To increase the efficiency of aggrephagy, specific scaffold proteins such as EPG-7 endow cargo specificity and link cargo–receptor complexes to the autophagic machinery.
Abstract
The mechanism by which protein aggregates are selectively degraded by autophagy is poorly understood. Previous studies show that a family of Atg8-interacting proteins function as receptors linking specific cargoes to the autophagic machinery. Here we demonstrate that during Caenorhabditis elegans embryogenesis, epg-7 functions as a scaffold protein mediating autophagic degradation of several protein aggregates, including aggregates of the p62 homologue SQST-1, but has little effect on other autophagy-regulated processes. EPG-7 self-oligomerizes and is degraded by autophagy independently of SQST-1. SQST-1 directly interacts with EPG-7 and colocalizes with EPG-7 aggregates in autophagy mutants. Mutations in epg-7 impair association of SQST-1 aggregates with LGG-1/Atg8 puncta. EPG-7 interacts with multiple ATG proteins and colocalizes with ATG-9 puncta in various autophagy mutants. Unlike core autophagy genes, epg-7 is dispensable for starvation-induced autophagic degradation of substrate aggregates. Our results indicate that under physiological conditions a scaffold protein endows cargo specificity and also elevates degradation efficiency by linking the cargo–receptor complex with the autophagic machinery.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a lysosome-mediated degradation process that involves the formation of a closed double-membrane autophagosome and its subsequent fusion with lysosomes for degradation (Xie and Klionsky, 2007; Nakatogawa et al., 2009). A group of Atg proteins has been identified in yeast, members of which form distinct complexes that act at different steps of autophagosome formation. These Atg protein complexes include the Atg1 serine–threonine kinase complex, the Vps34 class III PtdIns(3)P kinase complex, the Atg2–Atg18 complex for Atg9-recycling, and the two ubiquitin-like conjugation systems (Atg8–phosphatidylethanolamine conjugates and Atg5–Atg12 conjugates; Nakatogawa et al., 2009). In yeast, all Atg proteins are recruited in a hierarchical order to the preautophagosomal structure (PAS), where autophagosomes are generated (Nakatogawa et al., 2009). The multi-membrane spanning protein Atg9 concentrates in vesicles and tubules that traffic to the PAS and trigger the hierarchical recruitment of other Atg proteins as well as supplying the membrane for autophagosome formation (Suzuki et al., 2007; He et al., 2008; Mari et al., 2010). The autophagy process in higher eukaryotes involves more complex membrane dynamics, and requires the concerted action of highly conserved Atg proteins and also metazoan-specific autophagy proteins (Longatti and Tooze, 2009; Yang and Klionsky, 2010; Tian et al., 2010). The endoplasmic reticulum (ER), Golgi apparatus, endosomes, plasma membrane, and Atg9-positive vesicles have been shown to contribute to autophagosomal membranes in mammalian cells (Young et al., 2006; Axe et al., 2008; Ravikumar et al., 2010; Orsi et al., 2012). Among these membrane sources, PtdIns(3)P-enriched subdomains of the ER, called omegasomes, function as platforms for recruiting Atg proteins and act as cradles for generating autophagosomes (Axe et al., 2008). Atg9-positive vesicles dynamically and transiently interact with DFCP1-positive structures (Orsi et al., 2012). How Atg proteins act coordinately in autophagosome formation remains largely unknown.
Autophagy acts as a quality control system by selectively removing protein aggregates (a process known as aggrephagy) and damaged organelles. A family of Atg8/LC3 (mammalian Atg8 homologue)-interacting proteins act as receptors that mediate delivery of specific cargoes to the autophagic machinery via Atg8/LC3 binding (Noda et al., 2010; Johansen and Lamark, 2011). Among them, p62/sequestosome 1 (SQSTM1) acts as a cargo receptor for accumulation and autophagic degradation of ubiquitinated protein aggregates (Bjørkøy et al., 2005; Komatsu et al., 2007; Pankiv et al., 2007). p62 contains a self-polymerization PB1 domain, a conserved LC3-interacting region (LIR), and a ubiquitin-associating (UBA) domain. p62 itself is also a selective autophagy substrate (Bjørkøy et al., 2005; Pankiv et al., 2007). Oligomerization-defective and LIR motif mutants of p62 are severely inhibited in their ability to be degraded by autophagy (Ichimura et al., 2008). Under normal physiological conditions, in which autophagy occurs at a basal level, the interaction between the receptor and Atg8 appears not to be sufficient for the degradation of the cargo–receptor complex. During Caenorhabditis elegans embryogenesis, the germline-specific P granule components PGL-1 and PGL-3 are degraded by autophagy in somatic cells (Zhang et al., 2009). The self-oligomerization protein SEPA-1 acts as the receptor for the formation of PGL-1 and PGL-3 granules and also for their autophagic degradation. SEPA-1 directly binds to LGG-1 (the C. elegans Atg8 homologue) and is itself also removed by autophagy during embryogenesis (Zhang et al., 2009). Degradation of cargo (PGL-1 and PGL-3)–receptor (SEPA-1) complexes (known as PGL granules) also depends on EPG-2, loss of function of which results in separate localization of PGL granules and LGG-1–labeled structures (Tian et al., 2010). Very little is known about the mechanism by which protein aggregates such as p62 bodies are selectively recognized under physiological conditions and how autophagosomal membranes are formed closely surrounding protein aggregates.
The C. elegans SeQueSTosome-related protein, SQST-1, is degraded by autophagy (Tian et al., 2010). Here we identified that epg-7 is essential for degradation of several protein aggregates, including SQST-1–containing aggregates, during C. elegans embryogenesis. EPG-7, however, is dispensable for starvation-induced autophagic degradation of these protein aggregates. EPG-7 directly interacts with SQST-1 and also with multiple Atg proteins, including ATG-9, and colocalizes with ATG-9 puncta in various autophagy mutants. Our results suggest that under physiological conditions, EPG-7 functions as a platform for recruiting Atg proteins for autophagosome formation and thus augments the autophagic degradation efficiency of protein substrates.
Results
Mutations in epg-7 cause defective degradation of SQST-1–containing protein aggregates
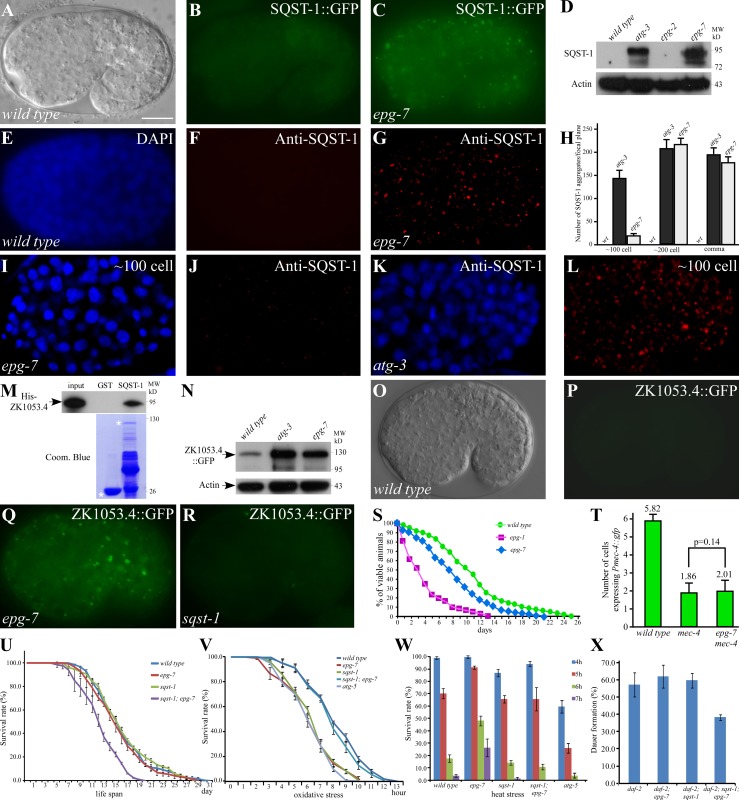
In wild-type embryos, sqst-1 is very weakly expressed and diffusely localized in the cytoplasm, whereas in autophagy mutants it is expressed at dramatically increased levels and accumulates into a large number of aggregates (Fig. 1, A, B, and D; Tian et al., 2010). Mutations in epg-7 (ectopic PGL granules or p62 aggregates) were identified from screens that caused defective degradation of SQST-1::GFP (Fig. 1 C). In epg-7 mutants, SQST-1::GFP and endogenous SQST-1 were detected at dramatically elevated levels and accumulated into a large number of aggregates during embryogenesis (Fig. 1, C–H; Fig. S1 A). sqst-1 mRNA levels remained unchanged in epg-7 mutants (Fig. S1 B). Compared with atg-3 mutants, the number of SQST-1 aggregates in epg-7 mutants was much less before the ∼100-cell stage, but was similar at the ∼200-cell and comma stages and onwards during embryogenesis (Fig. 1, H–L). Thus, epg-7 is required for removal of SQST-1 at specific embryonic developmental stages.
Figure 1.
Mutations in epg-7 cause defective degradation of SQST-1 aggregates. (A and B) In wild-type embryos, SQST-1::GFP is weakly expressed and diffusely localized in the cytoplasm. (A) DIC image of the embryo shown in B. (C) Elevated expression level and accumulation of many SQST-1::GFP aggregates in epg-7 mutant embryos. (D) Compared with wild type and epg-2 mutants, levels of SQST-1 are dramatically elevated in atg-3 and epg-7 mutant embryos in an immunoblotting assay. (E–G) Compared with wild-type embryos (E and F), endogenous SQST-1 shows dramatically elevated levels and accumulates into a large number of aggregates in epg-7 mutant embryos (G). ∼200-cell stage embryos are shown in E–G. (E) DAPI image of the embryo shown in F. (H) Number of SQST-1 aggregates per focal plane in wild type, atg-3 mutants, and epg-7 mutants. (I–L) At the ∼100-cell stage, far fewer SQST-1 aggregates are formed in epg-7 mutants than in atg-3 mutants. (I and K) DAPI images of the embryos shown in J and L, respectively. (M) SQST-1 directly interacts with ZK1053.4 in a GST pull-down assay. (N) Compared with wild-type animals, levels of ZK1053.4::GFP are dramatically elevated in atg-3 and epg-7 mutants. (O–R) ZK1053.4::GFP is weakly expressed in wild-type embryos (O–P) and sqst-1 mutants (R), but forms a large number of aggregates in epg-7 mutants (Q). (O) DIC image of the embryo shown in P. (S) Survival curve of L1 larvae under food depletion conditions in wild type, epg-1 mutants, and epg-7 mutants. This experiment was completed once (wild type: n = 2,964; epg-7: n = 602; epg-1: n = 215). Statistical analysis of L1 survival shows that epg-7 mutants exhibit significant difference to wild type and also to epg-1 mutants (log-rank test, P = 0.000). (T) Loss of function of epg-7 does not ameliorate the degeneration of mec-4::gfp–labeled touch neurons caused by mec-4(u231). (U) Survival curves of wild type and various mutants. sqst-1;epg-7 mutants have a shortened mean and maximum life span. (P = 0.000 for epg-7 or wild type vs. sqst-1;epg-7 in all three repeats; log-rank test). (V) Survival of hydrogen peroxide–treated animals. (In all three repeats, P = 0.000 for epg-7 vs. sqst-1;epg-7; P > 0.2 for wild type vs. sqst-1;epg-7; log-rank test). (W) Survival of animals at the indicated time after heat shock treatment. (P < 0.05 for wild type vs. epg-7 or epg-7 vs. sqst-1;epg-7 at 5, 6, and 7 h). (X) Dauer formation of daf-2(e1370) mutants in different backgrounds. Bars: (A–C, E–G, I–L, and O–R) 10 µm. C. elegans embryos remain the same size during embryogenesis and loss of autophagy activity has no effect on the embryo size. Thus, bars for embryos are only shown once in each figure.
A group of coiled-coil domain–containing proteins, termed the SEPA-1 family, are selectively removed by autophagy during embryogenesis (Zhang et al., 2009). Among them, ZK1053.4 and C35E7.6 colocalized with SQST-1 aggregates in autophagy mutants (Fig. S1 F; and unpublished data). SQST-1 was identified as a ZK1053.4-interacting protein in a yeast two-hybrid assay and directly interacted with ZK1053.4 in a pull-down assay (Fig. 1 M). In epg-7 mutants, ZK1053.4 and C35E7.6 were present at dramatically increased levels and accumulated into a large number of aggregates that colocalized with SQST-1 aggregates (Fig. 1, N–Q; Fig. S1, G and H). Deletion of sqst-1 affected neither the degradation of ZK1053.4 and C35E7.6 nor their aggregation in autophagy mutants (Fig. 1 R; Fig. S1, E, I, and J).
Degradation of the autophagy substrates SEPA-1 and EPG-2 was examined in epg-7 mutants. In wild-type embryos, EPG-2 aggregates and SEPA-1 aggregates are present at early stages and become almost undetectable at the comma stage and onwards (Fig. S1, K, L, O, and P; Zhang et al., 2009; Tian et al., 2010). A large number of EPG-2 aggregates and SEPA-1 aggregates accumulate and persist throughout embryogenesis in autophagy mutants (Fig. S1, M and Q; Zhang et al., 2009; Tian et al., 2010). Loss of epg-7 activity did not evidently affect the degradation of EPG-2 and SEPA-1 (Fig. S1, N and R). In epg-7 mutants, SQST-1 and SEPA-1 aggregates were located in close proximity, but did not colocalize, at early embryonic stages (Fig. S1 S). Other members of the SEPA-1 family, including T04D3.1, T04D3.2, ZK1053.3, and F44F1.6, form aggregates distinct from SQST-1 aggregates in autophagy mutants (Fig. S1 T). The expression pattern and level of these proteins was the same as wild type in epg-7 mutants (Fig. S1, U–W). Therefore, unlike mutations in essential autophagy genes, epg-7 mutations cause defects in degradation of only a subset of protein substrates.
The role of epg-7 in other autophagy-regulated processes and stress conditions
We determined whether epg-7 plays an essential role in other autophagy-regulated processes. Loss of autophagy activity severely impairs the survival of newly hatched L1 animals in the absence of food and also shortens the life span of adult animals under food repletion conditions (Tian et al., 2009; Kovács and Zhang, 2010). However, loss of epg-7 function only mildly impaired the life span of L1 larvae under food depletion conditions and had no effect on the adult life span (Fig. 1, S and U). Loss of function of epg-7, unlike mutations in essential autophagy genes, did not ameliorate the necrosis-like degradation of six touch neurons caused by the mec-4 gain-of-function allele, u231 (Fig. 1 T). These results show that epg-7 is not generally required for autophagy.
Compared with wild-type animals, epg-7 mutants produced fewer embryos, a larger percentage of which failed to hatch (Fig. S1, X and Y). epg-7 mutants also grew more slowly (Fig. S1 Z). sqst-1 mutant animals also had a lower brood size and hatch rate (Fig. S1, X and Y). sqst-1;epg-7 double mutants showed an even lower hatch rate than either single mutant, but had a wild-type brood size (Fig. S1, X and Y). The prolonged developmental time was not further extended in sqst-1;epg-7 mutants (Fig. S1 Z).
We investigated the role of epg-7 and sqst-1 under various stress conditions. Loss of function of epg-7 and sqst-1 increased the sensitivity of animals to oxidative stress, but sqst-1;epg-7 double mutants exhibit a wild-type resistance (Fig. 1 V). Compared with wild-type animals, epg-7 animals showed an increased resistance to heat stress, which was suppressed by simultaneous depletion of sqst-1 (Fig. 1 W). Environmental stress triggers animals entering the dauer diapause. A fraction of daf-2(e1370) mutant animals developed into dauers at a semi-permissive temperature. Loss of function of epg-7 or sqst-1 had no effect on the dauer formation of daf-2(e1370) animals (Fig. 1 X). However, under the same conditions, a smaller fraction of daf-2;sqst-1;epg-7 mutants entered the dauer (Fig. 1 X). Taken together, epg-7 mutants behave differently in response to different stresses and show a context-specific interaction with sqst-1.
epg-7 encodes a protein with similarity to FIP200 and Atg11 and is widely expressed
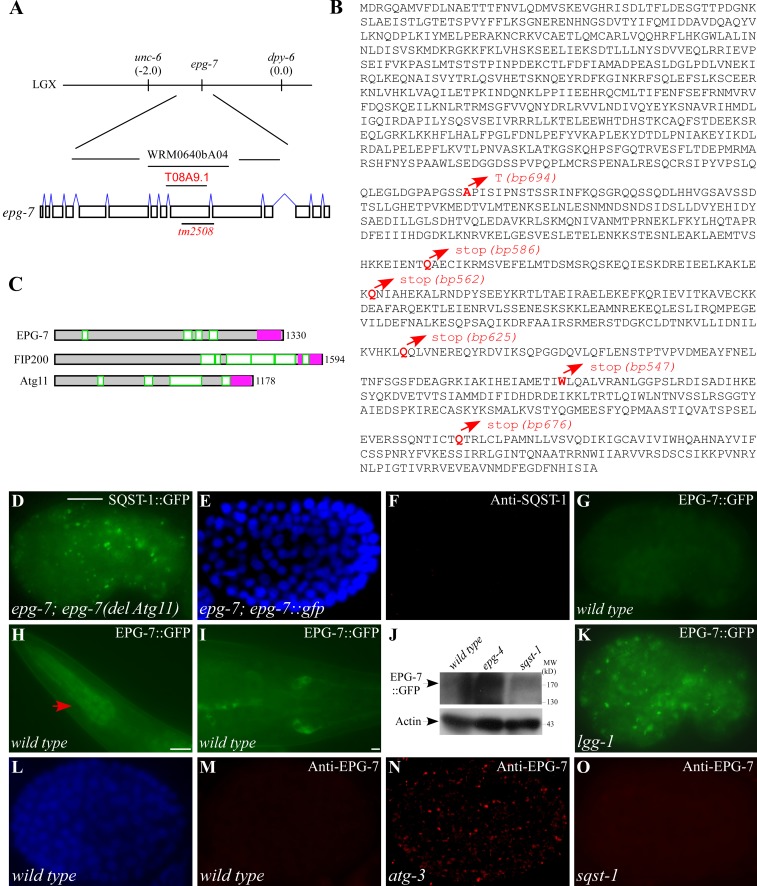
Transformation rescue experiments revealed that a transgene carrying a single gene t08a9.1 rescued the defective degradation of SQST-1 aggregates in epg-7 mutants (Fig. 2 A). epg-7 encodes a protein with 1,330 amino acids (Fig. 2 B). The molecular lesions were detected in epg-7(bp694, bp586, bp562, bp625, bp547, or bp676) mutants (Fig. 2 B). epg-7(tm2508) has a frame-shifting deletion of 502 bp in the coding region of epg-7, resulting in a putative protein with 765 amino acids (Fig. 2, A and B). Thus, epg-7(tm2508) is likely to be a genetic null and was used in this study.
Figure 2.
epg-7 encodes a protein with similarity to mammalian FIP200 and yeast Atg11 and is degraded by autophagy. (A) Cloning of epg-7. (B) Protein sequence of EPG-7. The molecular lesions in identified mutant alleles are highlighted in red. (C) Schematic structures of EPG-7, FIP200, and Atg11. The coiled-coil domains are highlighted in green. The Atg11 domain is depicted in pink. (D) A large number of SQST-1::GFP aggregates accumulate in epg-7 mutant embryos carrying an epg-7(del Atg11) transgene. (E and F) No SQST-1 aggregates are detected in epg-7 mutants carrying an epg-7::gfp reporter. (E) DAPI image of the embryo shown in F. (G) EPG-7::GFP is weakly expressed and diffusely localized in the cytoplasm in wild-type embryos. (H–I) EPG-7::GFP is expressed in pharyngeal muscles (H, arrow) and cells in the tail region (I) at larval stages. (J) Compared with wild-type animals, levels of EPG-7::GFP are dramatically elevated in epg-4 mutants, but remain unchanged in sqst-1 mutants. (K) The expression level of EPG-7::GFP is dramatically elevated and many EPG-7::GFP aggregates are formed in lgg-1 mutant embryos. (L and M) No EPG-7 aggregates, detected by anti-EPG-7 antibody, are formed in wild-type embryos. (L) DAPI image of the embryo shown in M. (N) Levels of EPG-7 are dramatically elevated and a large number of EPG-7 aggregates are formed in atg-3 mutants. (O) No EPG-7 aggregates are formed in sqst-1 mutant embryos. Bars: (D–G and K–O) 10 µm; (H) 10 µm; (I) 10 µm.
EPG-7 exhibits similarity to human FIP200 through most of its sequence (blast value 4e−19). FIP200 is a 1,594–amino acid protein which, like EPG-7, contains coiled-coil domains (Fig. 2 C; Hara et al., 2008). Some regions of EPG-7 also display similarity to yeast Atg11 (Fig. 2 C). The C termini of EPG-7, FIP200, and Atg11 contain the Pfam motif PF10377 (also known as the Atg11 motif). An epg-7(del Atg11) transgene, in which the Atg11 motif was deleted, failed to rescue the defective degradation of SQST-1 aggregates in epg-7(tm2508) mutants (Fig. 2 D), suggesting that the Atg11 motif is essential for epg-7 function.
A translational fusion reporter with gfp inserted at the C terminus of EPG-7, which was functional, showed that EPG-7::GFP was ubiquitously expressed and diffusely localized in the cytoplasm during embryogenesis (Fig. 2, E–G). At post-embryonic stages, EPG-7 was widely expressed, including in pharyngeal muscles, neurons, and cells in the tail region (Fig. 2, H and I).
EPG-7 is degraded by autophagy in a SQST-1–independent manner
We next determined whether EPG-7 is itself removed by autophagy. In autophagy mutants, EPG-7::GFP was detected at dramatically increased levels and accumulated into a large number of aggregates (Fig. 2, J and K). Endogenous EPG-7 showed weak expression in the cytoplasm in wild-type embryos (Fig. 2, L and M), but accumulated into a large number of cytoplasmic aggregates that persisted throughout embryogenesis in autophagy mutants (Fig. 2 N; Fig. S2 G). Levels of epg-7 mRNA were not elevated in autophagy mutants (Fig. S2 A).
SQST-1 aggregates exhibit distinct morphologies and distribution patterns in different autophagy mutants (Tian et al., 2010; Lu et al., 2011), which reflects the fact that loss of function of these genes causes defects at distinct steps of the aggrephagy pathway. Like SQST-1 aggregates, EPG-7 aggregates were spherical and dispersed in the cytoplasm in unc-51/Atg1, epg-1/Atg13, atg-3, epg-8/Atg14, bec-1, lgg-1/Atg8, atg-9, atg-18, and epg-5 mutants, but were enlarged and formed clusters in epg-3, -4, and -6 mutants (Fig. 2 N; Table S1; Fig. S2 G). Depletion of sqst-1 had no effect on EPG-7 degradation and formation of aggregates in autophagy mutants (Fig. 2, J and O; Fig. S2 I). In summary, EPG-7 is degraded by autophagy during embryogenesis in a SQST-1–independent manner.
EPG-7 colocalizes with SQST-1 in autophagy mutants and directly interacts with SQST-1
The above results raise the possibility that EPG-7 recruits SQST-1 for autophagic degradation. We performed coimmunostaining assays and found that EPG-7 aggregates colocalized with SQST-1 aggregates in atg-3, epg-8, atg-18, epg-5, and atg-9 mutants, but were separable from SEPA-1 aggregates (Fig. 3, A and B; Fig. S2, K and L). In epg-3, -4, and -6 mutants, EPG-7 aggregates and SQST-1 aggregates were enlarged and largely colocalized (Fig. 3 B; Fig. S2 M). In unc-51/Atg1 and epg-1/Atg13 mutants, EPG-7 aggregates only partially colocalized with SQST-1 aggregates, and some small EPG-7 aggregates were closely associated with, but did not overlap, the SQST-1 aggregates (Fig. 3 B; Fig. S2, N and O), suggesting that the UNC-51–EPG-1 complex regulates the association of EPG-7 with SQST-1.
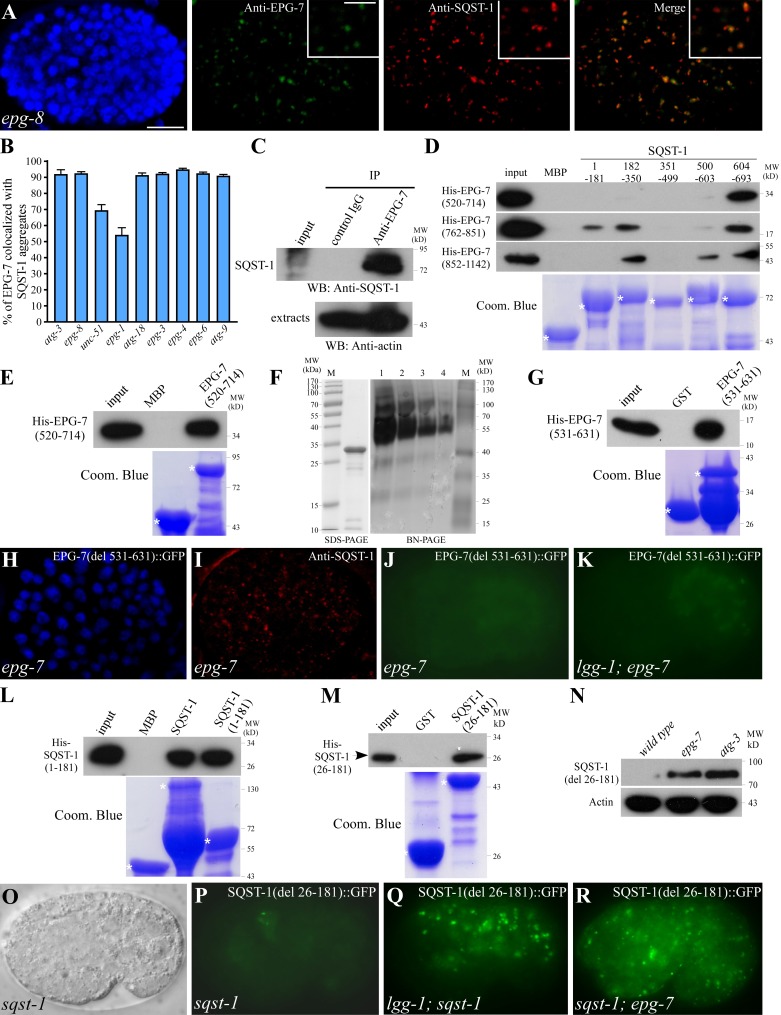
Figure 3.
EPG-7 colocalizes with SQST-1 aggregates in autophagy mutants and directly interacts with SQST-1. (A) EPG-7 colocalizes with SQST-1 aggregates in epg-8 mutant embryos. Colocalization was defined as two aggregates showing >70% overlap in their fluorescence signals. Insets show a magnified view. (B) Percentage of EPG-7 aggregates colocalized with SQST-1 in indicated autophagy mutants. (C) A coimmunoprecipitation assay reveals that EPG-7 associates with SQST-1. (D) EPG-7 interacts with SQST-1 in a pull-down assay. (E) EPG-7 self-associates in a pull-down assay. (F) SDS-PAGE and blue-native PAGE of purified EPG-7(520–714) proteins. Lane M shows molecular weight marker. Lanes 1, 2, 3, and 4 contain 10, 5, 2.5, and 1.25 µg of proteins, respectively. (G) An EPG-7 fragment containing amino acids 531–631 self-interacts. (H and I) SQST-1 aggregates accumulate in epg-7 mutant embryos carrying an epg-7(del 531–631)::gfp transgene. (H) DAPI image of the embryo shown in I. (J and K) EPG-7(del 531–631)::GFP is weakly expressed and diffusely localized in epg-7(tm2508) embryos. Its expression is not elevated in lgg-1(RNAi) animals. H–K show embryos carrying the same transgene. Images in J and K were taken using the same exposure time. (L and M) SQST-1 self-interacts (L) and the self-interaction is mediated by a fragment containing amino acids 26–181 (M). (N) Levels of SQST-1(del 26–181)::GFP are dramatically elevated in epg-7 and atg-3 mutants. (O and P) SQST-1(del 26–181)::GFP is weakly expressed and diffusely localized in sqst-1(ok2892) mutant embryos. (O) DIC image of the embryo shown in P. (Q and R) SQST-1(del 26–181)::GFP is expressed at dramatically elevated levels and accumulates into a large number of aggregates in lgg-1 (Q) and epg-7 (R) mutants. Bars: (A, H–K, and O–R), 10 µm; (A, insets) 5 µm.
Colocalization of EPG-7 with SQST-1 aggregates in autophagy mutants prompted us to investigate whether EPG-7 directly associates with SQST-1. Embryonic extracts were precipitated with anti–EPG-7 antibody and co-purified proteins were detected by Western blotting using anti–SQST-1 antibody. SQST-1 was specifically coimmunoprecipitated by anti–EPG-7, but not by control IgG (Fig. 3 C), suggesting that EPG-7 associates with SQST-1 in vivo.
Recombinant maltose-binding protein (MBP)–fused SQST-1 pulled down His-tagged EPG-7, indicating that EPG-7 directly interacts with SQST-1. By using a series of EPG-7 fragments, we found that multiple regions in EPG-7 possessed strong SQST-1 binding activity (Fig. 3 D). Similarly, several fragments of SQST-1 contained EPG-7 interacting activity (Fig. 3 D). Interactions among fragments of EPG-7 and SQST-1 were confirmed in yeast two-hybrid assays (Fig. S2 E). These results indicate that EPG-7 and SQST-1 have multiple interacting surfaces.
Self-association of EPG-7, but not SQST-1, is required for degradation of the SQST-1–EPG-7 complex
As EPG-7 forms aggregates in autophagy mutants, we determined whether EPG-7 self-associates. Bacterially expressed His-tagged EPG-7 was specifically pulled down by MBP-tagged EPG-7 in a pull-down assay (Fig. 3 E; and unpublished data). A size-exclusion chromatography and static light scattering assay and also blue-native PAGE showed that EPG-7(520–714) formed oligomers (Fig. 3 F; Fig. S2 B). The self-association domain of EPG-7 was further mapped to a fragment containing amino acids 531–631 (Fig. 3 G), which includes the codon that is mutated from alanine to threonine in EPG-7(bp694). Consistent with this, EPG-7(bp694) mutant proteins failed to self-associate in the pull-down assay (Fig. S2 C).
The epg-7(del 531–631)::gfp transgene, in which the self-association domain was deleted, was not functional in rescuing the defective degradation of SQST-1 aggregates in epg-7 mutants (Fig. 3, H and I). In atg-3;epg-7(bp694) mutants, no EPG-7(bp694) aggregates were detected (Fig. S2 J), suggesting that self-association is required for aggregate formation. Consistent with this, expression of EPG-7(del 531–631)::GFP was not elevated in autophagy mutants (Fig. 3, J and K; Fig. S2 D). These results indicate that the self-association domain of EPG-7 is essential for its function and also for its autophagic degradation.
In vitro pull-down assays showed that SQST-1 also self-interacted through a fragment containing amino acids 26–181 (Fig. 3, L and M). A GFP-tagged SQST-1 reporter with a deletion of the self-association domain, SQST-1(del 26–181)::GFP, was widely expressed and diffusely localized in the cytoplasm (Fig. 3, O and P). SQST-1(del 26–181)::GFP was detected at dramatically elevated levels and accumulated into numerous aggregates in autophagy and epg-7 mutants (Fig. 3, N, Q, and R). SQST-1(del 26–181)::GFP colocalized with EPG-7 in lgg-1 mutants (Fig. S2 P). Thus, self-association of EPG-7, but not SQST-1, is required for autophagic degradation of the SQST-1–EPG-7 complex.
EPG-7 regulates association of SQST-1 aggregates with LGG-1 puncta
Different autophagy mutants show distinct levels and distribution patterns of LGG-1 (Tian et al., 2010; Lu et al., 2011). In wild-type embryos, LGG-1 forms small punctate structures at early embryonic stages, mostly at the ∼100- to ∼200-cell stage, and the number of LGG-1 puncta decreases as development proceeds, with only few LGG-1 puncta detectable in fourfold stage embryos (Fig. 4, A–D; Tian et al., 2010). Compared with wild-type animals, levels of LGG-1-I and LGG-1-II (lipidated form) in epg-7 mutants remained unaltered (Fig. 4 E). Before the ∼100-cell stage in epg-7 mutants, some LGG-1 puncta were larger in size and stronger in intensity than those in wild-type embryos (Fig. 4 F). After the ∼100-cell stage, the size, formation, and distribution of LGG-1 puncta resembled those in wild-type embryos (Fig. 4, G and H). As in wild-type embryos, SEPA-1 aggregates partially colocalized with LGG-1 puncta in epg-7 mutants (Fig. S3 A). However, the LGG-1 puncta in epg-7 mutants were separable from SQST-1 aggregates (Fig. 4 I), consistent with that loss of function of epg-7 causes defective degradation of SQST-1, but not SEPA-1.
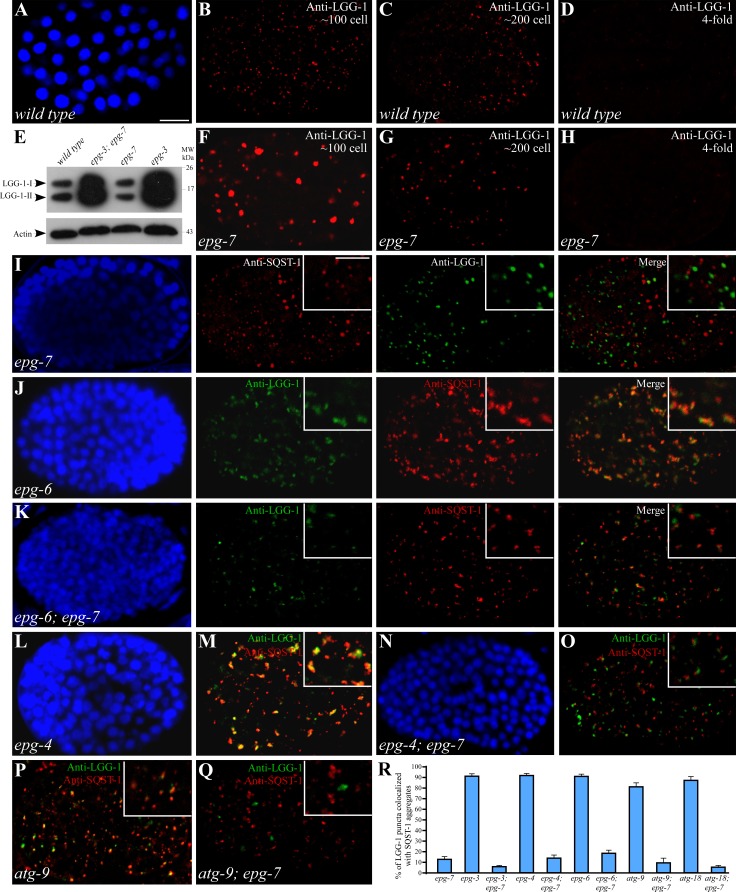
Figure 4.
Loss of epg-7 activity impairs the colocalization of SQST-1 aggregates with LGG-1 puncta in autophagy mutants. (A–D) Expression of LGG-1 in wild-type embryos at the ∼100-cell (A and B), the ∼200-cell (C), and the fourfold (D) stages. (A) DAPI image of the embryo shown in B. (E) Loss of function of epg-7 does not increase levels of LGG-1 and also does not suppress accumulation of LGG-1 in epg-3 mutant embryos. (F–H) Expression of LGG-1 in epg-7 mutant embryos before the ∼100-cell stage (F), at the ∼200-cell stage (G), and at the fourfold stage (H). Images in B–D and F–H were taken using the same exposure time. (I) SQST-1 aggregates are separable from LGG-1 puncta in epg-7 mutant embryos. Insets show a magnified view. (J) In epg-6 mutant embryos, LGG-1 puncta and SQST-1 aggregates are enlarged and colocalized. (K) LGG-1 and SQST-1 aggregates are smaller in size and largely separable in epg-6;epg-7 double mutants. (I–K) DAPI images of the embryos shown in the same row. (L–O) Enlarged LGG-1 and SQST-1 aggregates are colocalized in epg-4 mutants (L and M). Simultaneously depleting epg-7 reduces the size of LGG-1 puncta and SQST-1 aggregates, which become largely separable (N and O). (L and N) DAPI images of the embryos shown in M and O, respectively. (P) In atg-9 mutant embryos, LGG-1 puncta largely colocalize with SQST-1 aggregates at the comma stage. Notably, there are more SQST-1 aggregates than LGG-1 puncta. (Q) SQST-1 aggregates are separable from LGG-1 puncta in atg-9;epg-7 mutants at the comma stage. (R) Percentage of LGG-1 puncta colocalized with SQST-1 aggregates in indicated autophagy mutants. Comma-stage embryos were examined for atg-9 and atg-9;epg-7 mutants. ∼200-cell stage embryos were examined for other autophagy mutants. Bars: (A–D and F–Q) 10 µm; (insets in I–K, M, O, and P–Q) 5 µm.
Mutations in different autophagy genes that act at distinct steps of the aggrephagy pathway cause characteristic morphologies and distribution patterns of LGG-1 puncta and SQST-1 aggregates, which have been used to analyze the interactions among autophagy genes (Tian et al., 2010; Lu et al., 2011). We next performed epistasis analysis to examine the relationship between epg-7 and other autophagy genes. Mutations in epg-3, -4, and -6 cause a defect in the progression of isolation membranes to autophagosomes (Tian et al., 2010; Lu et al., 2011). Enlarged LGG-1 puncta colocalize with SQST-1 aggregates in epg-3, -4, and -6 mutants (Fig. 4, J, L, M, and R; Table 1; Tian et al., 2010; Lu et al., 2011). In epg-3;epg-7, epg-6;epg-7, and epg-4;epg-7 mutants, SQST-1 aggregates and LGG-1 puncta were smaller in size and largely separable, resembling those in epg-7 single mutants (Fig. 4, K, N, O, and R; Table 1). In atg-18 mutants, LGG-1 forms small puncta that colocalize with SQST-1 aggregates at the ∼200-cell stage (Fig. 4 R; Table 1; Fig. S3 B; Lu et al., 2011). In atg-18;epg-7 mutants, LGG-1 puncta were separable from SQST-1 aggregates at the ∼200-cell stage (Fig. 4 R; Table 1; Fig. S3 C). atg-9 mutant embryos have larger but fewer LGG-1 puncta than wild-type embryos. The large LGG-1 puncta before the ∼100-cell stage resembled those in epg-7 mutants. SQST-1 aggregates were separable from LGG-1 puncta at early embryonic stages, but partially colocalized from the ∼200-cell stage onwards (Fig. 4, P and R; Table 1; Fig. S3 D; and unpublished data). In atg-9;epg-7 mutants, SQST-1 aggregates were separable from LGG-1 puncta in comma-stage embryos (Fig. 4, Q and R; Table 1; Fig. S3 E). These genetic analyses indicate that epg-7 regulates the association of LGG-1 puncta and SQST-1 aggregates and functions upstream of all examined autophagy genes in the SQST-1 aggregate degradation pathway.
Table 1.
Genetic interactions of epg-7 with other autophagy mutants
| Genotype | Percentage of SQST-1 aggregates forming clusters (n)a | Percentage of LGG-1 puncta forming clusters (n)a | Percentage of LGG-1 puncta colocalizing with SQST-1 aggregates (n)ab |
| wild type | NAd | 2.3 (389) | NA |
| epg-7 | 3.4 (498) | 2.2 (311) | 13.9 (311) |
| epg-6 | 36.1 (368) | 24.5 (290) | 91.5 (290) |
| epg-6;epg-7 | 5.0 (382) | 3.1 (225) | 19.4 (225) |
| epg-3 | 26.0 (373) | 23.0 (280) | 91.6 (280) |
| epg-3;epg-7 | 5.3 (415) | 5.9 (273) | 6.9 (273) |
| epg-4 | 27.4 (415) | 24.4 (290) | 91.9 (290) |
| epg-4;epg-7 | 5.5 (367) | 3.1(256) | 14.6 (256) |
| atg-9c | 7.5 (477) | 4.7 (303) | 82.4 (303) |
| atg-9;epg-7 | 14.3 (343) | 0 (32) | 10.2 (32) |
| atg-18 | 13.6 (315) | 9.7 (259) | 87.7 (259) |
| atg-18;epg-7 | 9.4 (436) | 3.7 (326) | 6.2 (326) |
Confocal images of embryos at the comma stage were determined for atg-9 and atg-9;epg-7 mutants. ∼200-cell stage embryos were examined for other autophagy mutants. A cluster is defined as two or more punctate structures interconnected.
Colocalization refers to two aggregates showing >70% overlap in their fluorescence signals.
LGG-1 accumulates into large aggregates in some cells in atg-9 mutant. Those aggregates are not classified as clusters.
NA: SQST-1 aggregates are absent in wild-type embryos.
EPG-7 directly associates with multiple ATG proteins
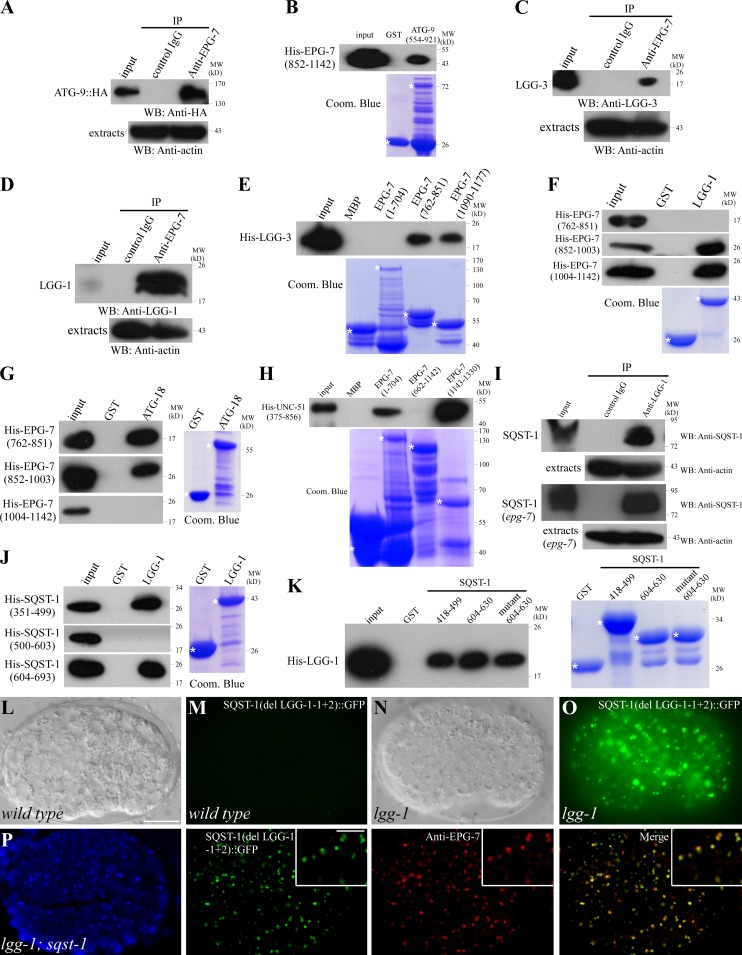
We hypothesized that the EPG-7–SQST-1 complex acts as a platform to recruit Atg proteins for autophagosome formation. Atg9 acts upstream in the hierarchy in recruiting other Atg proteins to the PAS, and also provides membranes for autophagosome formation (Suzuki et al., 2007; Mari et al., 2010). We thus investigated the interaction between EPG-7 and ATG-9. Extracts of embryos carrying an atg-9::HA transgene were precipitated by anti–EPG-7 antibody and the coprecipitants were analyzed by anti-HA antibody. We found that ATG-9 was specifically coprecipitated by anti–EPG-7 (Fig. 5 A). In vitro pull-down assays demonstrated that an EPG-7 fragment containing amino acids 852–1142 directly bound to ATG-9 (Fig. 5 B).
Figure 5.
EPG-7 directly interacts with multiple Atg proteins. (A) EPG-7 coimmunoprecipitates with ATG-9::HA. (B) EPG-7 directly interacts with ATG-9 in a GST pull-down assay. (C and D) Coimmunoprecipitation reveals that EPG-7 associates with LGG-3 (C) and LGG-1 (D). (E) EPG-7 directly interacts with LGG-3. (F) EPG-7 fragments containing amino acids 852–1003 or 1004–1142, but not 762–851, interact with LGG-1. (G) Fragments of EPG-7 containing amino acids 762–851 or 852–1003 directly interact with ATG-18. (H) Fragments of EPG-7 containing amino acids 1–704 or 1143–1330 directly interact with UNC-51. (I) SQST-1 associates with LGG-1 in vivo. Embryonic extracts from epg-7 mutants show that interaction of SQST-1 with LGG-1 is EPG-7 independent. (J) Fragments of SQST-1 containing amino acids 351–499 and 604–693, but not 500–603, interact with LGG-1. (K) Fragments of SQST-1–containing amino acids 418–499 and 604–630 bind to LGG-1. Mutating the putative LIR in the 604–630 fragment (605YPDL608 to APDA) does not affect the binding. (L and M) SQST-1(del LGG-1-1+2)::GFP, in which the two LGG-1–binding fragments (amino acids 418–499 and 604–630) are deleted, is weakly expressed and diffusely localized in the cytoplasm in sqst-1(ok2892) mutants. (N and O) Loss of function of lgg-1 dramatically elevates the expression level of SQST-1(del LGG-1-1+2)::GFP, which forms a large number of aggregates. (L and N) DIC images of the embryos shown in M and O, respectively. (P) SQST-1(del LGG-1-1+2)::GFP aggregates colocalize with EPG-7 in lgg-1;sqst-1 mutants. Insets show a magnified view. Bars: (L–P) 10 µm; (P, insets) 5 µm.
The interaction between EPG-7 and the LGG-3/Atg12 and LGG-1 conjugation systems was also examined. We found that both LGG-3 and LGG-1 were specifically coprecipitated with EPG-7 in a coimmunoprecipitation assay (Fig. 5, C and D). Fragments of EPG-7 containing amino acids 762–851 and 1090–1177 pulled down His-tagged LGG-3 (Fig. 5 E). Multiple fragments of EPG-7, including 852–1003 and 1004–1142, directly associated with LGG-1 in pull-down assays (Fig. 5 F; and unpublished data). These regions, however, do not contain the LIR motif. EPG-7 also directly interacted with ATG-18 and UNC-51 in pull-down assays (Fig. 5, G and H). Yeast two-hybrid assays confirmed the interactions among fragments of EPG-7 and ATG proteins (Fig. S2 F). No direct interactions were detected between EPG-7 and ATG-3, ATG-7, or ATG-5 in the pull-down assay (unpublished data).
To investigate whether interaction with ATG proteins is essential for the function of EPG-7, we constructed mutant epg-7 reporters with a deletion of ATG protein-binding fragments. The mutant reporters, including epg-7(del 762–851)::gfp, epg-7(del 852–1003)::gfp, and epg-7(del 1004–1142)::gfp, failed to rescue the defective degradation of SQST-1 aggregates in epg-7(tm2508) mutants (Fig. S3, F–K), suggesting that the ATG-binding domain is required for EPG-7 function. The expression level of epg-7(del 852–1003)::gfp and epg-7(del 1004–1142)::gfp was not elevated by loss of function of lgg-1 (Fig. S3, N–U; and unpublished data), indicating that these regions are also important for the autophagic removal of EPG-7.
We further determined whether SQST-1 directly binds to Atg proteins. Coimmunoprecipitation assays demonstrated that SQST-1 associates with LGG-1 that was independent of EPG-7 (Fig. 5 I). In vitro pull-down assays revealed that SQST-1(418–499) and SQST-1(604–630) strongly bound to LGG-1 (Fig. 5, J and K). SQST-1(418–499) contains no LIR motif. Mutating the LIR motif in SQST-1(604–630) from 605YPDL608 to APDA did not alter LGG-1 binding (Fig. 5 K), indicating that the LIR motif is not required for binding of SQST-1 to LGG-1. SQST-1 did not bind to ATG-9 and LGG-3 in the pull-down assay (unpublished data). sqst-1 mutant reporters with deletions of either one or both LGG-1 binding domains showed diffuse expression in the cytoplasm (Fig. 5, L and M; Fig. S3 L). In autophagy mutants, the mutant SQST-1 proteins were present at dramatically elevated levels and accumulated into numerous aggregates that colocalized with EPG-7 aggregates (Fig. 5, N–P; Fig. S3 M), suggesting that the LGG-1 binding domain is not required for autophagic degradation of SQST-1.
EPG-7 colocalizes with ATG-9 puncta in autophagy mutants
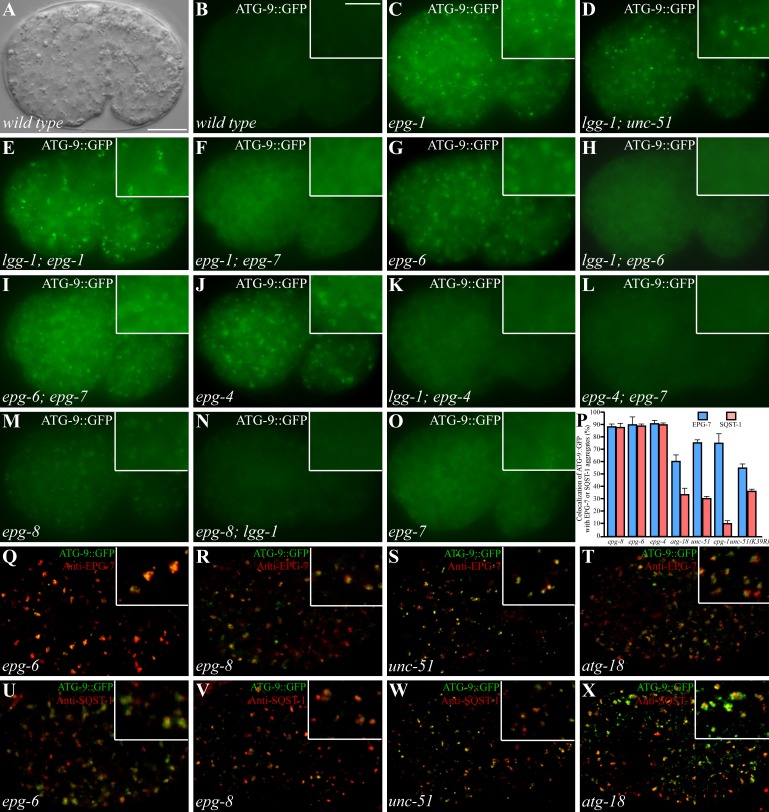
We next examined the distribution pattern of ATG-9 in various autophagy mutants. ATG-9::GFP diffusely localizes in the cytoplasm in wild-type embryos and forms distinct puncta in various autophagy mutants (Fig. 6, A and B; Lu et al., 2011). In mutants for the UNC-51–EPG-1 complex, ATG-9::GFP forms numerous small punctate structures (Fig. 6 C; Lu et al., 2011). In epg-6 and epg-4 mutants, ATG-9 forms many big aggregates (Fig. 6, G and J; Lu et al., 2011). atg-18 and epg-8 mutants also show accumulation of ATG-9::GFP puncta that are weaker in intensity than in the above autophagy mutants (Fig. 6 M, and unpublished data; Lu et al., 2011). Mutations in epg-7 showed no evident accumulation of ATG-9::GFP (Fig. 6 O).
Figure 6.
Formation of ATG-9 puncta in various autophagy mutants. (A and B) In wild-type embryos, ATG-9::GFP is diffusely localized in the cytoplasm. (A) DIC image of the embryo shown in B. Insets show a magnified view. (C) ATG-9::GFP forms a large number of small, intense puncta in epg-1 mutants. (D and E) Accumulation of ATG-9::GFP puncta in unc-51(D) and epg-1 (E) mutants is independent of lgg-1. (F) The ATG-9::GFP puncta largely disappear in epg-1;epg-7 mutants. (G) ATG-9::GFP accumulates into large punctate structures in epg-6 mutants. (H) Loss of function of lgg-1 suppresses the accumulation of ATG-9::GFP puncta in epg-6 mutants. (I) Loss of function of epg-7 has no effect on the formation of ATG-9::GFP puncta in epg-6 mutants. The ATG-9::GFP puncta are smaller in epg-6;epg-7 mutants than those in epg-6 single mutants. (J–L) ATG-9::GFP accumulates into large punctate structures in epg-4 mutants (J) and the accumulation is suppressed by simultaneously depleting the activity of lgg-1 (K) or epg-7 (L). (M and N) In epg-8 mutants, ATG-9 accumulates into aggregates (M) that are suppressed by loss of lgg-1 activity (N). (O) epg-7 mutants exhibit the same distribution pattern of ATG-9::GFP as wild-type embryos. (P) Percentage of ATG-9::GFP puncta colocalized with EPG-7 and SQST-1 aggregates in indicated autophagy mutants. (Q–T) The ATG-9::GFP puncta in epg-6 (Q), epg-8 (R), unc-51 (S), and atg-18 (T) mutants colocalize with EPG-7 aggregates. (U and V) ATG-9::GFP puncta in epg-6 (U) and epg-8 (V) mutants colocalize with SQST-1 aggregates. (W and X) In unc-51 (W) and atg-18 (X) mutants, ATG-9::GFP partially colocalizes with SQST-1 aggregates. Bars: (A–O and Q–X) 10 µm; (insets in B–O and Q–X) 5 µm.
Accumulation of ATG-9::GFP in epg-4, epg-6, epg-8, and atg-18 mutants was abolished by loss of function of lgg-1 (Fig. 6, H, K, and N; Table 2), which is consistent with the hypothesis that the LGG-1 conjugation system acts at an early step in autophagosome formation in the aggrephagy pathway (Lu et al., 2011). In epg-1 and unc-51 mutant embryos, LGG-1 puncta are absent from most embryonic cells, but accumulate into large aggregates in a few cells (Tian et al., 2010). We found that loss of lgg-1 activity did not affect the accumulation of ATG-9::GFP puncta and their colocalization with EPG-7 aggregates in unc-51 and epg-1 mutants (Fig. 6, D and E; Table 2). We further determined the role of EPG-7 in the accumulation of ATG-9::GFP in autophagy mutants. Simultaneously depleting epg-7 did not affect the formation of ATG-9::GFP puncta in epg-6 and epg-8 mutants (Fig. 6 I; Table 2), whereas ATG-9::GFP was largely diffusely localized in epg-1;epg-7 and epg-4;epg-7 mutants (Fig. 6, F and L; Table 2), suggesting that epg-7 acts upstream of epg-1 and epg-4.
Table 2.
Formation of ATG-9::GFP puncta and their colocalization with EPG-7 and SQST-1 aggregates
| Genotype | Number of ATG-9::GFP puncta per focal planea | Percentage of ATG-9::GFP puncta forming clusters (n)a | Percentage of ATG-9 puncta colocalizing with EPG-7 aggregates (n)ab | Percentage of ATG-9 puncta colocalizing with SQST-1 aggregates (n)ab |
| wild typec | 10.7 | 0.0 (32) | NA | NA |
| epg-7d | 31.7 | 5.2 (95) | NA | ND |
| epg-6 | 129.3 | 28.1 (388) | 90.5 (284) | 89.7 (388) |
| lgg-1;epg-6 | 16.3 | 1.8 (49) | ND | ND |
| epg-6;epg-7 | 126.7 | 9.7 (380) | NA | 81.2 (380) |
| epg-4 | 129.3 | 26.4 (388) | 91.4 (318) | 90.2 (388) |
| lgg-1;epg-4 | 24.3 | 2.6 (73) | ND | ND |
| epg-4;epg-7 | 23.3 | 4.4 (70) | NA | ND |
| epg-1 | 117.3 | 2.6 (352) | 75.5 (240) | 11.0 (352) |
| lgg-1;epg-1 | 118.0 | 2.8 (354) | 76.6 (354) | ND |
| atg-18 | 118.0 | 14.1 (354) | 60.8 (751) | 34.3 (354) |
| lgg-1;atg-18 | 22.7 | 1.3 (68) | ND | ND |
| epg-8 | 87.0 | 13.0 (261) | 88.5 (261) | 88.3 (241) |
| epg-8;lgg-1 | 19.0 | 1.7 (57) | ND | ND |
| epg-8;epg-7 | 92.3 | 9.0 (283) | NA | 88.0 (283) |
Confocal images of embryos at the ∼200-cell stage to comma stage were determined. A cluster is defined as two or more punctate structures interconnected.
Colocalization is defined as two aggregates showing >70% overlapped fluorescence signals.
NA: EPG-7 and SQST-1 aggregates are absent in wild-type embryos.
EPG-7 aggregates are absent in epg-7 and epg-7 double-mutant embryos.
ND, not determined.
EPG-7 aggregates colocalized with ATG-9::GFP puncta in epg-4, epg-6, and epg-8 mutants, and to a lesser extent in unc-51, epg-1, and atg-18 mutants, corroborating the association of EPG-7 with ATG-9 (Fig. 6, P–T; Table 2; Fig. S4, A–D). ATG-9::GFP and SQST-1 aggregates also colocalized in epg-8, epg-4, and epg-6 mutants (Fig. 6, P, U, and V; Fig. S4, E and F). In unc-51, epg-1, and atg-18 mutants, many ATG-9::GFP puncta were very closely associated with, but appeared not to overlap, SQST-1 aggregates at the ∼200-cell and comma stages (Fig. 6, P, W, and X; Table 2; Fig. S4, G and H). unc-51 mutants carrying a transgene expressing UNC-51(K39R), a putative kinase-deficient mutant, still showed partial colocalization of ATG-9::GFP puncta with EPG-7 and SQST-1 aggregates (Fig. 6 P). Loss of function of epg-7 caused no evident effect on the expression and distribution of EPG-1::GFP (Fig. S4, I–L). These results indicate that the UNC-51–EPG-1 complex regulates the association of EPG-7 with ATG-9 puncta and SQST-1 aggregates.
EPG-7 mediates autophagic degradation of several other types of protein aggregate
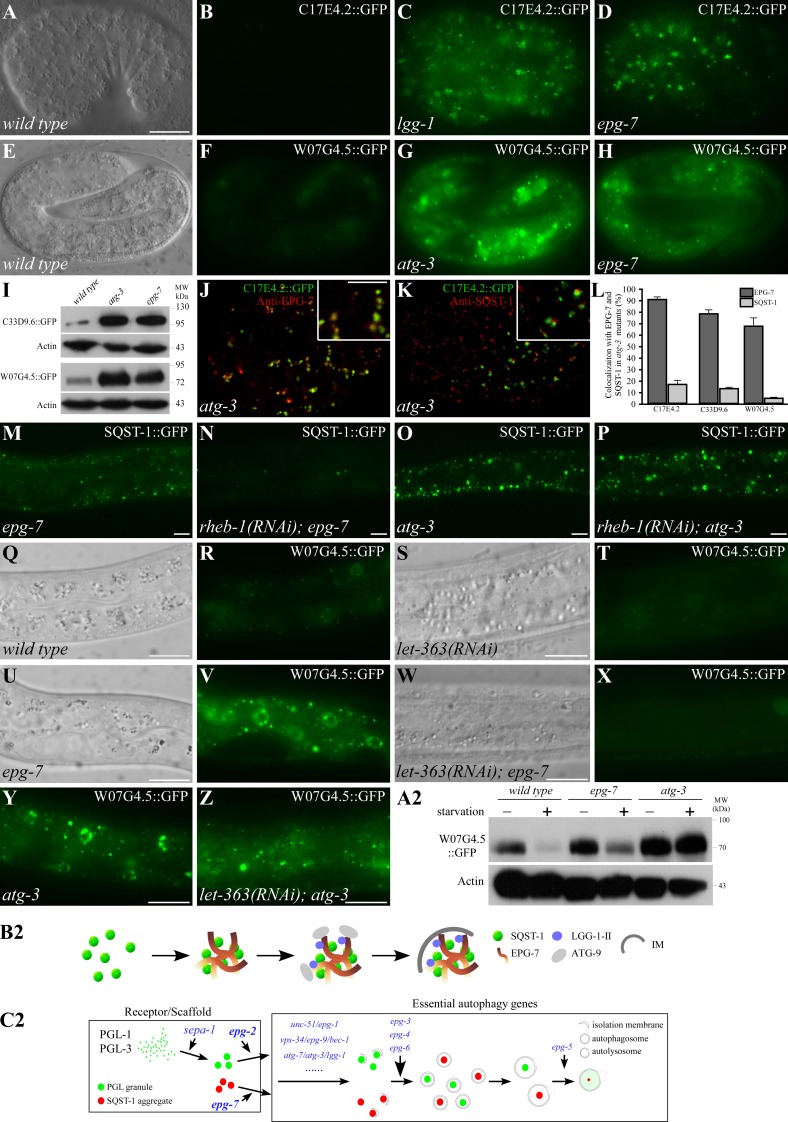
To investigate whether EPG-7 mediates degradation of substrates other than SQST-1 aggregates, we performed yeast two-hybrid screens and identified C17E4.2, C33D9.6, and W07G4.5 as EPG-7–interacting proteins. Homologues of C17E4.2 and C33D9.6 are not found in other organisms. w07g4.5 encodes a nematode-specific protein that is conserved in C. brenneri, C. remanei, and C. briggsae. Bioinformatic analysis indicated that these three proteins contain no known motifs. Loss of function of c17e4.2, c33d9.6, and w07g4.5 did not cause a defect in degradation of SEPA-1 and SQST-1 (Fig. S5, A–D; and unpublished data), indicating that they are not essential components of the autophagy pathway.
Translational fusion reporters were constructed to examine their expression patterns. C17E4.2::GFP and C33D9.6::GFP were weakly expressed and diffusely localized in the cytoplasm in early stage embryos (Fig. 7, A and B; Fig. S5 E). C17E4.2::GFP formed a few aggregates in the head region at the fourfold stage. W07G4.5::GFP was weakly expressed and diffuse in the cytoplasm before the comma stage. At late embryonic stages, W07G4.5 was present at elevated levels and formed aggregates in the intestine (Fig. 7, E and F). At post-embryonic stages, C17E4.2::GFP was weakly expressed in the intestine, vulva muscles, and a few cells in the head region (Fig. S5 G). C33D9.6::GFP was weakly expressed in multiple tissues, including muscles, hypodermal cells, and neurons in the ventral cord (Fig. S5 F). W07G4.5::GFP formed aggregates localized in the cytoplasm and also in the nuclei of intestinal cells at larval stages (Fig. S5 H).
Figure 7.
EPG-7 is not required for starvation-induced autophagic degradation of protein substrates. (A and B) C17E4.2::GFP is weakly expressed and diffusely localized in a wild-type embryo. (A) DIC image of the embryo shown in B. (C and D) C17E4.2::GFP is expressed at dramatically elevated levels and accumulates into many aggregates in lgg-1 (C) and epg-7 (D) mutant embryos. (E and F) W07G4.5::GFP is expressed in the intestine and a few aggregates are formed in a wild-type embryo at the fourfold stage. (E) DIC image of the embryo shown in F. (G and H) W07G4.5::GFP is expressed at dramatically elevated levels and accumulates into a large number of aggregates in atg-3 (G) and epg-7 (H) mutants. Images in B–D or F–H were taken using the same exposure time. (I) Levels of C33D9.6::GFP and W07G4.5::GFP are dramatically elevated in atg-3 and epg-7 mutant embryos. (J and K) C17E4.2 aggregates in atg-3 mutants overlap with EPG-7 aggregates (J), but are separable from SQST-1 aggregates (K). Insets show a magnified view. (L) Colocalization frequency of C17E4.2, C33D9.6, and W07G4.5 aggregates with EPG-7 and SQST-1 aggregates in atg-3 mutants. (M) SQST-1::GFP accumulates in hypodermal cells in epg-7 mutants at the larval stage. (N) SQST-1::GFP aggregates disappear in rheb-1(RNAi);epg-7 mutant larvae. (O) Dramatic accumulation of SQST-1::GFP aggregates in hypodermal cells in atg-3 mutant larvae. (P) SQST-1::GFP aggregates persist in rheb-1(RNAi);atg-3 mutant larvae. (Q–T) At larval stages, W07G4.5::GFP forms aggregates in intestinal cells (Q and R). These aggregates disappear upon RNAi knockdown of let-363 (S and T). (U–X) W07G4.5::GFP aggregates accumulate in intestinal cells in epg-7 mutant larvae (U and V). These aggregates disappear when let-363 is simultaneously depleted (W and X). (Y and Z) In atg-3 mutants, W07G4.5::GFP aggregates accumulate in intestinal cells (Y) and persist after RNAi knockdown of let-363 (Z). (A2) Levels of W07G4.5::GFP in wild type, epg-7 mutants, and atg-3 L1 larvae before and after 6 h starvation treatment. (B2) Model for the role of epg-7 in degradation of SQST-1 aggregates. SQST-1 directly interacts with EPG-7 and is recruited into EPG-7 aggregates. EPG-7 also associates with multiple Atg proteins, which may be recruited to the SQST-1–EPG-7 complex in a hierarchical order. IM, isolation membrane. (C2) Hierarchical relationship of the scaffold proteins EPG-2 and EPG-7 and other essential ATG and EPG proteins in the aggrephagy pathway. Bars: (A–H, J, and K) 10 µm; (J and K, insets) 5 µm; (M–Z) 10 µm.
The same transgenic lines were crossed into autophagy and epg-7 mutants. In lgg-1 and epg-7 mutants, C17E4.2::GFP was strongly expressed in most cells and formed numerous aggregates during embryogenesis (Fig. 7, C and D). C33D9.6::GFP was strongly expressed in multiple tissues including hypodermal cells, muscles, and neurons (Fig. S5, I–L). W07G4.5::GFP formed numerous aggregates in the intestine (Fig. 7, G and H). C33D9.6 and W07G4.5 expression levels were dramatically elevated in atg-3 and epg-7 mutants in an immunoblotting assay (Fig. 7 I). C17E4.2::GFP, C33D9.6::GFP, and W07G4.5::GFP aggregates largely colocalized with EPG-7, but were separable from SQST-1 aggregates in atg-3 mutants (Fig. 7, J–L; Fig. S5, M–Q). These results indicate that EPG-7 mediates degradation of protein aggregates distinct from SQST-1 aggregates.
epg-7 is dispensable for starvation-induced autophagic degradation of protein substrates
C. elegans embryogenesis does not rely on external nutrients. Autophagic degradation of SQST-1 during C. elegans embryogenesis is not regulated by mTOR signaling (Fig. S5, R–Y). Inactivation of let-363 (the C. elegans mTOR homologue) and rheb-1 had no effect on the accumulation of SQST-1 aggregates and W07G4.5 aggregates in epg-7 mutant embryos (Fig. S5, Z–C2). The development of larval animals relies on external nutrients, availability of which regulates autophagy activity (Kovács and Zhang, 2010). SQST-1 is weakly expressed and no aggregates are formed in wild-type larvae. SQST-1 aggregates dramatically accumulated in hypodermal and muscle cells in epg-7 and autophagy mutant larvae (Fig. 7, M and O). Starvation treatment or RNAi inactivation of let-363 and rheb-1 caused SQST-1 aggregates to largely disappear in epg-7 mutants (Fig. 7 N), but not in atg-3 mutants (Fig. 7 P). In wild-type animals, W07G4.5 formed aggregates in the larval intestine and the number of aggregates dramatically increased in epg-7 and atg-3 mutants (Fig. 7, Q, R, U, V, and Y). Upon starvation treatment or inactivation of Tor signaling, W07G4.5::GFP aggregates disappeared in wild type and epg-7 mutants, but remained abundant in atg-3 mutants (Fig. 7, S, T, W, X, and Z). Consistent with this, an immunoblotting assay showed that levels of W07G4.5::GFP in wild type and epg-7 mutants dramatically decreased after starvation but remained unchanged in atg-3 mutants (Fig. 7 A2). Similarly, starvation treatment resulted in the disappearance of C17E4.2::GFP and C33D9.6::GFP aggregates in epg-7 mutants, but had no effect in atg-3 mutants (Fig. S5, D2–K2; and unpublished data). In summary, EPG-7 is dispensable for starvation-induced autophagic degradation of protein substrates.
Discussion
Here we demonstrated that EPG-7 mediates the autophagic degradation of a set of protein aggregates during C. elegans embryogenesis. Compared with autophagy mutants, the number of SQST-1 aggregates is less in epg-7 mutants at the early embryonic stages, which could because early stage embryos contain a high level of autophagy contributed by the oocyte (Fig. S1, C and D) and SQST-1 is removed by autophagy in an EPG-7–independent manner. epg-7 is dispensable for starvation-induced autophagic degradation of protein substrates, which could be degraded nonselectively or via receptor–Atg8 binding. We propose that EPG-7 functions as a scaffold protein that augments degradation efficiency by the basal level of autophagy activity.
EPG-7 forms a complex with the cargo–receptor to provide a platform for recruiting Atg proteins for the formation of surrounding autophagosomal membranes, resembling the PAS in yeast and omegasomes in mammalian cells (Fig. 7, B2 and C2). In yeast, Atg11 and Atg17 function as scaffold proteins that directly interact with Atg1 and Atg9 to organize the PAS in the Cvt pathway and under autophagy-inducing conditions, respectively (Suzuki et al., 2007; Kawamata et al., 2008). Yeast Atg9 concentrates in vesicles and tubules, which move to the PAS and trigger the hierarchical recruitment of other Atg proteins that mediate the fusion of Atg9 compartments, leading to the formation of isolation membranes (Mari et al., 2010). The Atg17-dependent PAS localization of Atg9 and interaction between Atg17 and Atg9 requires Atg1 (Sekito et al., 2009). Atg1 kinase activity also affects the dynamics of Atg9 recruitment to the PAS (Cheong et al., 2008; Sekito et al., 2009). EPG-7 interacts with multiple Atg proteins, including ULK1 and ATG-9. During C. elegans embryogenesis, the early fusion of ATG-9–positive structures during autophagosome formation may be mediated by lipidated LGG-1 associated with the SQST-1–EPG-7 aggregate. Loss of function of the LGG-1 conjugation system abolishes the accumulation of ATG-9 puncta in epg-4, epg-6, atg-18, and epg-8 mutants. In the SQST-1 degradation pathway, the UNC-51–EPG-1 complex regulates association of EPG-7 with SQST-1 aggregates and ATG-9::GFP.
Scaffold proteins are used for selective transport of cargoes into autophagosome-like vesicles in other systems under growth conditions. Atg11 mediates the delivery of precursors of aminopeptidase I (Ape1) into the autophagosome-like Cvt vesicle (Nair and Klionsky, 2005). Atg11 is incorporated into the prApe1–Atg19 (receptor) complex (the Cvt complex) by interacting with Atg19 (Shintani et al., 2002). Atg11 transports both the Cvt complex and Atg9 to the PAS, where they recruit Atg proteins (Shintani et al., 2002; Yorimitsu and Klionsky, 2005; He et al., 2006). Loss of Atg11 function disrupts the association of Atg9 tubulo-vesicular clusters with the Cvt complex (Mari et al., 2010). Unlike EPG-7, Atg11 itself does not form aggregates and is released from the Cvt complex before it is transported to the vacuole (Kim et al., 2001; Yorimitsu and Klionsky, 2005). Similar to the role of EPG-7 in degradation of SQST-1 aggregates, Atg11 is dispensable for Cvt complex delivery under starvation conditions (Kim et al., 2001). In mammalian cells, p62 and NBR1 function as receptors for aggregation and degradation of polyubiquitinated proteins (Johansen and Lamark, 2011). The phosphatidylinositol 3-phosphate (PI(3)P)–binding protein Alfy has been suggested to act as a specific adaptor that creates a scaffold between p62-positive ubiquitinated protein aggregates and the autophagy proteins Atg5/Atg12/Atg16L and LC3 (Filimonenko et al., 2010). Loss of Alfy activity impairs autophagic clearance of ubiquitin-containing aggregates (Filimonenko et al., 2010; Clausen et al., 2010). Alfy differs from EPG-7 in several aspects. First, autophagic degradation of p62 itself is not dependent on Alfy (Clausen et al., 2010). Second, EPG-7 is degraded by autophagy in a manner independent of SQST-1, whereas Alfy is not a selective autophagy substrate and is only degraded by autophagy when associated with p62 bodies (Clausen et al., 2010). Third, EPG-7 is ubiquitously and diffusely localized in the cytoplasm, whereas Alfy forms aggregates, which in response to cellular stress, redistribute from the nucleus to cytoplasmic ubiquitin-containing protein aggregates in a p62-dependent manner (Simonsen et al., 2004; Clausen et al., 2010; Filimonenko et al., 2010). The physiological function of Alfy remains unknown. Our study indicates that under normal physiological conditions, a scaffold protein linking the cargo–receptor complex with the autophagic machinery endows target selectivity and also promotes autophagic degradation efficiency.
Materials and methods
Strains
The following strains were used in this work: epg-7(tm2508), atg-2(bp576), atg-3(bp412), epg-1(bp414), unc-51(e1189), atg-18(gk378), atg-9(bp564), atg-5(bp546), epg-8(bp251), epg-3(bp405), epg-4(bp425), epg-5(tm3425), epg-6(bp242), sqst-1(ok2892), mec-4(u231), bpIs151(sqst-1::gfp, unc-76), bpIs236(epg-7::gfp, rol-6), bpIs126(f44f1.6::gfp, rol-6), bpIs120(zk1053.3::gfp, rol-6), bpIs238(epg-7::gfp, unc-76), bpIs239(w07g4.5::gfp, unc-76), bpIs242(c33d9.6::gfp, unc-76), bpIs244(c17e4.2::gfp, unc-76), bpIs129(t04d3.1::gfp, rol-6), bpIs128(zk1053.4::gfp, rol-6), bpIs132(c35e7.6::gfp, rol-6), bzIs8(Pmec-4::gfp), and bpIs211(Pnfya-1::atg-9::gfp, unc-76). All experiments were performed at 20°C unless otherwise noted.
Isolation, mapping, and cloning of epg-7
Animals carrying the sqst-1::gfp (bpIs151) reporter were mutagenized with ethyl methanesulfonate. Embryos derived from F2 animals were examined. From ∼10,000 genomes screened, six epg-7 mutant alleles were isolated, in which SQST-1::GFP accumulated into a large number of aggregates from the ∼100-cell stage and the aggregates persisted throughout embryogenesis.
The genetic position of epg-7 was determined by three-factor mapping. From the cross between epg-7 and unc-6 (−2.08) dpy-6 (−0.00) (carrying the sqst-1::gfp reporter) animals, 23 out of 38 Dpy non-Unc and 17 out of 52 Unc non-Dpy animals carried the epg-7 mutation. A transgene containing fosmid WRM0640bA04, subsequently delineated to the single gene t08a9.1 (T08A9, nt 20964–28120, including 2110 bp promoter), rescued the defective degradation of SQST-1 aggregates in epg-7(tm2508) mutants. The molecular lesions in epg-7(bp694, bp586, bp562, bp625, bp547, or bp676) were determined by genomic sequencing. bp586, bp562, bp625, and bp676 mutants have a glutamine to stop codon mutation at residues 809, 852, 1006, and 1213, respectively. bp547 contains a tryptophan to stop codon mutation at amino acid 1076. The alanine at amino acid 614 is mutated to threonine in bp694 mutants.
Indirect immunofluorescence
Embryos were collected by cutting 30–40 gravid hermaphrodites, permeabilized by freeze-cracking, and fixed with methanol/acetone (−20°C methanol for 15 min, −20°C acetone for 15 min). For immunostaining, the slides were blocked with PBS (containing 1% BSA + 1% bovine serum) for 1 h and then incubated with primary antibodies for 1–2 h at room temperature (RT), followed by 1 h incubation with FITC or rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). After final wash, samples were mounted with Vectashield (Vector Laboratories). The following primary antibodies were used: polyclonal rat anti-SQST-1, polyclonal rat or rabbit anti–EPG-7, polyclonal rat or rabbit anti–LGG-1, and polyclonal rabbit anti–SEPA-1. Slides were viewed using a confocal microscope (LSM 510 Meta; Carl Zeiss) with a 63×/1.40 oil-immersion objective lens (Plan-Apochromat; Carl Zeiss) and a camera (AxioCam HRm; Carl Zeiss) at RT. Images were processed and viewed using LSM Image Browser software (Carl Zeiss).
Live embryos and worms were examined by light microscopy (Imager A1; Carl Zeiss) a with 40×/0.75 objective lens (Plan-NeoFluar; Carl Zeiss), a 100×/1.40 oil-immersion objective lens (Plan-Apochromat; Carl Zeiss), and a camera (AxioCam MRm; Carl Zeiss) at RT. Images were processed and viewed using AxioVision v4.6.3.0 software (Carl Zeiss).
Reporter construction
Reporters for epg-7, sqst-1, c17e4.2, c33d9.6, and w07g4.5 with gfp inserted at the C-terminal end were constructed by a PCR fusion-based approach or by subcloning. The promoter region of epg-7 (T08A9, nt 20975–23073) was inserted between the SphI and XbaI sites of the pPD95.79 vector, then the full-length epg-7 cDNA was inserted between the BamHI and KpnI sites to construct the translational fusion reporter with gfp inserted at the C terminus. A translational fusion reporter for sqst-1 was constructed by cloning the promoter region of sqst-1 (T12G3, nt 5195–7043) between the SalI and BamHI sites of pPD95.79. The full-length sqst-1 cDNA was further inserted into this vector between the AscI and XmaI sites. The constructs were co-injected with pRF4(rol-6[su1006]) into wild-type animals.
epg-7(del 531–631)::gfp, epg-7(del 762–851)::gfp, epg-7(del 852–1003)::gfp, epg-7(del 1004–1142)::gfp, sqst-1(del 26–181)::gfp, sqst-1(del 418–499)::gfp, sqst-1(del 604–630)::gfp, and sqst-1(del 418–499+604-630)::gfp were made using a PCR-based method to introduce truncations into the epg-7 or sqst-1 reporter. These constructs were injected into wild-type, epg-7, or sqst-1 animals at the concentration of 5 ng/ul.
The DNA sequences for constructing reporters for c17e4.2, c33d9.6, and w07g4.5 were: c17e4.2 (C17E4, nt 3696–11510, including 1923 bp promoter), c33d9.6 (C33D9, nt 7602–3305, including 1886 bp promoter), and w07g4.5 (W07G4, nt 25540–29151, including 1919 bp promoter). The PCR fusion products with gfp inserted at the C terminus were co-injected with unc-76 expression vector into unc-76(e911) animals.
At least two stable transgenic lines were analyzed for each reporter.
RNAi experiments
The DNA template used for RNA synthesis was lgg-1 (C32D5: nt 35232–35666).
Yeast two-hybrid assay
Coding cDNA fragments of EPG-7 (amino acids 1–634 or 596–1330) were fused to the GAL4-binding domain in the vector pPC97 between SalI and NotI sites and then transformed into the yeast host strain Mav20 before screening a C. elegans cDNA library cloned in pPC86. From 800,000 clones screened, 36 clones interacting with EPG-7 were identified. Among them, 5 clones corresponded to C17E4.2, 5 clones corresponded to C33D9.6, and 18 clones corresponded to W07G4.5.
In vitro pull-down assay
cDNAs encoding full-length or truncated EPG-7, SQST-1, LGG-1, LGG-3, ATG-9, ATG-18, and UNC-51 were cloned into pET-28a (for His tagging), pGEX-6p-1 (for GST fusion), or pMal-C2X (for MBP fusion). GST fusion proteins or MBP fusion proteins were incubated with His-tagged proteins and glutathione Sepharose beads (for GST fusion proteins) or Amylose resin (for MBP fusion proteins). Bound proteins were analyzed by Western blot using an anti-His antibody. 10% of the fusion protein used for pull-down was shown as input.
In vivo coimmunoprecipitation assay
Extracts of embryos or embryos carrying an atg-9::HA transgene were immunoprecipitated with anti-EPG-7 polyclonal antibody or anti-LGG-1 antibody, then incubated with 30 µl protein G–Sepharose beads. After extensive washes, the immunoprecipitates were analyzed by Western blotting using an appropriate antibody. Anti–SEA-2 antibody was used as control IgG. Levels of actin show that equal amounts of extracts were used for immunoprecipitation.
Antibody preparation
To generate antibodies, cDNA encoding a fragment of EPG-7 (amino acids 705–1003) was cloned into the pET-28a vector, expressed as a His-tagged fusion protein in E. coli BL21 and purified for use as an immunogen in rats. In epg-1;epg-7(tm2508) or atg-3;epg-7(bp694) double mutants no EPG-7 aggregates were detected, confirming the specificity of the antibody.
Starvation assay
Embryos were collected by bleaching old adult hermaphrodites and were then incubated in 3 ml of sterilized water at 20°C. At the indicated time point, an aliquot from each sample tube was placed on a plate seeded with E. coli OP50. After 3 d, the number of L4 larvae or adults was counted. The number of animals on day 0 of starvation was used as the denominator to calculate the percentage of animals that survived starvation.
Brood size
The total number of embryos produced by one hermaphrodite was scored as brood size. A single L4 animal was transferred to a fresh plate twice a day during the reproductive period. Three animals were examined for each genotype.
Hatch rate
Adult animals were transferred to a new plate to produce embryos for 1 h and then removed. The number of embryos was used as the denominator to calculate the percentage of animals that hatched to L1 larvae.
Developmental time
Adult animals were transferred to a new plate to produce embryos for 1 h and then removed. Time for embryos developing into L1 larvae or growing into adults was determined as 90% of worms growing into the first-larval stage (L1) and the young adult stage, respectively. The experiment was performed at least three times.
Life span
Adult animals were transferred to a new plate to produce embryos for 1 h and then removed. Lifespan of the progeny was examined. Day 0 was defined as the day when the adults were removed. Animals were transferred to fresh plates every day during the reproductive period and every 4–5 d thereafter. Animals were scored as dead if they failed to respond to a gentle tap on the head and tail with a platinum wire. Worms with exploded vulva, or that had bagged (died from internal hatching) or crawled off the plate were excluded. Three independent assays were tested for each experiment.
Oxidative stress assay
1-d-old adults were immersed in M9 buffer containing 5 mM hydrogen peroxide and incubated at 20°C. Worms that did not move in 10 s in M9 solution were scored as dead. The number of dead worms was scored every hour until all worms were dead. 50 worms from each strain were treated, and the experiment was performed three times.
Heat stress assay
2-d-old adults were shifted to 35°C for 4, 5, 6, or 7 h. At the indicated time, plates were returned to 20°C to allow recovery for 1 h and animals were examined for touch-provoked movement and pharyngeal pumping. Worms not displaying these traits were scored as dead. 50 worms from each strain were examined, and the experiment was performed three times.
Dauer formation assay
Adult animals were transferred to a new plate to produce embryos for 1 h and then removed. The plate was kept at 20°C until embryos hatched to L1 larvae. The L1 population was then reared at 21°C. The plates were scored after 72 h. The number of total animals was used as the denominator to calculate the percentage of animals that entered the dauer stage.
Statistical analysis
Student’s t test was performed for statistical analysis of mec-4::gfp-labeled touch neurons, brood size, hatch rate, dauer formation, heat stress, and developmental time. The difference in L1 survival, life span, and oxidative stress was compared using the Kaplan-Meier method and log-rank tests. All statistical tests are two-sided and are performed using SPSS version 16.0.
Online supplemental material
Fig. S1 shows defective degradation of SQST-1–containing aggregates in epg-7 mutants. Fig. S2 shows colocalization of EPG-7 with SQST-1 aggregates in autophagy mutants. Fig. S3 shows localization of SQST-1 aggregates and LGG-1 puncta in various autophagy mutants. Fig. S4 shows the relationship of ATG-9::GFP with EPG-7 aggregates and SQST-1 aggregates in various autophagy mutants. Fig. S5 shows that EPG-7 is dispensable for starvation-induced autophagic degradation of protein substrates. Table S1 shows autophagy genes in yeast, C. elegans, and mammals. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201209098/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201209098.dv.
Supplementary Material
Acknowledgments
We are grateful to X. Wang and members of Hong Zhang’s laboratory for their helpful comments on the manuscript, NIBS Antibody Center for generating anti-EPG-7 antibodies, and I. Hanson for editing work. We thank S. Matani for the tm2508 allele.
This work was supported by the National Basic Research Program of China (2013CB910100, 2011CB910100) and also a grant from the National Natural Science Foundation of China (31225018) to Hong Zhang. The research of Hong Zhang was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- LIR
- LC3-interacting region
- MBP
- maltose-binding protein
- PAS
- preautophagosomal structure
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D.J. 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 19:668–681 10.1091/mbc.E07-08-0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.H., Lamark T., Isakson P., Finley K., Larsen K.B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., Johansen T. 2010. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 6:330–344 10.4161/auto.6.3.11226 [DOI] [PubMed] [Google Scholar]
- Filimonenko M., Isakson P., Finley K.D., Anderson M., Jeong H., Melia T.J., Bartlett B.J., Myers K.M., Birkeland H.C., Lamark T., et al. 2010. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell. 38:265–279 10.1016/j.molcel.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.L., Legakis J.E., Klionsky D.J. 2006. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 175:925–935 10.1083/jcb.200606084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Baba M., Cao Y., Klionsky D.J. 2008. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell. 19:5506–5516 10.1091/mbc.E08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Johansen T., Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:279–296 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 19:2039–2050 10.1091/mbc.E07-10-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kamada Y., Stromhaug P.E., Guan J., Hefner-Gravink A., Baba M., Scott S.V., Ohsumi Y., Dunn W.A., Jr, Klionsky D.J. 2001. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153:381–396 10.1083/jcb.153.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Kovács A.L., Zhang H. 2010. Role of autophagy in Caenorhabditis elegans. FEBS Lett. 584:1335–1341 10.1016/j.febslet.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Longatti A., Tooze S.A. 2009. Vesicular trafficking and autophagosome formation. Cell Death Differ. 16:956–965 10.1038/cdd.2009.39 [DOI] [PubMed] [Google Scholar]
- Lu Q., Yang P.G., Huang X.X., Hu W.Q., Guo B., Wu F., Lin L., Kovács A.L., Yu L., Zhang H. 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 21:343–357 10.1016/j.devcel.2011.06.024 [DOI] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., Reggiori F. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U., Klionsky D.J. 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 280:41785–41788 10.1074/jbc.R500016200 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584:1379–1385 10.1016/j.febslet.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Orsi A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., Tooze S.A. 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 23:1860–1873 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T., Kawamata T., Ichikawa R., Suzuki K., Ohsumi Y. 2009. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 14:525–538 10.1111/j.1365-2443.2009.01299.x [DOI] [PubMed] [Google Scholar]
- Shintani T., Huang W.P., Stromhaug P.E., Klionsky D.J. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 3:825–837 10.1016/S1534-5807(02)00373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Birkeland H.C., Gillooly D.J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. 2004. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 117:4239–4251 10.1242/jcs.01287 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Tian E., Wang F.X., Han J.H., Zhang H. 2009. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 5:608–615 10.4161/auto.5.5.8624 [DOI] [PubMed] [Google Scholar]
- Tian Y., Li Z.P., Hu W.Q., Ren H.Y., Tian E., Zhao Y., Lu Q., Huang X.X., Yang P.G., Li X., et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 141:1042–1055 10.1016/j.cell.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Yang Z., Klionsky D.J. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22:124–131 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D.J. 2005. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 16:1593–1605 10.1091/mbc.E04-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhang Y.X., Yan L.B., Zhou Z., Yang P.G., Tian E., Zhang K., Zhao Y., Li Z.P., Song B., Han J.H., et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.