.T., Toretsky J.A., Rosenberg S.A., Shukla N., Ladanyi M., et al. 2011. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 333:1039–1043 10.1126/science.1203619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R., Hernando E., Díaz-Rodríguez E., Teruya-Feldstein J., Cordón-Cardo C., Lowe S.W., Benezra R. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 11:9–23 10.1016/j.ccr.2006.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Charles J., Hamilton M.L., Petes T.D. 2010. Meiotic chromosome segregation in triploid strains of Saccharomyces cerevisiae. Genetics. 186:537–550 10.1534/genetics.110.121533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 144:27–40 10.1016/j.cell.2010.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Bloom M.S., Solanki-Patil T., Smith S., Sipahimalani P., Li Z., Kofoed M., Ben-Aroya S., Myung K., Hieter P. 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7:e1002057 10.1371/journal.pgen.1002057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., van Osch M.H., Bos F.L., Hanks S., Rahman N., Kops G.J. 2010. Molecular causes for BUBR1 dysfunction in the human cancer predisposition syndrome mosaic variegated aneuploidy. Cancer Res. 70:4891–4900 10.1158/0008-5472.CAN-09-4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.C., Williams B.R., Siegel J.J., Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell. 144:499–512 10.1016/j.cell.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188:369–381 10.1083/jcb.200905057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E.M., Sokolsky T., Tucker C.M., Chan L.Y., Boselli M., Dunham M.J., Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 317:916–924 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- Upender M.B., Habermann J.K., McShane L.M., Korn E.L., Barrett J.C., Difilippantonio M.J., Ried T. 2004. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 64:6941–6949 10.1158/0008-5472.CAN-04-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree J.H., Jeganathan K.B., Malureanu L., van Deursen J.M. 2010. Overexpression of the E2 ubiquitin–conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 188:83–100 10.1083/jcb.200906147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H., Mayer B., Stemmann O., Nigg E.A. 2010. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 123:806–813 10.1242/jcs.058255 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yu R., Melmed S. 2001. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 15:1870–1879 10.1210/me.15.11.1870 [DOI] [PubMed] [Google Scholar]
- Warren C.D., Brady D.M., Johnston R.C., Hanna J.S., Hardwick K.G., Spencer F.A. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 13:3029–3041 10.1091/mbc.E02-04-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Wijshake T., Malureanu L.A., Baker D.J., Jeganathan K.B., van de Sluis B., van Deursen J.M. 2012. Reduced life- and healthspan in mice carrying a mono-allelic BubR1 MVA mutation. PLoS Genet. 8:e1003138 10.1371/journal.pgen.1003138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.R., Prabhu V.R., Hunter K.E., Glazier C.M., Whittaker C.A., Housman D.E., Amon A. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 322:703–709 10.1126/science.1160058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo R.A., Poon R.Y. 2004. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 18:1317–1330 10.1101/gad.1165204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjöblom T., Leary R.J., Shen D., Boca S.M., Barber T., Ptak J., et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science. 318:1108–1113 10.1126/science.1145720 [DOI] [PubMed] [Google Scholar]
- Xu H., Balakrishnan K., Malaterre J., Beasley M., Yan Y., Essers J., Appeldoorn E., Tomaszewski J.M., Vazquez M., Verschoor S., et al. 2010. Rad21-cohesin haploinsufficiency impedes DNA repair and enhances gastrointestinal radiosensitivity in mice. PLoS ONE. 5:e12112 10.1371/journal.pone.0012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.H., Kaushal D., Rehen S.K., Kriedt K., Kingsbury M.A., McConnell M.J., Chun J. 2003. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J. Neurosci. 23:10454–10462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Foreman O., Wigle D.A., Kosari F., Vasmatzis G., Salisbury J.L., van Deursen J., Galardy P.J. 2012. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Invest. 122:4362–4374 10.1172/JCI63084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Flesken-Nikitin A., Chen P.L., Lee W.H. 2002. Deficiency of Retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res. 62:2498–2502 [PubMed] [Google Scholar]
- Zhu J., Pavelka N., Bradford W.D., Rancati G., Li R. 2012. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet. 8:e1002719 10.1371/journal.pgen.1002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimonjic D., Brooks M.W., Popescu N., Weinberg R.A., Hahn W.C. 2001. Derivation of human tumor cells in vitro without widespread genomic instability. Cancer Res. 61:8838–8844 [PubMed] [Google Scholar]