Accurate cell division requires timely kinetochore disassembly, which is promoted by CDK phosphorylation of target genes, including CENP-T, and nuclear exclusion of the Ndc80 complex.
Abstract
Accurate chromosome segregation requires assembly of the multiprotein kinetochore complex. Prior work has identified more than 100 different kinetochore components in human cells. However, little is known about the regulatory processes that specify their assembly upon mitotic entry and disassembly at mitotic exit. In this paper, we used a live-cell imaging–based assay to quantify kinetochore disassembly kinetics and systematically analyze the role of potential regulatory mechanisms in controlling kinetochore assembly state. We find that kinetochore assembly and disassembly was driven primarily by mitotic phosphorylation downstream of cyclin-dependent kinase (CDK). In addition, we demonstrate that nuclear exclusion of the Ndc80 complex helped restrict kinetochore formation to mitosis. Combining constitutive CDK-dependent phosphorylation of CENP-T and forced nuclear localization of the Ndc80 complex partially prevented kinetochore disassembly at mitotic exit and led to chromosome segregation defects in subsequent divisions. In total, we find that the coordinated temporal regulation of outer kinetochore assembly is essential for accurate cell division.
Introduction
Chromosome segregation requires a physical attachment between centromere DNA and spindle microtubule polymers that is mediated by the multiprotein kinetochore (Cheeseman and Desai, 2008). A striking feature of the vertebrate kinetochore is its dramatic cell cycle–coupled reorganization. In vertebrates, a subset of DNA-proximal kinetochore proteins, termed the constitutive associated centromere network, is present at centromeres throughout the cell cycle to provide a platform for outer kinetochore assembly (Perpelescu and Fukagawa, 2011). However, the majority of the other ∼100 different kinetochore components only associate with centromeres during mitosis (Gascoigne and Cheeseman, 2011). Recent work has defined two parallel pathways for outer kinetochore assembly via the constitutive centromere proteins CENP-C and CENP-T (Hori et al., 2008; Carroll et al., 2010; Gascoigne et al., 2011; Guse et al., 2011; Nishino et al., 2013). However, how these pathways are regulated to restrict outer kinetochore formation to mitosis and the consequences of uncoupling kinetochore assembly from cell cycle progression remain unknown.
We have previously proposed several mechanisms by which kinetochore assembly could be temporally regulated (Gascoigne and Cheeseman, 2011): (a) Changes in protein stability or targeted degradation, (b) changes in the physical structure or forces that act on kinetochores throughout the cell cycle, such as interactions with microtubules, (c) changes in the subcellular localization of kinetochore proteins, or (d) cell cycle–coupled posttranslational modifications of kinetochore proteins. To define the contributions of these potential regulatory mechanisms to timely kinetochore assembly, we performed a systematic analysis of kinetochore assembly and disassembly in human cells. Using a live-cell imaging–based assay, we quantified the assembly and disassembly kinetics of 10 representative kinetochore proteins and used this assay to systematically evaluate the aforementioned regulatory mechanisms. Our results indicate that protein phosphorylation downstream of CDK and controlled nuclear localization coordinately control kinetochore assembly state. Disrupting these regulatory pathways to partially trap mitotic kinetochore assembly leads to defects in subsequent cell divisions.
Results and discussion
A live-cell assay to monitor kinetochore assembly dynamics
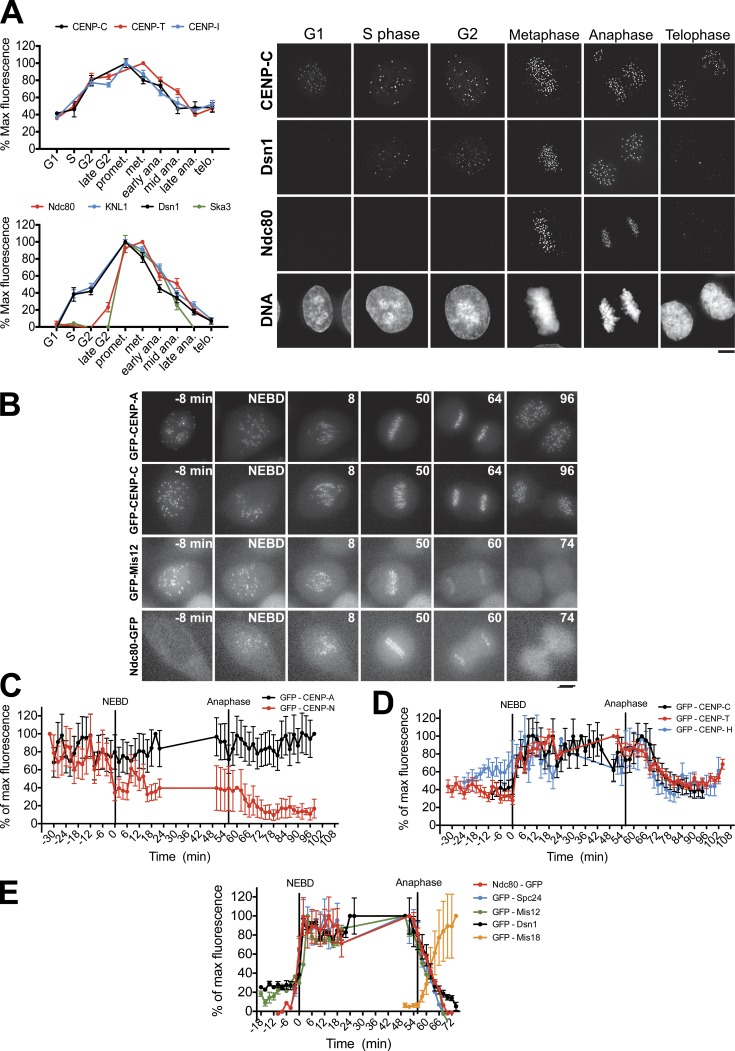
To probe the mechanisms that control kinetochore assembly state, we sought to monitor the kinetics of kinetochore assembly and disassembly in human cells. We first quantified the endogenous levels of representative kinetochore proteins during different cell cycle stages. For these experiments, we costained with anti-PCNA antibodies to define the specific cell cycle stage for interphase cells (Fig. 1 A). The constitutive associated centromere network proteins CENP-C, CENP-T, and CENP-I were present at centromeres throughout the cell cycle with levels peaking in G2 and mitosis, consistent with previous observations (McClelland et al., 2007; Eskat et al., 2012). In contrast, components of the KNL1–Mis12 complex–Ndc80 complex (KMN) network, which forms the core of the outer kinetochore–microtubule interface (Cheeseman et al., 2006), were absent from centromeres in G1. The Mis12 complex and KNL1 were recruited to centromeres during S phase, whereas the Ndc80 complex was recruited to kinetochores in late G2, consistent with previous fixed cell analysis (Cheeseman et al., 2008). All components of the KMN network disassembled from kinetochores at anaphase. The localization of the Ska1 complex mirrored that of Ndc80 (Fig. 1 A), consistent with their coordinated functions (Schmidt et al., 2012).
Figure 1.
Kinetochore proteins show distinct assembly and disassembly behaviors. (A, left) Quantification of centromere intensity in immunofluorescence images for the indicated proteins. promet., prometaphase; met., metaphase; ana., anaphase; telo., telophase. (right) Representative immunofluorescence images of kinetochore protein localization after detergent preextraction. (B) Images from time-lapse videos of cells expressing GFP-tagged proteins. Numbering indicates minutes relative to nuclear envelope breakdown (NEBD). (C–E) Quantification of centromeric GFP signals in time-lapse images. n ≥ 8 cells per protein. Error bars indicate SEM. Bars, 5 µM.
To understand the nature of these differing temporal localization patterns, we next developed a live-cell assay to monitor kinetochore protein localization with high temporal resolution. For this analysis, we generated clonal HeLa cell lines stably expressing GFP fusions for 10 representative inner and outer kinetochore proteins. We imaged each cell line and quantified GFP fluorescence at 2-min intervals starting 30 min before mitotic entry until 30 min after mitotic exit, during which the bulk of kinetochore assembly and disassembly occurs. All analyzed cell lines entered and exited mitosis with apparently normal kinetics during imaging. Using these imaging conditions, cells expressing GFP alone showed a fluorophore bleaching rate of ∼5% per hour, which would not significantly affect the assays described here (see Materials and methods) because of the short duration of these experiments (Fig. S1 A).
Quantification of kinetochore-localized GFP signals during mitosis revealed a striking diversity in the assembly and disassembly kinetics of both inner and outer kinetochore proteins (Fig. 1, B–D). Consistent with previous work (Jansen et al., 2007), the centromeric histone H3 variant CENP-A displayed constant levels at kinetochores from mitotic entry to exit (Fig. 1, B and C). Surprisingly, the constitutive centromere proteins CENP-C, CENP-T, and CENP-H showed a distinct assembly and disassembly behavior during mitosis, with levels at kinetochores increasing 50% in the first 10 min after nuclear envelope breakdown (NEBD) and decreasing equivalently beginning 10 min after anaphase onset (Fig. 1, B and D). In contrast, CENP-N displayed inverse assembly kinetics, with levels decreasing by ∼40% at NEBD and falling an additional 20% beginning 10 min after anaphase onset (Fig. 1, B and C). This is consistent with previous work indicating that CENP-N levels at centromeres are low during mitosis (McClelland et al., 2007; Hellwig et al., 2011). Thus, although these proteins are present at centromeres constitutively, their levels vary dramatically throughout the cell cycle.
We next analyzed the assembly and disassembly kinetics of the outer kinetochore KMN network. Consistent with our fixed cell analysis, the Mis12 complex components Dsn1 and Mis12 were detectable at centromeres before NEBD (Fig. 1, B and E). However, their levels were only ∼30% of those after NEBD, indicating that their assembly occurs primarily upon mitotic entry. In contrast, the Ndc80 complex components Ndc80/Hec1 and Spc25 were undetectable at centromeres until prophase. In each case, we observed identical assembly kinetics for distinct subunits of a stably associated complex. The disassembly of KMN network components from centromeres began 6 min before the first visible signs of anaphase chromosome movement, and the levels of all KMN network proteins were undetectable 20 min after anaphase onset (Fig. 1, B and E). The timing of KMN network disassembly precisely coincided with the recruitment of the CENP-A targeting factor Mis18-α to kinetochores (Fig. 1 E). We note that these results are consistent with similar changes in kinetochore assembly observed recently in Drosophila melanogaster (Venkei et al., 2012). Collectively, these data indicate that kinetochore protein localization changes dramatically throughout the cell cycle, particularly at time points immediately surrounding mitotic entry and exit.
Kinetochore disassembly occurs independently of protein degradation
Previously, we proposed several potential regulatory mechanisms that could control kinetochore assembly and disassembly (see Introduction; Gascoigne and Cheeseman, 2011). To identify the contribution of these molecular mechanisms to timely kinetochore formation, we sought to systematically assess the effects of perturbations to each potential mechanism on the kinetics of kinetochore assembly and disassembly.
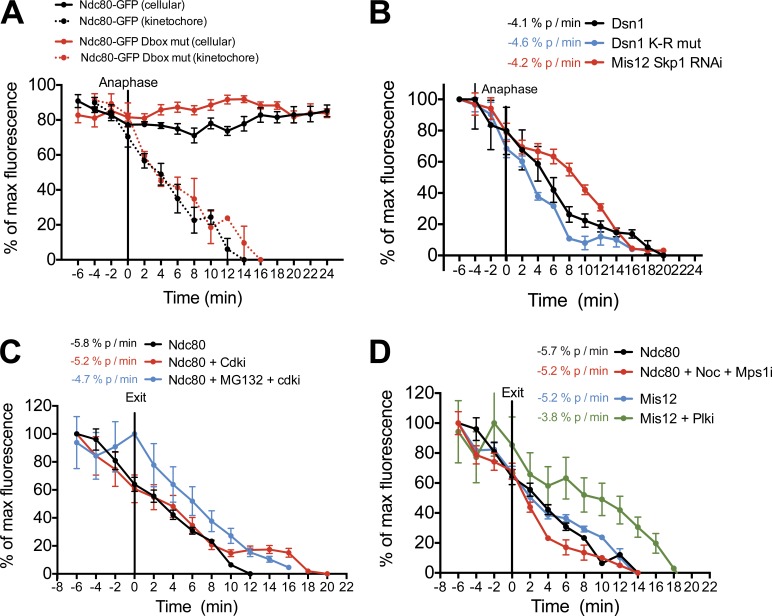
We first considered the contribution of protein degradation to kinetochore disassembly. Anaphase-promoting complex/cyclosome (APC/C)–mediated ubiquitination- and proteasome-dependent degradation of mitotic substrates is a key regulatory mechanism controlling the transition from metaphase to anaphase (Peters, 2006). Kinetochore disassembly begins at anaphase onset when APC/C activity is high. To test the requirements for kinetochore protein degradation to direct kinetochore disassembly, we focused on two outer kinetochore proteins for which ubiquitination has been reported previously (Kim et al., 2011): the Ndc80 complex subunit Ndc80/Hec1 and the Mis12 complex subunit Dsn1. Ndc80 contains a destruction box (D box) at residues 372–380 similar to those targeting substrates for APC/C—Cdc20-mediated degradation (Glotzer et al., 1991). A previous study indicated that removal of this sequence altered the function of Ndc80 (Li et al., 2011). However, we found that the kinetochore levels of either Ndc80-GFP or Ndc80-GFP mutated for the D box sequence (Ndc80ΔDbox; residues 372 and 375—RxxL to AxxA) disassembled from kinetochores during anaphase with similar kinetics (Fig. 2 A). Similarly, the cellular levels of both Ndc80-GFP and Ndc80ΔDbox-GFP remained unchanged from metaphase to G1 based on fluorescence imaging, indicating that APC-dependent degradation of Ndc80 does not drive its disassembly from kinetochores.
Figure 2.
Protein degradation and morphological changes at anaphase do not drive kinetochore disassembly. (A–D) Quantification of centromeric GFP intensities in cells exiting mitosis. At the onset of imaging, cells were treated with 5 µM flavopiridol (CDKi), 20 µM MG132, 200 ng/ml nocodazole (Noc), 10 µM BI2536 (Plki), or 2 µM AZ3146 (Mps1i) as indicated. n ≥ 8 cells, error bars represent SEM, and the rate of fluorescence change over time is indicated in percentage points per minute for each protein. mut, mutant.
We next tested the role of Mis12 complex degradation in timely kinetochore disassembly. The localization of the Mis12 complex to kinetochores has been reported to depend on the balanced action of the Hsp90 chaperone to stabilize the complex and the SCF (Skp1, Cullin, and F-box) ubiquitin ligase to target it for degradation (Davies and Kaplan, 2010). When the SCF complex component Skp1 was depleted by RNAi, we observed a subtle increase in levels of the Mis12 complex component Dsn1 at kinetochores, but no significant changes in its disassembly kinetics compared with controls (Fig. 2 B and Fig. S1 C). Similarly, a Dsn1 mutant in which all lysine residues were changed to arginine (Dsn1K-R), preventing its ubiquitination, localized to kinetochores (Fig. S1 C) and showed similar kinetochore disassembly kinetics to wild-type Dsn1 (Fig. 2 B).
Finally, to assess the global requirement for protein degradation for timely kinetochore disassembly, we monitored the levels of Ndc80-GFP and GFP-Mis12 at kinetochores in the presence of the proteasome inhibitor MG132 (Fig. 2 C and Fig. S2 B). To initiate mitotic exit in the absence of proteasome activity, we treated cells with the CDK inhibitor (CDKi) flavopiridol. The disassembly kinetics of Ndc80 and Mis12 from kinetochores were identical in the presence or absence of MG132, indicating that global proteasome-mediated protein degradation is not required for kinetochore disassembly (Fig. 2 C and Fig. S2 B). Collectively, these data strongly suggest that kinetochore disassembly is not driven by protein degradation at the metaphase to anaphase transition in human cells.
Kinetochore disassembly can occur independently of cytoskeletal changes at anaphase
We next sought to assess the contribution of changes in microtubule dynamics and cytokinesis to kinetochore disassembly. When kinetochore–microtubule interactions were prevented by treatment with the microtubule-depolymerizing drug nocodazole, Ndc80 disassembly from kinetochores occurred with normal kinetics in cells induced to exit mitosis by inhibition of the Mps1 spindle assembly checkpoint kinase (Fig. 2 D). These data indicate that kinetochore disassembly is not driven by changes in microtubule dynamics at mitotic exit or via a dynein-mediated stripping mechanism as has been proposed for proteins involved in spindle assembly checkpoint function (Howell et al., 2001). Similarly, when cytokinesis was prevented by inhibition of Plk1 (Polo-like kinase 1) activity during metaphase, disassembly of Mis12 was only subtly delayed (Fig. 2 D). Collectively, these data indicate that kinetochore disassembly can occur independently of the morphological changes that occur during metaphase and anaphase.
Interphase subcellular localization of kinetochore proteins contributes to kinetochore assembly dynamics
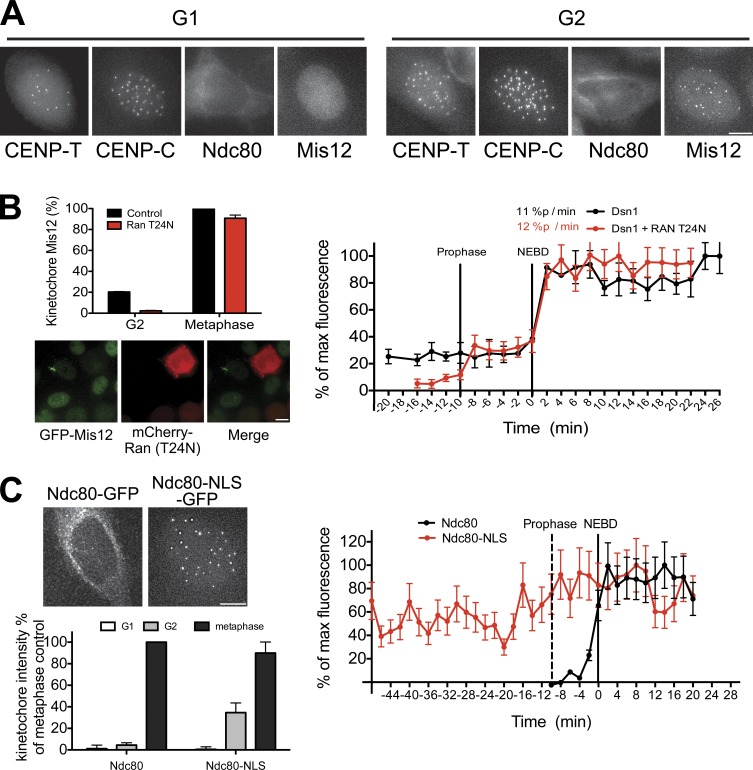
Kinetochore proteins show diverse subcellular localization during interphase. For example, the Mis12 complex resides in the nucleus throughout interphase even when not localized to kinetochores and is present at centromeres from S phase though anaphase (Fig. 3 A). In contrast, the Ndc80 complex is cytoplasmic and is excluded from the nucleus until prophase (Fig. 3 A). To assess the importance of the nuclear localization of the Mis12 complex before mitosis, we monitored the effects of disrupting nuclear import on kinetochore assembly. Expression of a dominant-negative mutant of the Ran GTPase (Ran T24N) inhibits nuclear import (Kornbluth et al., 1994). After expression of mCherry-Ran T24N in HeLa cells, GFP-Mis12 (Fig. 3 B) and GFP–CENP-T (Fig. S1 D) no longer localized to centromeres in G2, indicating that nuclear import is required for their premitotic loading. In contrast, the levels of Mis12 and CENP-T at metaphase kinetochores were normal in the presence of Ran T24N, indicating that kinetochore assembly can occur after NEBD even in the absence of premitotic loading. Intriguingly, in the presence of Ran T24N, GFP-Mis12 exhibited biphasic loading kinetics: ∼20% of Mis12 loaded onto kinetochores during prophase and the remaining 80% loaded at NEBD. This suggests that additional factors act immediately before mitosis to contribute to maximal kinetochore assembly.
Figure 3.
Interphase localization of kinetochore proteins contributes to kinetochore assembly dynamics. (A) GFP-tagged protein localization for the indicated proteins and cell cycle stages. (B, bottom left) Images of cells expressing GFP-Mis12 and mCherry-Ran-T24N. (top) Quantification of centromeric GFP-Mis12. (C, top left) Images of Ndc80-GFP or Ndc80-NLS GFP in G2 cells. (bottom) Quantification of centromeric GFP intensity. (B and C, right) Graphs show quantification of centromeric GFP signals in cells entering mitosis. Bars, 10 µm. n ≥ 10 cells. Error bars represent SDs.
To test the role of Ndc80 complex nuclear exclusion in timely kinetochore assembly, we artificially targeted Ndc80-GFP to the nucleus throughout the cell cycle using a fusion to the SV40 NLS (Fig. 3 C). Ndc80-NLS-GFP localized to kinetochores during mitosis, indicating that it was correctly incorporated into the Ndc80 complex. Both Ndc80-GFP and Ndc80-NLS-GFP were absent from centromeres in G1 (Fig. 3 C). However, Ndc80-NLS-GFP localized prematurely to centromeres during G2, indicating that the centromere is competent to recruit Ndc80 at this time. Thus, nuclear exclusion prevents the premature loading of the Ndc80 complex onto kinetochores.
CDK-mediated phosphoregulation of kinetochore assembly
In Fig. 3, we demonstrated that artificially targeting Ndc80 to the nucleus induces premature Ndc80 complex localization to kinetochores during interphase. However, loading of Ndc80-NLS-GFP at centromeres during G2 was only 40% of that observed at metaphase, indicating that additional factors control mitotic kinetochore assembly (Fig. 3). Several mitotic kinases are known to play crucial roles in kinetochore function (Malumbres, 2011). To broadly assess the role of mitotic phosphorylation in kinetochore assembly, we monitored kinetochore protein localization after individual inhibition of the mitotic kinases Aurora B, Plk1, Mps1, or CDK1 (Fig. S2 A). Inhibition of Aurora B, Plk1, or Mps1 caused only minor effects on the kinetochore localization of CENP-T, CENP-C, CENP-I, Dsn1, KNL1, Ndc80, or Ska3 (Fig. S2 A). However, we observed a strong reduction in the localization of multiple kinetochore components after inhibition of CDK activity (Fig. S2 A).
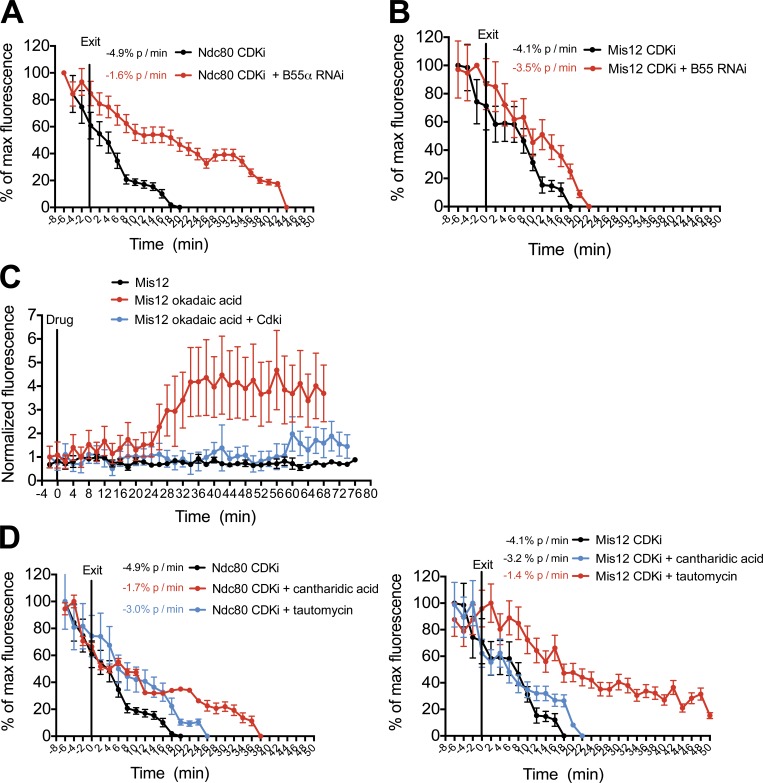
To test the role of CDK activity in kinetochore assembly and disassembly, we next monitored outer kinetochore disassembly kinetics after inhibition of CDK activity using the CDKi flavopiridol in mitotic HeLa cells expressing Ndc80-GFP or GFP-Mis12. Disassembly of Ndc80 or Mis12 from kinetochores occurred within 5 min of treatment with 5 µM flavopiridol and occurred with similar kinetics to cells exiting an unperturbed mitosis. This indicates that inhibition of CDK activity alone is sufficient for outer kinetochore disassembly (Fig. 4, A and B). Interestingly, when CDK activity was only partially inhibited by treatment with a low dose of flavopiridol (2 µM), Ndc80 disassembly occurred with slower kinetics than in unperturbed cells (Fig. S2 B). Similarly, when we induced mitotic exit by CDK inhibition in the presence of the general phosphatase inhibitor okadaic acid, the disassembly kinetics of Ndc80 and Mis12 were also slowed, indicating a high sensitivity to the levels of CDK-dependent phosphorylation (Fig. S2 B). When interphase cells were treated with okadaic acid, we observed a rapid assembly of GFP-Mis12 and Ndc80-NLS-GFP at kinetochores in G2 cells (Fig. 4 C and Fig. S2 C). This okadaic acid–induced assembly was prevented by pretreatment with CDKi.
Figure 4.
CDK activity drives kinetochore assembly and disassembly. (A–E) Quantification of centromeric GFP intensity in cells exiting (A, B, D, and E) or entering (C) mitosis. Cells were imaged 48 h after depletion of PP2R2A–B55-α and immediately after treatment with 5 µM flavopiridol (CDKi; A and B), 30 min after treatment with 5 µM flavopiridol (CDKi) and immediately after treatment with 1 µM okadaic acid (C), and 30 min after treatment with 50 µM cantharidic acid or 3 µM tautomycin and immediately after treatment with 5 µM flavopiridol (CDKi; D and E). n ≥ 8 cells. Error bars represent SEM.
The observation that outer kinetochore assembly is acutely sensitive to CDK activity suggests that the action of counteracting phosphatases may be crucial for timely disassembly of these proteins at mitotic exit. The PP2A (protein phosphatase 2A) and its regulatory subunit B55-α have recently been identified as critical factors for the dephosphorylation of CDK1 substrates during anaphase (Schmitz et al., 2010). When we induced mitotic exit by CDK inhibition in cells depleted of B55-α, the kinetics of Ndc80 disassembly from kinetochores were significantly reduced (Fig. 4 A and Fig. S2 D), indicating a crucial role for PP2A–B55-α in kinetochore disassembly. In contrast, Mis12 complex disassembly was only subtly affected by B55-α depletion (Fig. 4 B). These data hint at differing sensitivities to the CDK kinase/phosphatase balance for the assembly/disassembly of different kinetochore subunits. Consistent with this, treatment of HeLa cells with diverse phosphatase inhibitory compounds had differing effects on the disassembly of kinetochore subcomplexes (Fig. 4, D and E). Collectively, these data indicate that the kinetochore assembly state is highly sensitive to the balance of CDK kinase and phosphatase activity within the cell.
Phosphorylation of CENP-T by CDK controls assembly of a functional pool of Ndc80 at kinetochores
The observation that outer kinetochore disassembly is exquisitely sensitive to CDK activity suggests that kinetochore proteins could be direct CDK substrates, with phosphorylation controlling protein interactions at kinetochores. We have previously reported that CENP-T can interact directly with the Ndc80 complex via its N terminus, an interaction that is enhanced by phosphorylation of CENP-T by CDK (Gascoigne et al., 2011; Nishino et al., 2013). Biochemical and structural analysis of CENP-T has demonstrated that mutation of these phosphorylated residues to aspartic acid effectively mimics phosphorylation of the protein (Nishino et al., 2013). Surprisingly, although preventing phosphorylation of CENP-T blocked recruitment of the Ndc80 complex to kinetochores (Gascoigne et al., 2011), phosphomimetic mutation of 10 CDK sites in CENP-T (CENP-TSD) had only a minor effect on kinetochore disassembly (Fig. 5, A and B). When monitored in our live-cell assay, the disassembly kinetics of Ndc80-GFP in the presence of CENP-TSD were similar to control cells, with only a small additional pool of Ndc80 (20%) persisting 10 min after anaphase onset. Disassembly of the Mis12 complex subunit GFP-Dsn1 was also unaffected in CENP-TSD mutants (Fig. 5 B).
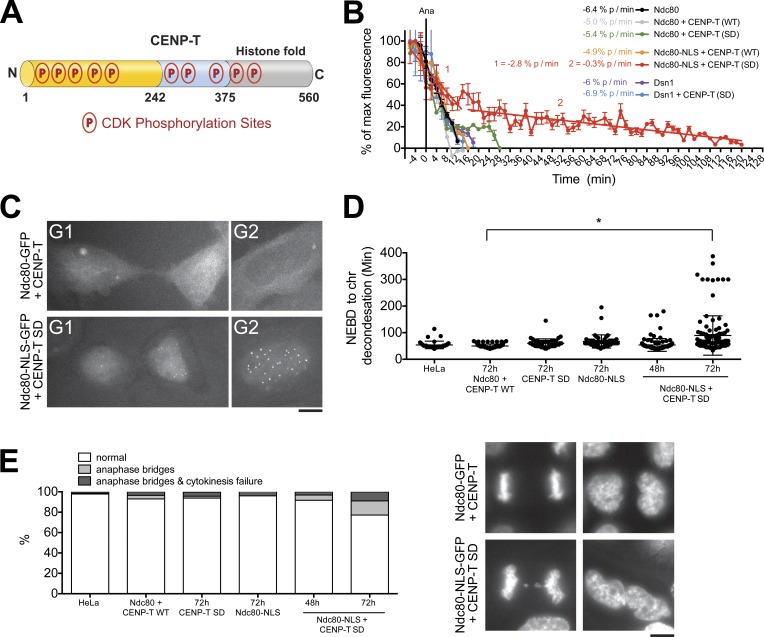
Figure 5.
CENP-T phosphorylation controls assembly of the Ndc80 complex at kinetochores. (A) Diagram of CDK-directed phosphorylation sites in CENP-T. The Ndc80 interaction region is shown in yellow. N, N terminus; C, C terminus. (B) Quantification of centromeric GFP intensity in cells exiting mitosis for cells expressing the indicated RNAi-resistant fluorescently tagged proteins after depletion of the endogenous proteins. n ≥ 8 cells. Error bars represent SEM. (C) Representative images of cells expressing the indicated GFP-tagged protein. (D and E) Quantification of chromosome segregation defects in cells treated as in B showing the time from nuclear envelope breakdown (NEBD) to chromatin (chr) decondensation (D) and percentage of cells showing anaphase abnormalities (E). Horizontal lines indicate the means. n ≥ 100 cells per condition. Asterisk indicates a significant difference as determined by the Mann–Whitney U test, P < 0.005. WT, wild type. Bars, 5 µm.
In addition to CENP-T dephosphorylation, the aforementioned results suggested that exclusion of Ndc80 from the newly forming nucleus during anaphase might also be important for kinetochore disassembly. To test this, we prevented the nuclear exclusion of Ndc80 in the presence of constitutively phosphorylated CENP-T by coexpression of Ndc80-NLS-GFP and mCherry–CENP-TSD after RNAi-based depletion of the endogenous proteins. Ndc80-NLS-GFP disassembled from kinetochores with similar kinetics to Ndc80-GFP in the presence of wild-type CENP-T. Importantly, in the presence of CENP-TSD, the disassembly of Ndc80-NLS-GFP was greatly slowed and showed biphasic kinetics. In Ndc80-NLS-GFP/CENP-TSD double mutants, 40% of Ndc80 remained at kinetochores 40 min after anaphase onset, and Ndc80 was still detectable at centromeres 2 h after anaphase (Fig. 5, B and C). In contrast, under these conditions, disassembly of the Mis12 complex was unaffected (Fig. S3 A), suggesting that there are multiple independent pathways for kinetochore disassembly.
We next sought to test the consequences of this delayed outer kinetochore disassembly. Replacement of CENP-T with GFP-CENP-TSD alone or Ndc80 with Ndc80-NLS-GFP alone had no significant effect on chromosome segregation. In addition, when both Ndc80 and CENP-T proteins were replaced with Ndc80-NLS-GFP and mCherry-CENP-TSD, no phenotype was observable 48 h after siRNA addition. This is in stark contrast to replacement of CENP-T with GFP-CENP-TSA (phosphoinhibitory), in which a highly penetrant chromosome alignment defect was observed 48 h after depletion of the endogenous protein (Gascoigne et al., 2011). However, in subsequent cell divisions (72 h time point), Ndc80-NLS-GFP/mCherry–CENP-TSD cells spent twice as long in mitosis as control cells, with 25% of cells displaying chromosome bridges at anaphase (Fig. 5, D and E). In metaphase Ndc80-NLS/CENP-TSD cells, the levels of the representative kinetochore proteins CENP-A and Dsn1 were normal in the subsequent cell division after the failure of full kinetochore disassembly (72 h; Fig. S3 C). This suggests that the observed chromosome segregation defects are unlikely to be caused by abnormalities in kinetochore composition. In addition, the failure of Ndc80 to fully disassemble from centromeres did not lead to detectable persistent microtubule interactions at telophase or prevent nuclear envelope reformation (Fig. S3 B). Collectively, these data indicate a crucial role for CDK-dependent phosphorylation of CENP-T in the direct recruitment of the Ndc80 complex to mitotic kinetochores and suggest that removal of this phosphorylation is essential for full kinetochore disassembly and accurate chromosome segregation in subsequent divisions.
Here, we systematically tested the contributions of multiple cellular activities to timely kinetochore disassembly. We find that protein degradation and cell cycle–coupled cytoskeletal changes do not play critical roles in the regulation of the kinetochore assembly state in human cells. Instead, we find that CDK-dependent phosphorylation of kinetochore substrates, coupled with exclusion of the Ndc80 complex from the nucleus, directs the kinetics of kinetochore assembly and disassembly. Importantly, disrupting timely kinetochore disassembly prevents accurate chromosome segregation in subsequent cell cycles. As kinetochore composition appears normal in these subsequent mitoses, we speculate that chromosome bridges observed in such cells arise from problems with centromere replication in the presence of constitutively localized outer kinetochore components. Although this work defines the basic parameters that control kinetochore assembly state, our results suggest that additional aspects of kinetochore assembly state are also regulated in addition to CENP-T phosphorylation and Ndc80 subcellular localization. For example, the differential sensitivity of the Ndc80 and Mis12 complexes to phosphatase inhibition suggests that multiple regulatory pathways may act to control kinetochore assembly during mitotic entry and exit. Defining the other interactions and properties that coordinately control kinetochore assembly is an exciting challenge for future work.
Materials and methods
Cell culture and transfection
Human cell lines were maintained in DMEM supplemented with 100 U/ml streptomycin, 100 U/ml penicillin, 2 mM glutamine, and 10% (vol/vol) fetal calf serum. All cells were cultured at 37°C with 5% CO2. HeLa LacZeo/TO cells (a gift from S. Taylor, University of Manchester, Manchester, England, UK) were maintained in 2 µg/ml Blasticidin and 200 µg/ml Zeocin. Cells expressing tetracycline-inducible GFPLAP or mCherryLAP fusions were generated by Flp recombinase–mediated integration of a pCDNA5-FRT-TO–based plasmid (Invitrogen). Cells were then maintained in 2 µg/ml Blasticidin and 400 µg/ml Hygromycin B. Clonal cell lines constitutively expressing GFPLAP or mCherryLAP fusions were generated using retroviral infection of HeLa cells with a pBABE-blast– or pBABE-Puromycin–based vector. See Table S1 for a list of cell lines used in this study. Transient transfection of cells with plasmids encoding mCherry-Ran T24N and mCherry-Dsn1K-R was performed with transfection reagent (Effectene; QIAGEN) according to the manufacturer’s instructions.
Codon-optimized and RNAi-resistant CENP-T (Mr. Gene) was used in all experiments. RNAi-resistant Ndc80 was generated by site-directed mutation (Agilent Technologies) of siRNA-targeting nucleotides in the human coding sequence. siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were analyzed 48 h after siRNA transfection unless stated otherwise. Nontargeting control siRNAs and gene-specific siRNAs were obtained from Thermo Fisher Scientific. Single siRNAs were used against CENP-T (5′-CGGAGAGCCCUGCUUGAAA-3′), Ndc80/Hec1 (5′-GAAGUUCAAAAGCUGGAUGAUCUU-3′), and PP2R2A–B55-α (5′-CUGCAGAUGAUUUGCGGAUUAUU-3′; Schmitz et al., 2010), and a pool of siRNAs was used against Skp1A (5′-CUACUUGCAUGUAAAGAAU-3′, 5′-GGAGAAAUGUAACUGGACA-3′, 5′-CUAGUAUGAUGGAAAGUUU-3′, and 5′-CGCAAGACCUUCAAUAUCA-3′).
Immunofluorescence and microscopy
Where indicated, cells were incubated with 20 µM MG132 (Sigma-Aldrich), 5 µM Flavopiridol (CDKi; Sigma-Aldrich), 10 µM BI2536 (Plk1 inhibitor [Plki]; Tocris Bioscience), 2 µM AZ3146 (Mps1i; Tocris Bioscience), 2 µM ZM447439 (Aurora B inhibitor; Tocris Bioscience), or 0.66 µM nocodazole before fixation or live-cell imaging. For immunofluorescence analysis, cells were grown on glass coverslips. Where indicated, cytoplasmic contents were preextracted before fixation by incubation for 2 min in PBS + 1% (vol/vol) Triton X-100. Cells were fixed by incubation for 15 min in PBS + 4% (vol/vol) formaldehyde before incubation for 30 min in TBS + 3% (wt/vol) BSA + 0.1% (vol/vol) Triton X-100. Antibodies used for staining are listed in Table S2. Cy2-, Cy3-, and Cy5-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories, Inc. All antibodies were diluted in TBS + 3% BSA and incubated with cells for 1 h at room temperature. DNA was visualized by a 5-min incubation with 10 µg/ml Hoechst in PBS.
Images were acquired on a deconvolution microscope (DeltaVision Core; Applied Precision) equipped with a charge-coupled device camera (CoolSNAP HQ2; Photometrics). For fixed-cell analysis, 40 z sections were acquired at 0.2-µm steps using a 60×, 1.3 NA U-Plan Apochromat objective (Olympus). Images were acquired at room temperature through glycerol-based mounting media. Cy2, Cy3, Cy5, or Hoechst florescence was observed using appropriate filters. Images were deconvolved using 10 cycles of enhanced ratio deconvolution on DeltaVision software (Applied Precision).
For time-lapse imaging, cells were imaged in CO2-independent media (Invitrogen) at 37°C. Images were acquired every 2 min using six z sections at 0.7-µm intervals using a 40× U-Plan Apochromat/340 NA objective (Olympus). GFP or mCherry fluorescence was observed using appropriate filters. To quantify fluorescent intensity, maximum intensity projections were generated, and individual kinetochores were analyzed at each frame using MetaMorph software (Molecular Devices). Integrated fluorescence intensity was measured in a 7 × 7–pixel region containing a kinetochore. Background was subtracted by subtracting from this the integrated intensity from a 7 × 7–pixel region of cytoplasm in the same cell. At least 10 kinetochores were analyzed per cell. To visualize chromosome movement during live-cell imaging, cells were incubated for 30 min in 1 µg/ml Hoechst. Media were replaced with CO2-independent media without Hoechst before imaging.
Online supplemental material
Fig. S1 assesses factors controlling kinetochore assembly kinetics. Fig. S2 shows that inhibition of CDK, but not other mitotic kinases, affects kinetochore assembly. Fig. S3 shows that inhibition of Ndc80 complex disassembly does not cause immediate mitotic defects. Table S1 shows cell lines used in this study. Table S2 shows antibodies used to visualize kinetochore proteins. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201301006/DC1.
Supplementary Material
Acknowledgments
We thank Mike Lampson, Geert Kops, Tatsuo Fukagawa, and members of the Cheeseman laboratory for helpful discussions and critical reading of the manuscript.
This work was supported by awards to I.M. Cheeseman from the Searle Scholars Program and the Leukemia & Lymphoma Society and a grant from the National Institutes of Health/National Institute of General Medical Sciences to I.M. Cheeseman (GM088313). K.E. Gascoigne is supported by a Leukemia & Lymphoma Society Special Fellows award.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- CDKi
- CDK inhibitor
- KMN
- KNL1–Mis12 complex–Ndc80 complex
- NEBD
- nuclear envelope breakdown
- Plki
- Plk1 inhibitor
References
- Carroll C.W., Milks K.J., Straight A.F. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A.E., Kaplan K.B. 2010. Hsp90–Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore–microtubule binding sites. J. Cell Biol. 189:261–274 10.1083/jcb.200910036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskat A., Deng W., Hofmeister A., Rudolphi S., Emmerth S., Hellwig D., Ulbricht T., Döring V., Bancroft J.M., McAinsh A.D., et al. 2012. Step-wise assembly, maturation and dynamic behavior of the human CENP-P/O/R/Q/U kinetochore sub-complex. PLoS ONE. 7:e44717 10.1371/journal.pone.0044717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Cheeseman I.M. 2011. Kinetochore assembly: if you build it, they will come. Curr. Opin. Cell Biol. 23:102–108 10.1016/j.ceb.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138 10.1038/349132a0 [DOI] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., Straight A.F. 2011. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 477:354–358 10.1038/nature10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig D., Emmerth S., Ulbricht T., Döring V., Hoischen C., Martin R., Samora C.P., McAinsh A.D., Carroll C.W., Straight A.F., et al. 2011. Dynamics of CENP-N kinetochore binding during the cell cycle. J. Cell Sci. 124:3871–3883 10.1242/jcs.088625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.-H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L.E.T., Black B.E., Foltz D.R., Cleveland D.W. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., et al. 2011. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 44:325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S., Dasso M., Newport J. 1994. Evidence for a dual role for TC4 protein in regulating nuclear structure and cell cycle progression. J. Cell Biol. 125:705–719 10.1083/jcb.125.4.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhou Y., Wang G.F., Liao S.C., Ke Y.B., Wu W., Li X.H., Zhang R.L., Fu Y.C. 2011. Anaphase-promoting complex/cyclosome controls HEC1 stability. Cell Prolif. 44:1–9 10.1111/j.1365-2184.2010.00712.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. 2011. Physiological relevance of cell cycle kinases. Physiol. Rev. 91:973–1007 10.1152/physrev.00025.2010 [DOI] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., Meraldi P. 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 26:5033–5047 10.1038/sj.emboj.7601927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., Fukagawa T. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T. 2011. The ABCs of CENPs. Chromosoma. 120:425–446 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M.H., Held M., Janssens V., Hutchins J.R., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A.I., Poser I., et al. 2010. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12:886–893 10.1038/ncb2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkei Z., Przewloka M.R., Ladak Y., Albadri S., Sossick A., Juhasz G., Novák B., Glover D.M. 2012. Spatiotemporal dynamics of Spc105 regulates the assembly of the Drosophila kinetochore. Open Biol. 2:110032 10.1098/rsob.110032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.