To elucidate mechanisms by which RUNX1–ETO, the common acute myeloid leukemia t(8;21) fusion protein, primes hematopoietic cells for oncogenic transformation, we previously carried out Affymetrix gene expression profiling of RUNX1–ETO-expressing human CD34+ progenitor cells.1 We identified the MYC target gene, MYCT1, as a gene significantly upregulated at an early developmental stage that could be integral to RUNX1–ETO-mediated leukemic phenotype (Figure 1). To determine whether increased MYCT1 expression recapitulates the RUNX1–ETO phenotype, we performed retroviral transduction of MYCT1 as a single molecular abnormality into hematopoietic cell lines and normal human cord blood-derived CD34+ progenitor cells. In the absence of commercially available MYCT1 antibody, retroviral expression vectors, containing full-length MYCT1 coding sequence with (MYCT1-HA) or without (MYCT1) the addition of a C-terminal HA antigen tag, were also generated (Supplementary Materials and methods and Supplementary Figure S1), and transgenic MYCT1 over-expression was confirmed at the mRNA and protein levels (Supplementary Figure S2). Functional validation (Supplementary Figure S3) was provided by replication of the reduced viability of MYCT1-expressing murine myeloid 32D cells in the absence of WEHI-3B-conditioned medium reported by Yin et al.2
Figure 1.
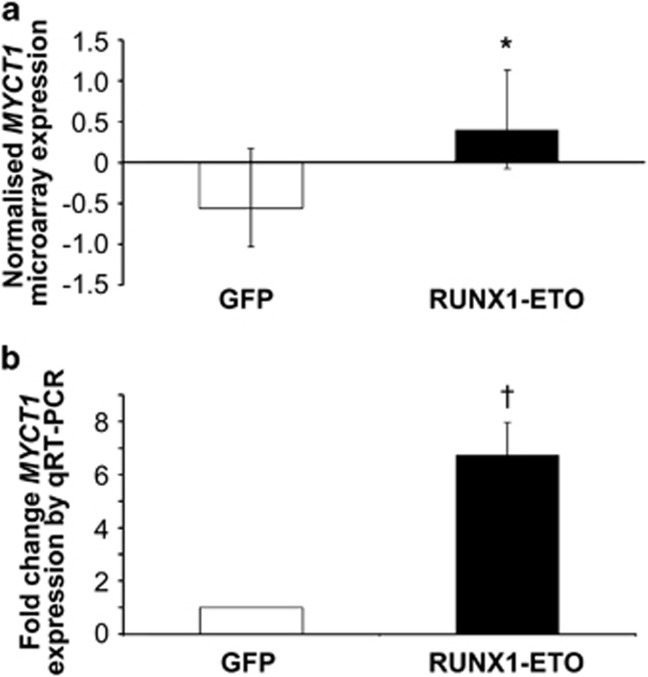
Up-regulation of MYCT1 expression by RUNX1–ETO in human CD34+ hematopoietic progenitor cells. Normal human CD34+ hematopoietic progenitor cells were purified from human cord blood and infected with RUNX1–ETO or control vector expressing GFP alone. (a) Total RNA from pair-matched RUNX1–ETO and GFP control transduced cells was extracted by lysis in Trizol on day 3 of culture and subjected to Affymetrix microarray analysis to determine differential MYCT1 gene expression. (b) Quantitative RT–PCR (qRT–PCR) analysis of MYCT1 expression in GFP control and RUNX1–ETO transduced cells. Normalised expression values were determined by crossing point analysis of MYCT1 signal compared with ABL housekeeping gene control in duplicate samples. Fold change was calculated using the 2−ΔΔCt method. Data represents mean±1 s.d. (n>3). Statistical significance was calculated by unpaired t-test, *P<0.05 and paired t-test, †P<0.01.
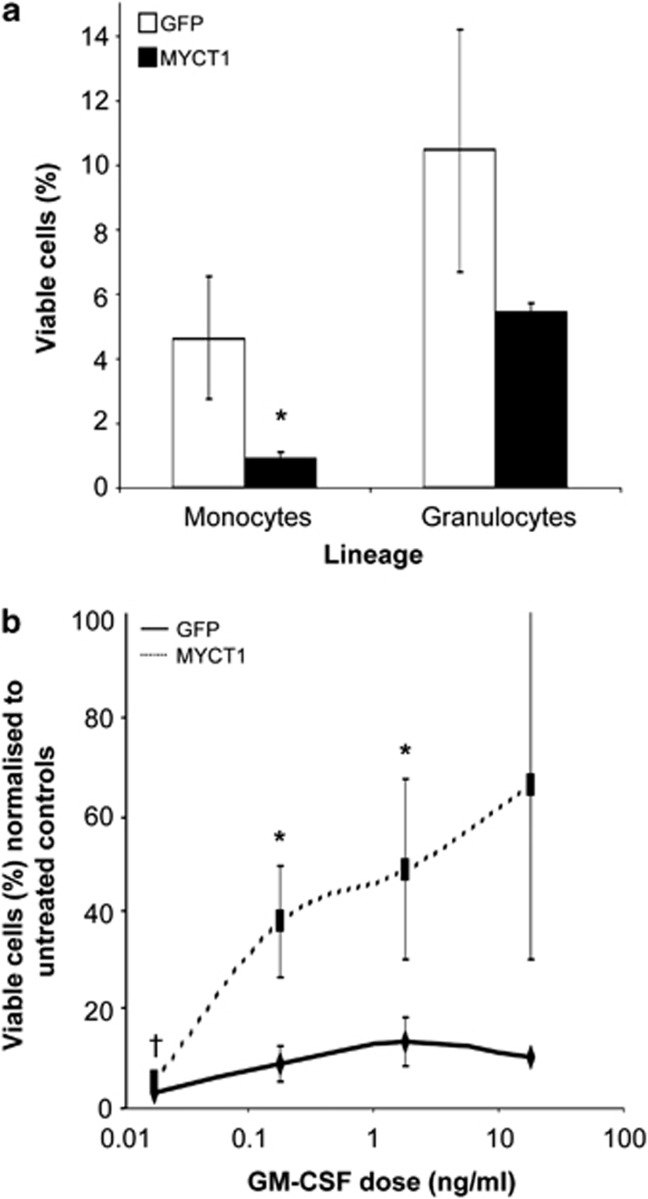
Increased sensitivity to apoptotic stimuli has been reported for both RUNX1–ETO-expressing cells and cells in which MYC signalling is deregulated.3 To determine the contribution of altered MYCT1 expression to primary myeloid progenitor cell viability, we performed cell survival assays using key hematopoietic cytokines. As shown in Figure 2a, cells transduced with MYCT1 showed significantly reduced intrinsic viability in the absence of serum and cytokine supplements: a five-fold reduction in number of viable MYCT1-expressing monocytes compared with GFP controls was measured after 48 h in culture. A lesser two-fold reduction in viable MYCT1-expressing granulocytes was also measured, suggesting that MYCT1 expression impacts on the survival of both lineages. Taken in combination with the reduced viability of MYCT1-expressing mouse 32D cells in the absence of IL-3, these results indicate that MYCT1 expression negatively impacts on survival or alters the threshold of trophic signalling required to prevent the activation of apoptosis in myeloid cells. MYCT1-expressing cells cultured in the presence of IL-3, SCF, G-CSF, IL-6, TNF-α or TPO experienced slightly reduced or unchanged viability compared with GFP control cells that only reached statistical significance at specific doses in cells of monocyte lineage (Supplementary Figure S3). In striking contrast, MYCT1-expressing monocytes demonstrated a notable increase in cell survival responses to GM-CSF over a wide range of doses (Figure 2b). The mean dose of GM-CSF required to support 50% maximal cell survival (defined as the number of live cells measured in the highest dose culture) was three-fold lower for MYCT1-expressing monocytes than GFP controls (data not shown). An increase in granulocytic cell survival was also observed but was not statistically significant (data not shown). These experiments unveil GM-CSF as unique amongst all cytokines assayed in its ability to counteract the substantially-diminished intrinsic viability of MYCT-expressing cells. MYCT1 may, therefore, be involved in the translation of anti-apoptotic signals from the GM-CSF receptor in myeloid cells and amplify the GM-CSF signalling that supports abnormal granulopoiesis in RUNX1–ETO-expressing cells,4 promoting monocyte lineage in the absence of this oncoprotein. The different magnitude in responses between granulocytes and monocytes may reflect the differential activation of STAT3 and STAT5 downstream signalling components in these lineages.5
Figure 2.
GM-CSF supports enhanced survival of MYCT1-expressing monocytes that show reduced intrinsic viability. MYCT1 and GFP control transduced day 4 hematopoietic progenitor cells were subjected to survival assays in the absence of serum. (a) Untreated MYCT1-expressing monocytes and granulocytes show reduced intrinsic viability compared with GFP control cells. The proportion of viable cells (%) was calculated as the number of viable monocytes or granulocytes recovered after 48 h/equivalent input populations at time zero. (b) MYCT1-expressing monocytes have increased viability in a broad range of GM-CSF doses compared with GFP control cells. The proportion of viable cells (%) in each cytokine dose was normalised to the proportion of viable cells in the absence of cytokine (untreated cells) after 48 h. Data represents mean plus±1 s.d. (n=3) and statistical significance was calculated by paired t-test, *P<0.05 and †P<0.01.
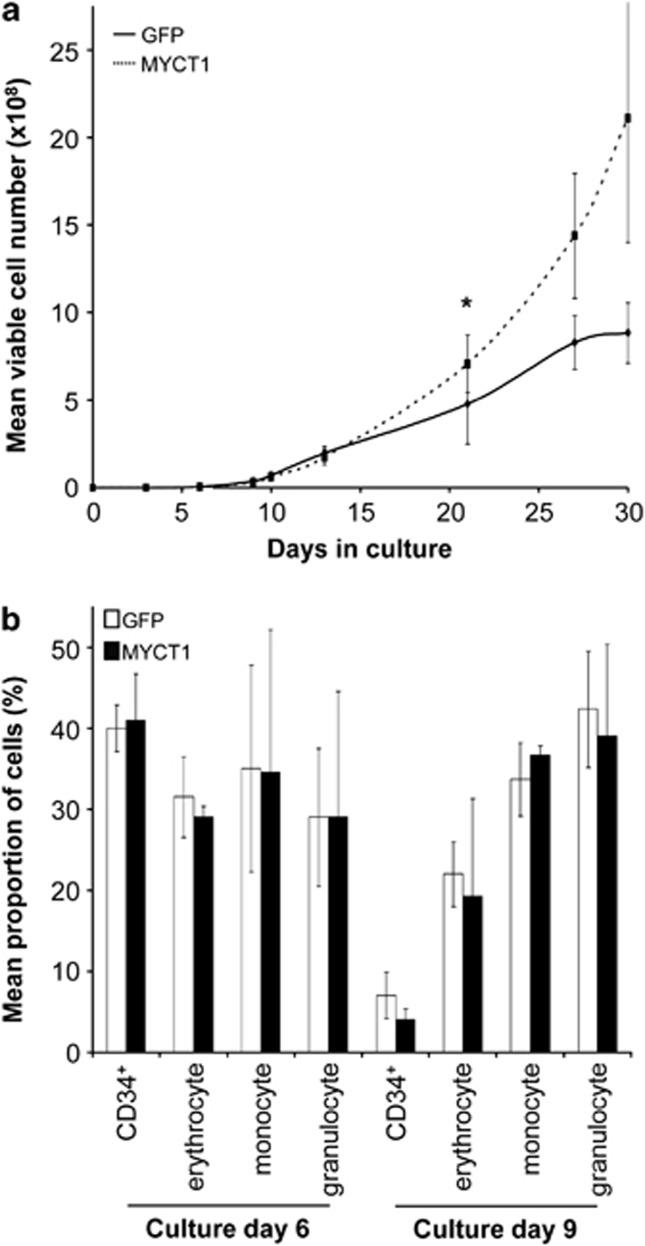
To determine whether MYCT1 expression alters hematopoietic cell proliferative capacity as well as intrinsic viability, we performed bulk culture of MYCT1-expressing and GFP control populations, measuring live cell numbers over 30 days. Interestingly, no differences in viability of MYCT1-expressing cells were detected under these conditions of cytokine and serum sufficiency and, instead, a moderate but significant increase in population growth was observed over time (1.5-fold compared with GFP control after 21 days; Figure 3a). This MYCT-driven expansion is in contrast to the absence of growth reported previously in MYCT1-expressing murine fibroblasts.2 This disparity may reflect the context-dependent differential activity of MYCT1, highlighting an important role for this transcription factor in hematopoiesis. However, the former report followed cell growth only as far as 5 days, so that late-onset or more modest influences of MYCT1 may have been missed. Distinct from RUNX1–ETO-expressing CD34+ cells,6 there was no transient growth inhibition detected for MYCT1-expressing cells, suggesting that MYCT1 exerts disparate effects on growth regulatory networks from RUNX1–ETO. The dual effects of increased growth and propensity to apoptosis promoted by MYCT1 expression are consistent with a role in MYC-mediated cellular transformation.
Figure 3.
MYCT1 expression promotes population expansion without altering lineage distributions in bulk liquid culture. Following retroviral transduction, MYCT1 and GFP control transgenic CD34+ cells were cultured in medium supplemented with 5 ng/ml IL-3, SCF, G-CSF and GM-CSF for 30 days at 37 °C. At the time-points indicated, liquid cultures were analysed by four-colour immunophenotypic analysis. (a) MYCT1-expressing hematopoietic progenitor cells demonstrate increased growth over 30 days in culture compared with GFP control cells. (b) MYCT1-expressing and GFP control cells generate analogous proportions of erythrocyte (CD13lo CD36hi), monocyte (CD13hi CD36hi) and granulocyte (CD13hi CD36lo) populations on days 6 and 9 of bulk culture. There is no differential retention of the CD34+ progenitor cell marker. Data represents mean plus±1 s.d. (n=3) and statistical significance was calculated by paired t-test, *P<0.05 and †P<0.01.
The phenotype of MYCT1-expressing cells in bulk culture was assessed by multicolour flow cytometry to discern any lineage-selective proliferative activity to parallel the increased survival of MYCT1-expressing monocytes observed in survival assays. We have previously demonstrated retention of CD34 hematopoietic progenitor cell marker expression by RUNX1–ETO-expressing cells, as well as a reduction in maturing granulocytes characteristic of t(8;21) leukaemia.6 However, we detected no significant shift in the distributions of monocyte, granulocyte and erythrocyte populations in bulk cultures of MYCT1-expressing cells (Figure 3b), and the downregulation of CD34+ expression between day 6 and 9 of culture was comparable with control populations and in accordance with normal programs of lineage maturation. Equivalent proportions of monocyte and granulocyte populations measured in MYCT1 and GFP control cultures were sustained beyond the time at which detectable differences in overall growth were detected (day 21; Figure 3a and data not shown), suggesting that the expansion of MYCT1-expressing cultures over time is not lineage-specific. These data indicate that MYCT1 over-expression does not maintain cells in an undifferentiated state or alter normal lineage commitment and maturation in these culture conditions.
To determine any selective promotion of monocyte self-renewal under more restrictive conditions, we performed colony-forming assays by limiting dilution. Although the expression of MYCT1 did not significantly affect the overall colony-forming efficiency, we found a consistent trend for increased frequency (1.2-fold) of monocyte colonies compared with control cells (Supplementary Figure S4A). Interestingly, we also measured an increase in mean size of MYCT1-expressing monocyte colonies, which were 1.5-fold bigger than GFP control monocyte colonies (Supplementary Figure S4B). There was little discernible effect on size in the other lineages examined, corroborating our findings of greater impact of aberrant MYCT1 expression for the monocyte lineage under conditions of restricted trophic support. These results are distinct from those we have obtained using RUNX1–ETO-expressing cells, where a suppression of granulocyte colony formation without concomitant increase in monocyte colony-forming capacity was determined.6 However, we did observe a more than two-fold increase in the numbers of monocyte colonies forming from RUNX1–ETO-expressing cells plated at later hematopoietic stages of development (days 9 and 16) compared with GFP controls. The impact of MYCT1 expression for monocyte colony frequency and size also parallels the enhanced self-generative capacity of macrophage progenitor cells and monocytic tumour development reported in mice expressing low levels of a MYC transgene.7 Thus, effects on monocyte/macrophage renewal are seen with RUNX1–ETO and MYC as well as MYCT1 deregulation.
In summary, these results suggest MYCT1 transcription can be activated downstream of RUNX1–ETO as well as MYC, contributing to key aspects of the leukaemic phenotypes these oncoproteins promote, as well as the specific promotion of monocyte survival and sensitivity to GM-CSF. We have recently uncovered an analogous downregulation of the lineage-modulating transcription factor, GFI1B8 by both RUNX1–ETO and MYCT1 in human hematopoietic progenitor cells, which is mirrored in t(8;21) acute myeloid leukemia patients (Supplementary Figure S5 and Supplementary Table 1). This novel gene network highlights MYCT1 as a key node in aberrant RUNX1–ETO and MYC signalling and merits further investigation as a potential mechanism for the myeloid expansion and maturation block that characterises acute myeloid leukemia disease.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Tonks A, Pearn L, Musson M, Gilkes A, Mills KI, Burnett AK, et al. Transcriptional dysregulation mediated by RUNX1-RUNX1T1 in normal human progenitor cells and in acute myeloid leukaemia. Leukemia. 2007;21:2495–2505. doi: 10.1038/sj.leu.2404961. [DOI] [PubMed] [Google Scholar]
- Yin X, Grove L, Rogulski K, Prochownik EV. Myc target in myeloid cells-1, a novel c-Myc target, recapitulates multiple c-Myc phenotypes. J Biol Chem. 2002;277:19998–20010. doi: 10.1074/jbc.M200860200. [DOI] [PubMed] [Google Scholar]
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- Kita K, Shirakawa S, Kamada N. Cellular characteristics of acute myeloblastic leukemia associated with t(8;21)(q22;q22). The Japanese Cooperative Group of Leukemia/Lymphoma. Leuk Lymphoma. 1994;13:229–234. doi: 10.3109/10428199409056286. [DOI] [PubMed] [Google Scholar]
- Marvin J, Swaminathan S, Kraker G, Chadburn A, Jacobberger J, Goolsby C. Normal bone marrow signal transduction profiles: a requisite for enhanced detection of signaling dysregulations in AML. Blood. 2011;117:e120–e130. doi: 10.1182/blood-2010-10-316026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks A, Tonks AJ, Pearn L, Pearce L, Hoy T, Couzens S, et al. Expression of AML1-ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia. 2004;18:1238–1245. doi: 10.1038/sj.leu.2403396. [DOI] [PubMed] [Google Scholar]
- Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood. 2006;108:653–661. doi: 10.1182/blood-2006-01-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong B, Grimes HL, Yang TY, Bear SE, Qin Z, Du K, et al. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol Cell Biol. 1998;18:2462–2473. doi: 10.1128/mcb.18.5.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.