Abstract
Human securin, also known as human pituitary tumor-transforming gene 1 (pttg1), plays a key role in cell-cycle regulation. Two homologous genes, pttg2 and pttg3, have been identified although very little is known about their physiological function. In this study, we aimed at the characterization of these two pttg1 homologs. Real-time PCR analysis using specific probes demonstrated that Pttg2 is expressed at very low levels in various cell lines and tissues whereas Pttg3 was largely undetectable. We focused on the study of Pttg2 and found that, unlike PTTG1, PTTG2 lacks transactivation activity and does not bind to separase, making improbable a role in the control of sister chromatids separation. To further investigate the biological role of pttg2, we used short hairpin RNA inhibition of Pttg2 and found that cells with reduced Pttg2 levels assumed a rounded morphology compatible with a defect in cell adhesion and died by apoptosis in a p53- and p21-dependent manner. Using microarray technology, we generated a gene expression profile of Pttg2-depleted cells versus wild-type cells and found that knockdown of PTTG2 results in concomitant downregulation of E-cadherin and elevated vimentin levels, consistent with EMT induction. The observation of aberrant cellular behaviors in Pttg2-silenced cells reveals functions for pttg2 in cell adhesion and provides insights into a potential role in cell invasion.
Keywords: Pttg/securin, EMT, E-cadherin, Anoikis
The securin family of proteins play a key role in the regulation of sister chromatids separation during mitosis.1 These proteins have been identified in Drosophila (Pimples proteins), fission yeast (CUT2), budding yeast (PDS1) and vertebrates (Securin/Pttg1). In addition to its role during mitosis, PTTG1 has the capacity to transform murine fibroblasts in vitro, it promotes tumor formation in athymic mice and it is abundantly expressed in different cancer cell lines and tumors, including pituitary adenomas, primary epithelial neoplasms and hematopoietic malignancies.2
Human securin prevents premature chromosome segregation through the inhibition of separase, a protease that cleaves cohesin, which links sister chromatids.3 Deregulated securin may thereby result in chromosome missegregation leading to an increase in proto-oncogene dose or to loss of heterozygocity of tumor suppressors.4 The pttg1 gene locates at chromosome 5q33 and consists of five introns and six exons. In higher mammals, two homologous intronless genes, pttg2 and pttg3, which are located on chromosomes 4p12 and 8q13, respectively, have been reported.5, 6 The homology between the three genes is very high, being PTTG1 89% identical to PTTG3 and 91% identical to PTTG2, although PTTG2 and PTTG3 are only 84% identical at the amino-acid level. PTTG2 and PTTG3 contain several conserved motifs present in PTTG1 including two proline-rich domains at the C-terminus, suggesting that these genes might mediate their functions through the interaction with other proteins. In addition, the destruction box present in PTTG17 is also conserved in both proteins but not the KEN box in the case of PTTG2, which could affect the stability of this protein. RT-PCR studies have shown that Pttg1 is the most abundant and mainly expressed in normal adult testis and thymus.8 Pttg2 mRNA is expressed at low levels in various cell lines and in normal and tumor tissues.5, 6 In contrast, the expression of Pttg3 by northern blotting revealed no expression of Pttg3 in human tissues. In fact, pttg3 has been recently classified as a pseudogene in different studies.9
Epithelial-to-mesenchymal transition (EMT) is a key event in embryonic morphogenesis involving the expression of several EMT-associated genes. In addition, EMT occurs during the progression of some human cancer providing motility and invasiveness to cancer cells.10, 11 EMT-inducing genes with essential roles in EMT include twist,12 zeb1,13 or snail/slug.14, 15 These genes function as direct (snail/slug and zeb1) or indirect (twist) transcriptional repressors of E-cadherin whose functional loss is one of the hallmarks of EMT. Other changes associated to EMT include loss of cell–cell adhesion, altered polarity, rearrangement of cytoskeletal proteins (intermediate filaments revert from keratins to vimentin) and resistance to cell death-induced by loss of cellular attachment (anoikis).
Given the lack of studies of Pttg2, we sought to complete a detailed characterization of this gene in order to reveal its putative cellular functions. We found that depletion of PTTG2 led to cell rounding followed by p53- and p21-dependent apoptosis. Also, we observed that the lack of PTTG2 results in low E-cadherin and high vimentin levels, which are associated with the induction of EMT. This work reveals novel aspects of Pttg2 and attributes an important role for Pttg2 in cell adhesion and EMT induction.
Results
Characterization of Pttg2-silenced HCT116 cells
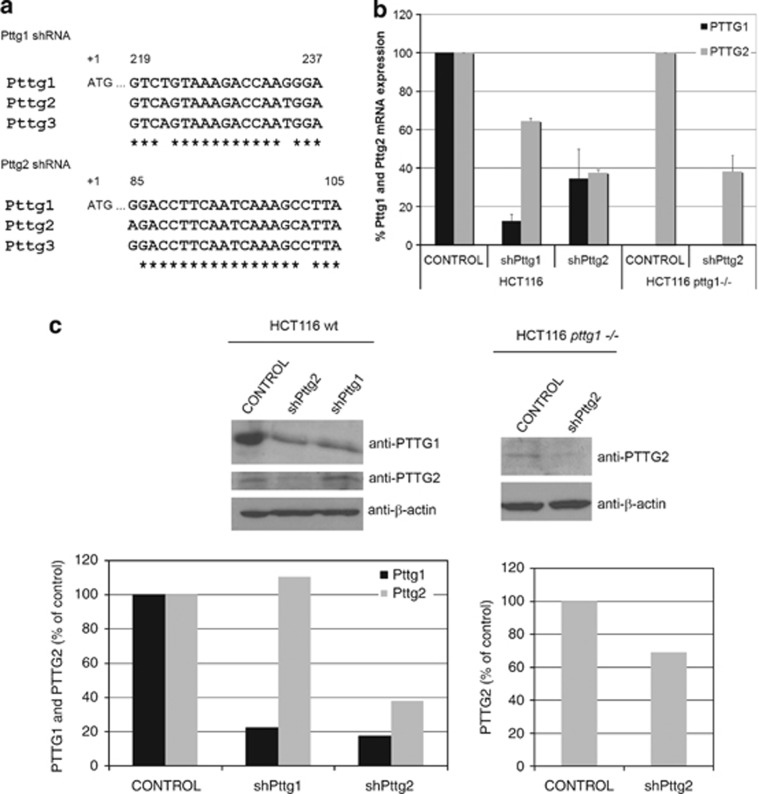
pttg1 shares high-sequence homology with the intronless genes, pttg2 and pttg3 of unknown biological functions. To dissect the cellular function of each isoform, we aimed to search for specific shRNAs against Pttg1, Pttg2 and Pttg3. However, during the course of this study, pttg3 was classified as a pseudogene (http://www.ncbi.nlm.nih.gov/nuccore/NR_002734.1) and therefore, excluded from this study. Importantly, pttg3 was also disqualified as a coding gene from the Intronless Genes Database (http://www.bioinfo-cbs.org). For this reason, here we focused on the characterization of the pttg2 gene. To find the most efficient and specific shRNA against Pttg2, we tested the capacity of five different shRNA lentiviral clones (Sigma, St. Louis, MO, USA), located at different regions within Pttg2 ORF, to reduce Pttg2 levels. Unfortunately, none of them (including a duplex siRNA containing five mismatches compared with Pttg1) were able to specifically silence Pttg2 without altering Pttg1 mRNA levels. Based on these results, we selected the shRNAs for Pttg1 (shPttg1) and Pttg2 (shPttg2) shown in Figure 1a, both containing two mismatches. Pttg2 mRNA levels were reduced by 62% or 35% using shPttg2 and shPttg1, respectively, while Pttg1 was reduced 65% by shPttg2 and 90% by shPttg1 (Figure 1b). A similar reduction in Pttg2 levels was observed in the absence of Pttg1 as determined by qPCR using HCT116 pttg1−/− cells (Figure 1b). The reduction of PTTG1 and PTTG2 at the protein level was checked by immunoblotting using extracts of HCT116 cells infected with lentivirus expressing shPttg1, shPttg2 or empty vector and specific PTTG1 and PTTG2 antibodies (Figure 1c, left panel). As observed, the reduction of both proteins at the protein level was consistent with the reduction of the mRNA levels shown in Figure 1b. Similarly, a decrease of PTTG2 levels in the HCT116 pttg1−/− cell line was detected (Figure 1c). Here, it should be noted, the low PTTG2 expression levels compared with PTTG1. This is not due to antibody sensitivity since three different PTTG2 antibodies raised against GST and His-tagged proteins or a synthetic peptide located at the C-terminal region of PTTG2, were used. This made unfeasible to routinely check PTTG2 protein levels during the study. Also, similar levels of Pttg2 mRNA expression were found in cells of different origin including human embryonic kidney HEK293T cells, hTERT-immortalized retinal pigment epithelial cell line RPE1, human breast adenocarcinoma MCF7 cells and the cervical cancer cell line HeLa (data not shown). In addition, low levels of Pttg2 were found in numerous human tissues as ascertained by normal tissue cDNA arrays (TissueScan, Origene, Rockville, MD, USA) and Pttg2-specific Taqman probes (Supplementary Figure S1).
Figure 1.
Characterization of Pttg2-depleted HCT116 cells. (a) Schematic diagram showing the sequences and location of shPttg1 and shPttg2. Multiple sequence alignments were made using the Clustal W software (www.clustal.org). (b) Relative quantification of Pttg1 and Pttg2 mRNA levels in Pttg1 and Pttg2-silenced HCT116 wild-type and HCT116 pttg1−/− cells by real-time PCR. Expression of each gene is shown as mean±S.E.M. of three independent experiments. Data are shown as 2−ΔCT normalized to HPRT1. (c) Analysis of PTTG1 and PTTG2 protein levels 72 h after shRNA-Pttg1 or shRNA-Pttg2 treatment in HCT116 wild-type and HCT116 pttg1−/− cells. β-Actin was used as loading control. Densitometric analysis for western blots of PTTG1 and PTTG2 protein expression (% of control) in control and shRNA-treated (shPttg1 or shPttg2) wild-type and Pttg1−/− HCT116 cells
Pttg2 and Pttg1 show different biochemical properties
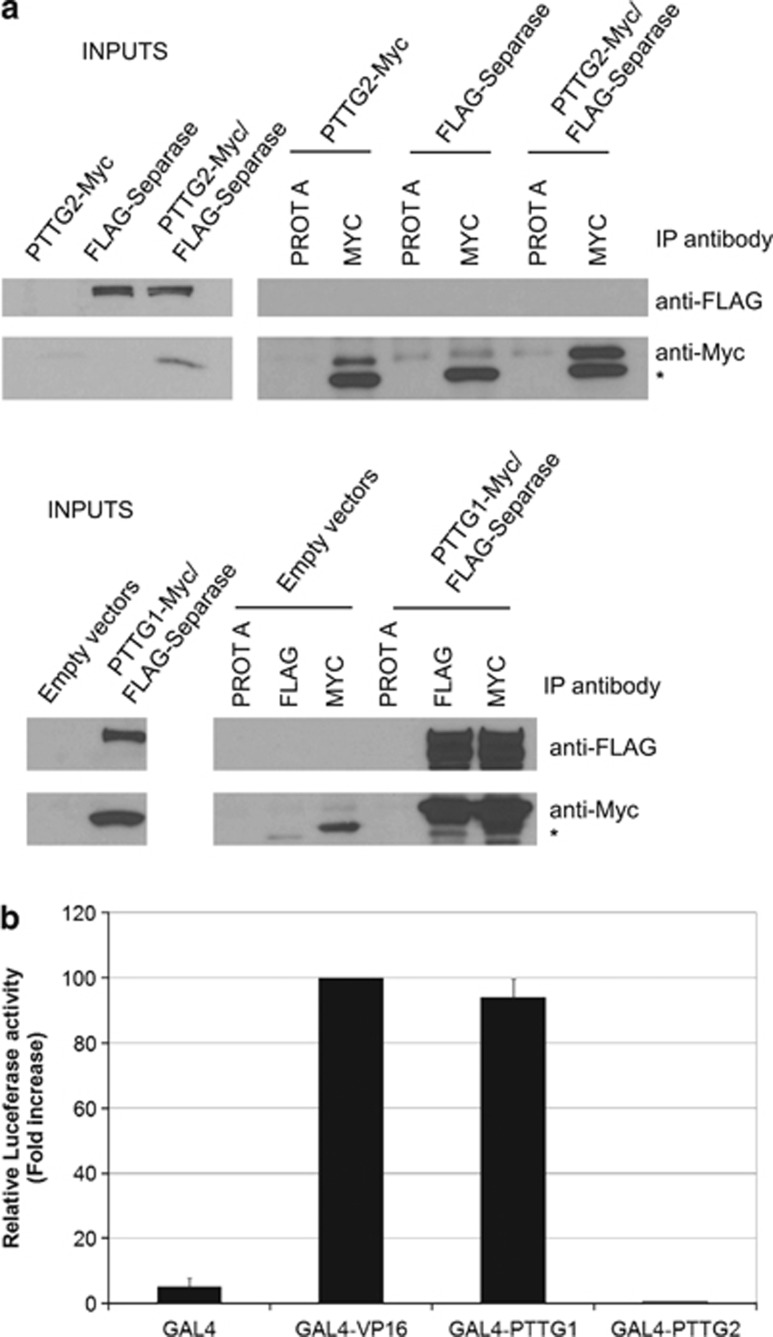
The high-sequence homology between Pttg1 and Pttg2 prompted us to test whether, similarly to Pttg1, Pttg2 could also regulate sister chromatid separation through binding to separase. We performed immunoprecipitation assays in HEK293T cells using C-terminal Myc-tagged PTTG2 and N-terminal FLAG-tagged separase and found that Pttg2 lacks the ability to interact with separase under the experimental conditions used (Figure 2a, upper panel). As a control, we show binding of PTTG1 to separase (Figure 2a, lower panel).16 This indicates that likely PTTG2 does not play a role in preventing premature chromatids separation, at least not through the interaction with separase. Moreover, we investigated the transactivation capacity of PTTG2, which represents another important feature of PTTG1. We generated fusions of GAL4-DNA-binding domain with full-length PTTG2 as well as with the VP16 transactivation domain and tested them in HEK293T cells for activation of transcription from a luciferase reporter construct. The results showed no significant increase of reporter gene activity by GAL4-PTTG2 over the background, confirming the lack of transcriptional activity (Figure 2b). As positive controls, we used GAL4-VP16 and GAL4-PTTG1 previously reported to upregulate luciferase reporter activity.8 Altogether, these results indicate that most likely, PTTG1 and PTTG2 participate in different biological responses.
Figure 2.
Pttg1 and Pttg2 show different biochemical properties. (a) PTTG2 does not interact with Separase. Extracts of HEK293T cells expressing N-terminally Flag-tagged separase, C-terminally Myc-tagged PTTG2, or both were used for immunoprecipitation with anti-Myc antibodies. The immunoprecipitates were analyzed by immunoblotting with anti-FLAG and anti-Myc antibodies. The asterisks indicate the immunoglobulin light chains. As positive control, the interaction between PTTG1 and separase was tested using Flag-separase and Myc-tagged PTTG1. (b) PTTG2 lacks transactivation capacity. The ORF of Pttg2 was fused to the GAL4 DNA-binding domain (pGAL4-DBD-Pttg2) and tested for transcription activation in HEK293T cells cotransfected with a GAL4 site-dependent reporter plasmid driving expression of the luciferase gene (pGAL4-4RE-Luc). pGAL4-PTTG1 and pGAL4-VP16 were used as positive controls. Luciferase values from at least three independent transfections are plotted as fold activation over control
Silencing of Pttg2 impairs normal cell adhesion, reduces cell proliferation and promotes apoptosis in HCT116 cells
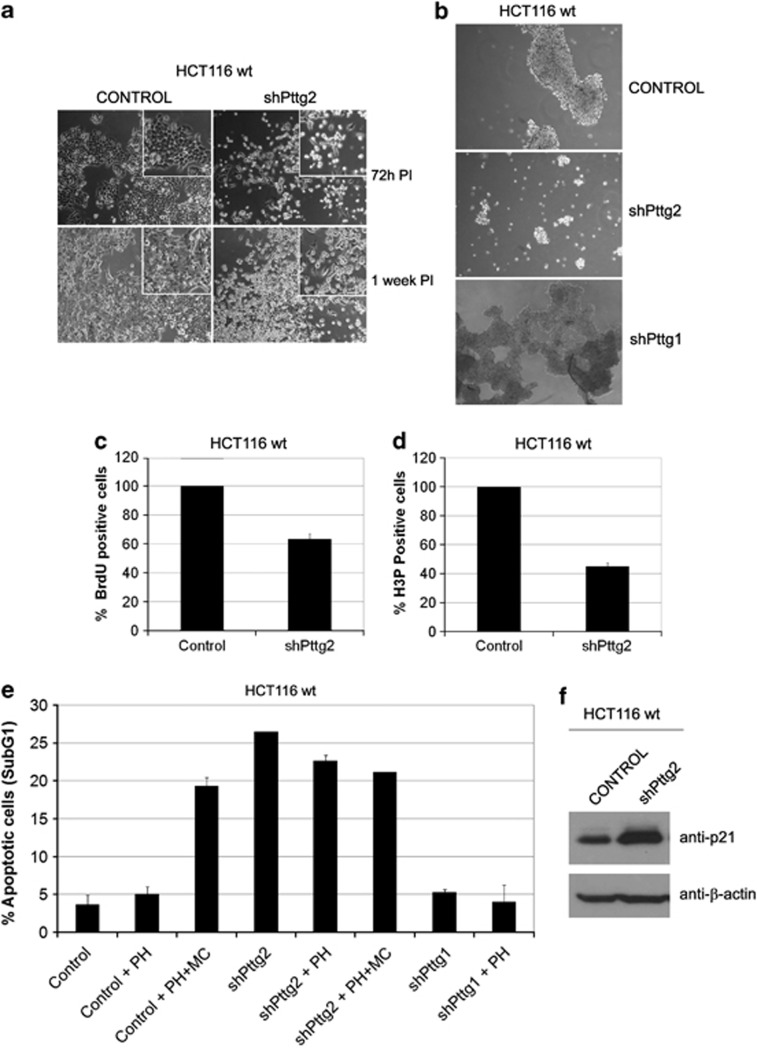
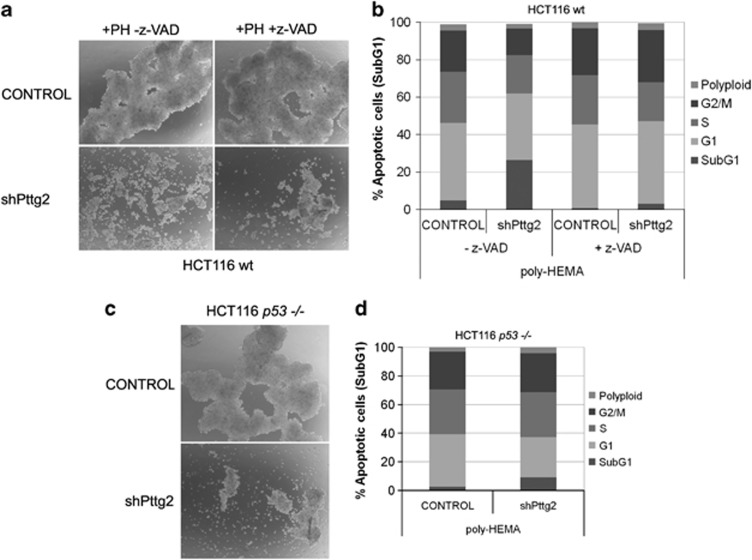
To elucidate Pttg2 function, we made use of the RNAi technology and infected HCT116 with lentiviral particles expressing the Pttg2 shRNA described in Figure 1a. As depicted in Figure 3a, the shape of Pttg2-depleted cells was rounder compared with control cells, morphology compatible with an apparent loss of cell adhesion. Importantly, the same phenotype was reproduced in human embryonic kidney HEK293T cells, indicating that this effect is a general property of Pttg2 downregulation (Supplementary Figure S2c). To study the adhesion capacity of these cells under anchorage-independent conditions, we transferred mock-infected cells and cells infected with shPttg2 onto poly-HEMA-coated plates. Poly-HEMA is known to prevent cell–matrix interactions while preserving cell–cell contacts. Under these conditions, control cells formed dense spheroidal aggregates while shPttg2-infected cells appeared more dispersed and formed only loose clumps of cells (Figure 3b, middle panel), indicating that the cell–cell interconnection is compromised. Given that the treatment with shPttg2 also results in a reduction of PTTG1 levels, we analyzed the morphology of cells with reduced PTTG1 levels (shPttg1-treated) growing on poly-HEMA plates. As shown in Figure 3, these cells formed dense typical round colonies with tightly associated cell–cell contacts similarly to control cells (Figure 3b, lower panel). This suggests that the observed phenotype is mainly due to PTTG2 deficiency. Further, when the proliferation of cells with diminished PTTG2 levels was measured, we found that Pttg2-depleted cells showed reduced BrdU incorporation (37%) and mitotic index (55% of nocodazole-treated cells) compared with control cells (Figures 3c and d, respectively), indicating that in our cells, the loss of cellular adherence is accompanied by reduced proliferative capacity.
Figure 3.
Silencing of Pttg2 impairs cell adhesion and proliferative capacity of HCT116 cells. (a) Phase-contrast images ( × 10 objective) of Pttg2-silenced HCT116 cells 72 h (upper panels) and 1 week post-infection (PI) (lower panels). Inserts show close-up views of representative cell culture areas. Pttg2-depleted cells show rounder morphology than control cells, indicating a defect in cell adhesion properties. (b) Spheroid formation of Pttg2 and Pttg1-silenced HCT116 (wt) cells in suspension. Seventy-two hours post-infection, cells were transferred to poly-HEMA-coated plates at 6 × 104 cells/ml. After 48 h in suspension, cells were photographed at 10 × . Control and shPttg1-treated cells formed dense spheroids whereas shPttg2-silenced cells adhere to each other very weakly. Representative areas of each culture were selected. (c) Percentage of BrdU-positive cells after shRNA-Pttg2 treatment. HCT116 cells were infected and assayed for BrdU incorporation 72 h post-infection. Bars represent the mean±S.E.M. (d) Analysis of mitotic index in Pttg2-depleted HCT116 cells. Cells were treated with shRNA-Pttg2 lentivirus and incubated with nocodazole 1 μM 72 h post-infection, for 24 h. Percentage of H3P-positive (mitotic) cells was measured by flow cytometry. Bars represent the mean±S.E.M. (e) Percentage of cell death following shRNA Pttg2 or shRNA Pttg1 treatment of HCT116 cells growing under adherent or suspension conditions. Seventy-two hours post-infection, cells were transferred onto control or poly-HEMA-coated (PH) plates in the absence or presence of methylcellulose (MC) and the levels of apoptosis was determined 48 h later by measuring the percentage of cells containing a subG1 DNA content by flow cytometry. Results represent the means of three independent experiments±S.E.M. (f) p21 induction in Pttg2-depleted HCT116 cells. HCT116 cells treated with shRNA-Pttg2 lentivirus for 72 h were harvested and the levels of p21 determined by immunoblotting using specific p21 antibodies. β-Actin was used as loading control
Notably, we observed that cell aggregation in suspension promoted cell survival while loss of adhesion of shPttg2-treated cells was followed by increased cell death (20–30%) as measured by the number of cells presenting fragmented DNA (subG1 peak) (Figure 3e). To reinforce the results indicating that downregulation of Pttg2 and not Pttg1 cause induction of apoptosis in HCT116 cells, we measured the apoptotic response of Pttg2-silenced cells in which Pttg1 levels have been restored by ectopic expression of Pttg1 cDNA. As shown in Supplementary Figure S3, Pttg1 was restored to levels similar as those observed in control cells while maintaining low Pttg2 levels (Supplementary Figure S3a). In this scenario, analysis of the apoptotic response in these cells showed that restoration of Pttg1 levels in Pttg2-silenced cells does not rescue the apoptotic phenotype (Supplementary Figure S3b). In addition, a similar apoptotic response was observed in HEK293T cells treated with shPttg2 (Supplementary Figures S2b and c) that specifically resulted in Pttg2 downregulation while leaving Pttg1 levels unaltered (Supplementary Figure S2a). Collectively, these data reinforce the idea that Pttg2 is an important regulator of apoptosis in both cell types.
As a control of the importance of cell aggregation in cell survival of HCT116 cells, we used poly-HEMA-coated plates and methylcellulose in the culture medium to minimize cell–cell contacts in suspension. As expected, cell aggregates were not formed in the methylcellulose suspension cultures (data not shown) in both control and Pttg2-depleted cells, resulting in augmented cell death in control cells at levels similar to Pttg2-depleted cells under all conditions. This indicates that the leading cause of death in Pttg2-depleted cells is the loss of cell adhesion.
As mentioned above, Pttg2 shRNA led to the reduction of both PTTG1 and PTTG2 levels. To clarify the role of Pttg1 depletion in the cell death seen in Pttg2-silenced HCT116 cells, we determined the cell-cycle profile of cells separately treated with shPttg1 or shPttg2 (Figure 3e). As shown, a reduction of Pttg1 mRNA levels (by 90%, data not shown) without a concomitant reduction of Pttg2 did not result in an increase in the number of dead cells. Consistent with earlier reports, we conclude that cell–cell adhesions under anchorage-independent conditions are crucial for the survival of HCT116 cells and confirm the important role that Pttg2 plays in the maintenance of cellular adhesion.
In order to characterize in detail the observed lack of adhesion phenotype, we examined the expression levels of p21, a protein previously reported to be upregulated under these conditions.17, 18 In fact, loss of adhesion following Pttg2 silencing in HCT116 cells was accompanied by an increase in p21 (Figure 3f). Similarly, Pttg2 silencing resulted in increased p21 levels in HCT116 whereas depletion of Pttg1 in these cells did not affect p21 protein levels (Supplementary Figure S4). These results further confirmed a defective cell adhesion phenotype in cells with reduced Pttg2 levels.
Pttg2-depleted cells die from p53- and p21-dependent apoptosis
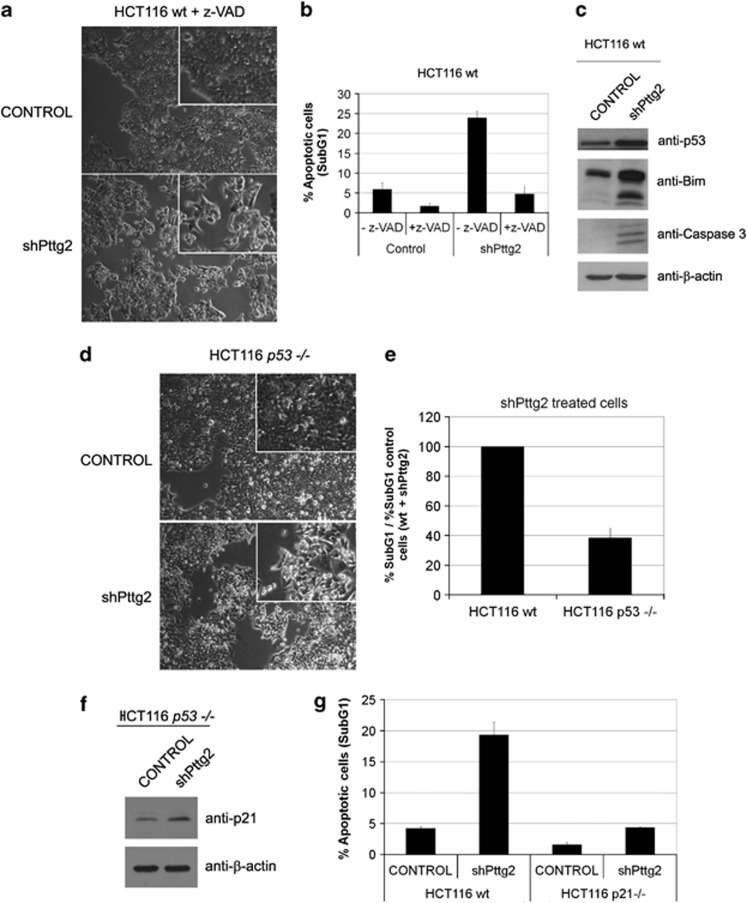
To further characterize the observed adhesion-dependent cell death, we tested the effect of the addition of the caspase inhibitor z-VAD to the cell media of control and Pttg2-silenced cells growing under adherent conditions. As depicted in Figure 4a, the cell death phenotype observed in shPttg2-treated cells, was partially reverted in the presence of z-VAD. In agreement, the number of dead cells after PTTG2 silencing was drastically reduced (Figure 4b), suggesting that cells lacking Pttg2 eventually die through caspase-dependent apoptosis following cell rounding, a phenomenon also known as anoikis. In addition, western blot analysis of extracts from HCT116 cells treated with shPttg2 displayed elevated p53 protein (Figure 4c, upper panel), as well as the apoptosis indicator active caspase-3 (Figure 4c, lower panel) or the detachment-induced pro-apoptotic factor Bim (Figure 4c, middle panel), reported to undergo alternative splicing to generate three variants: BimEL, BimL and BimS.19 To further analyze the role of p53 in Pttg2-dependent anoikis, we used HCT116 p53−/− cells infected with control lentivirus or lentivirus expressing shPttg2. The absence of p53 in shPttg2-treated cells decreased the extent of anoikis morphology (Figure 4d) and led to reduce number of apoptotic cells compared with shPttg2-treated wild-type cells (Figure 4e).
Figure 4.
Defective adhesion in Pttg2-depleted HCT116 cells results in p53- and p21-dependent apoptosis. (a) Phase-contrast images of Pttg2-silenced HCT116 cells after z-VAD treatment. A round morphology was still observed in Pttg2-depleted cells, suggesting a defective cell adhesion. (b) Apoptosis can be rescued in Pttg2-silenced HCT116 cells after caspase inhibition treatment. The percentage of apoptotic cells (subG1) after PTTG2 depletion was drastically reduced after treatment with the caspase inhibitor z-VAD. Bars represent the mean±S.E.M. (c) Levels of apoptotic indicators in Pttg2-silenced HCT116 cells. Cells were harvested after 72 h post-infection and assayed for p53, p21, BIM and cleaved-Capase 3 levels by western blot using specific antibodies. β-Actin was used as loading control. (d, e) Role of p53 in Pttg2-dependent anoikis. (d) Phase-contrast images of Pttg2-depleted HCT116 p53−/− cells. The absence of PTTG2 in HCT116 p53−/− results in cell rounding similar to wild-type HCT116 cells. (e) FACS analysis of Pttg2-depleted HCT116 p53−/− cells. In the absence of PTTG2, the percentage of apoptotic cells (subG1) was partially reduced in HCT116 p53−/− cells compared with HCT116 wild-type cells. Bars represent the mean±S.E.M. (f) The induction of p21 in Pttg2-treated cells occurs in a p53-dependent manner. Western blot analysis of p21 levels in HCT116 p53−/− cells 72 h post-infection with shPttg2 or control lentivirus. β-Actin was used as loading control. (g) Role of p21 in anoikis in Pttg2-depleted cells. The percentage of apoptotic cells in HCT116 p21−/− following treatment with shPttg2 was significantly reduced compared with HCT116 wild-type cells. Bars represent the mean±S.E.M.
As mentioned before, loss of cell adhesion led to the concomitant induction of p21 in a p53-dependent manner as cells lacking p53 and treated with shPttg2 did not present a significant increase in p21 levels (Figure 4f). In addition to its function as a cell-cycle regulator, a role in apoptosis has been attributed to p21.20, 21 To study the contribution of p21 to shPttg2-mediated apoptosis, we used HCT116 p21−/− cells and our shRNA against Pttg2. Infection of shPttg2 did not cause cell death in p21-deleted HCT116 human colon carcinoma cells (Figure 4g). These results indicate that reduced Pttg2 levels induce inhibition of cell growth through p53-dependent p21 induction, and p21 appears to be a critical effector of shPttg2-induced cell death. Taken together, these results extend previous studies showing that the disruption of cell–matrix interactions in murine cells activates the p53–p21 pathway.18
We asked further if the observed loss of cell adhesion precedes the onset of apoptosis. To this purpose, we used shPttg2-treated cells growing on poly-HEMA-coated plates and treated with z-VAD. As displayed in Figure 5a, the loss of adhesion phenotype of shPttg2-silenced cells was also found in the presence of z-VAD. Despite this, we observed reduced number of dead cells, indicating an apoptotic effect due to loss of appropriate adhesion (Figure 5b). The same effect was observed in HCT116 p53−/− cells expressing shPttg2 (Figures 5c and d).
Figure 5.
Loss of cell adhesion in shPttg2-treated cells precedes the onset of apoptosis. (a) Spheroid formation of Pttg2-depleted cells after z-VAD treatment. Wild-type HCT116 cells were infected for 72 h in the presence or absence of the caspases inhibitor z-VAD and the cell–cell adhesion capacity assayed on poly-HEMA (PH)-coated plates. The experiments were repeated three times, and representative areas of each culture are shown. (b) Cell-cycle profile of Pttg2-silenced HCT116 cells growing under poly-HEMA conditions after z-VAD treatment. The percentage of apoptotic cells (subG1) was significantly reduced in the presence of z-VAD, whereas the other cell-cycle phases (G1, S and G2/M) were not modified. Data are representative of two independent experiments. (c) Phase-contrast images to assess multicellular aggregates formation in shPttg2-treated HCT116 p53−/− cells growing on poly-HEMA-coated plates. Images show a representative region of each condition. (d) Analysis of the cell-cycle profile of Pttg2-depleted HCT116 p53−/− cells growing under poly-HEMA conditions. ShPttg2-treated cells lacking p53 showed a reduced percentage of cell death (subG1). The other cell-cycle phases (G1, S and G2/M) remained unaltered. Data are representative of two independent experiments
Pttg2 expression is required for normal tubulin distribution
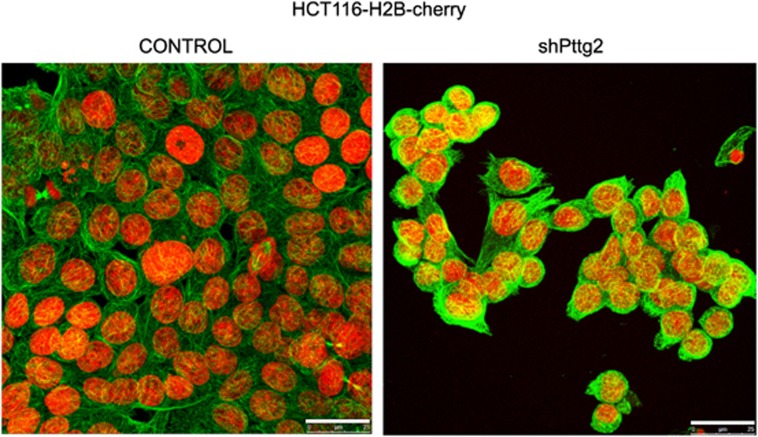
To test if the altered cellular morphology could be a consequence of cytoskeletal defects, we analyzed α-tubulin distribution by immunostaining in HCT116 cells infected with control lentivirus or lentivirus expressing Pttg2 shRNA. As shown in Figure 6, Pttg2-depleted cells showed an altered pattern of α-tubulin compared with control cells, suggesting that the reduced intercellular adhesions of Pttg2-depleted cells are coupled with abnormal distribution of cytoskeletal proteins such as α-tubulin. In addition, the levels of acetylated α-tubulin were elevated in shPttg2-depleted cells as ascertained by western blot (data not shown) indicative of impaired microtubules dynamics in cells with reduced Pttg2 levels.
Figure 6.
Pttg2-depleted cells exhibit cytoskeletal defects. Microtubule cytoskeleton of PTTG2 depleted cells. Confocal images of Pttg2-silenced HCT116 cells stably expressing Histone H2B (H2B)-cherry fusion gene. Seventy-two hours post-infection, cells were fixed with methanol at −20 °C and stained for α-tubulin (green). Scale bars, 25 μm
PTTG2 regulates E-cadherin and vimentin levels leading to EMT induction
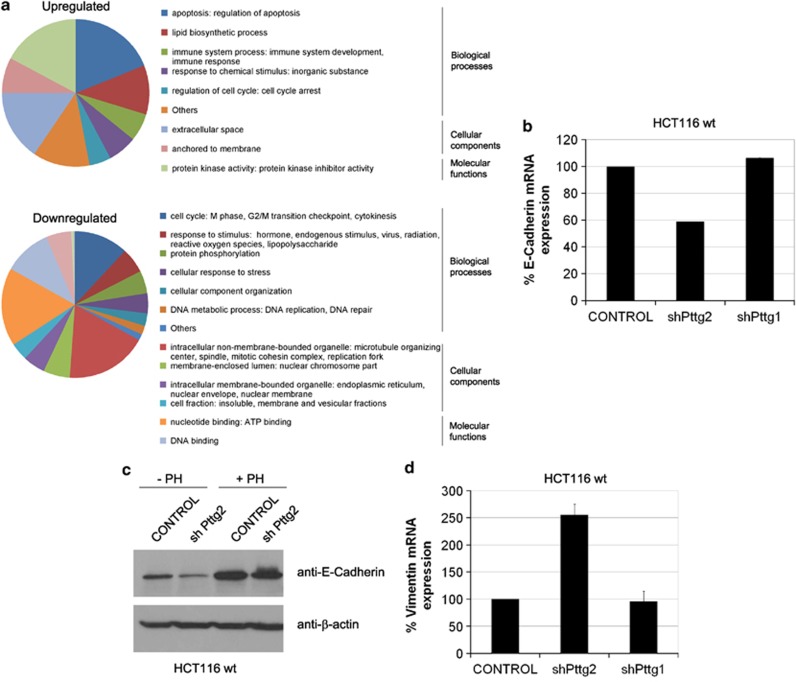
To elucidate the mechanisms by which Pttg2 regulates cell adhesion, we analyzed the gene expression profiles of cells depleted of Pttg2 compared with wild-type cells. Of the 28 869 genes represented on the GeneChip Human Gene 1.0 ST array, 439 genes were identified to be differentially expressed, with a significance P<0.01 and fold change >1.5 (Supplementary Table S2). Some examples of differentially regulated genes classified by fold change or biological interest have been listed in Table 1. Functional analysis of the 157 upregulated (138 annotated genes) and 323 downregulated probes (301 annotated genes) was performed using DAVID program (david.abcc.ncifcrf.gov). As revealed by the Gene Ontology (GO), among the upregulated probes, genes involved in apoptosis, protein kinase activity and related to the control of the extracellular space were specifically activated by Pttg2 downregulation (Figure 7a; Table 1). On the other hand, of the downregulated 323 probes, genes involved in cell cycle, nucleotide binding and to intracellular non-membrane-bounded organelles were particularly downregulated by shPttg2 treatment.
Table 1. Examples of differentially regulated genes in shPttg2-treated cells.
| Name | Symbol | mRNA accesion | Fold-change | Biological function |
|---|---|---|---|---|
| Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | NM_001548 | 5.27 | Cell proliferation |
| Vimentin | VIM | NM_003380 | 1.86 | Cell adhesion |
| Cyclin-dependent kinase inhibitor 1A | p21 | NM_078467 | 1.79 | Cell cycle |
| Bcl2-like 11 | BCL2L11 (Bim) | NM_138621 | 1.64 | Induction of apoptosis by extracellular signals |
| Bcl2-antagonist/killer 1 | BAK1 | NM_001188 | 1.64 | Apoptosis |
| Fas (TNF receptor superfamily member 6) | FAS | NM_000043 | 1.62 | Apoptosis |
| Homo sapiens zinc-finger E-box binding homeobox 1 (ZEB1), transcript variant 2 | ZEB1 | NM_030751 | 1.39 | Cell adhesion |
| Homo sapiens snail homolog 1 | SNAI1 | NM_005985 | 1.33 | Cell adhesion |
| Homo sapiens fibronectin 1 | FN1 | NM_212482 | 0.77 | Cell adhesion substrate adhesion-dependent cell spreading |
| Homo sapiens cadherin 1, type 1, E-cadherin (epithelial) | CDH1 | NM_004360 | 0.67 | Cell adhesion cell–cell adherens junction |
| microRNA 21 | MIR21 | NR_029493 | 0.54 | Cell migration |
| Homo sapiens integrin, α V (vitronectin receptor, α polypeptide, antigen CD51) (ITGAV), transcript variant 1, mRNA | ITGAV | NM_002210 | 0.47 | Cell adhesion cell–matrix adhesion |
| Pituitary tumor-transforming gene 1 | PTTG1 | NM_004219 | 0.43 | Cell cycle |
Figure 7.
Pttg2 depletion results in deregulated E-cadherin and vimentin levels and induction of the epithelial-to-mesenchymal transition. (a) Summary of gene expression profile according to Gene Ontology. Pie charts show the percentage of genes included in each category (molecular function, biological process and cellular component) and subcategories with a P-value <0.01. The GO analysis was performed using DAVID v.6.7 program. GO analysis of differentially upregulated or downregulated genes are shown in upper and lower panels, respectively. (b) Validation of E-Cadherin downregulation in Pttg2-silenced cells by quantitative PCR. HCT116 cells were infected with shPttg2 or control lentivirus and assayed for E-cadherin mRNA levels 72 h post-infection. Values were normalized using the hprt1 gene as an internal control. Columns represent the mean values of 2–3 independent experiments ±S.E.M. (c) Western blot analysis of E-Cadherin repression in Pttg2-depleted HCT116 cells. The expression levels of the E-Cadherin protein in shPttg2 cells was compared with control cells expressing an empty lentiviral plasmid. Cells were grown either as a monolayer (attached) or in suspension for 48 h on poly-HEMA-coated plates (+PH) and total lysates analyzed by immunoblotting. (d) Vimentin mRNA induction in shPttg2-treated cells. HCT116 cells were infected with shPttg2 or control lentivirus and assayed for vimentin levels 72 h post-infection by quantitative PCR. Values were normalized using the hprt1 gene as an internal control. Columns represent the mean values of 2–3 independent experiments±S.E.M.
Remarkably, consistent with our phenotypic observations, among the differentially expressed genes we found two previously reported genes, p21 and Bim, known to be upregulated following cell detachment. As previously shown in Figures 3f and c, we also found increased p21 and Bim levels following Pttg2 knockdown, which further validate our array data. Also, because of the impaired cell adhesion phenotype of shPttg2 cells, we chose to examine E-cadherin, another differentially regulated gene that showed 1.49-fold decrease in cells with low Pttg2 levels. To enhance the significance of the array analysis, the mRNA data of E-cadherin was validated by real-time PCR and the respective protein analyzed by western blot. As displayed in Figure 7b, quantitative PCR analysis showed a reduction of 40% in E-cadherin mRNA levels after shPttg2 transfection. Also, E-cadherin protein levels were reduced in cells with reduced PTTG2 levels (Figure 7c). Otherwise, it has been shown that in HCT116 cells, the amount of E-cadherin protein was strongly increased when cells were transferred from monolayer to suspension culture over a period of 48 h.22 Notably, Pttg2-depleted cells presented lower levels of E-cadherin when growing under non-adherent conditions on poly-HEMA-coated plates compared with control cells growing under similar conditions (Figure 7c).
Next, given the round morphology and the low E-cadherin levels of cells with reduced Pttg2 levels, we explored the association between lack of adhesion of shPttg2 cells and EMT induction. For this purpose, we chose to examine the levels of vimentin, another mesenchymal phenotypic marker. Both the array data and real-time PCR experiments showed that the loss of E-cadherin correlated with a significant increase in vimentin mRNA levels (1.87-fold-induction in the arrays experiments and 2.55-fold-induction in quantitative PCR analysis) (Figure 7d). In conclusion, our data show a phenotype resembling EMT, characterized by repression of E-cadherin and increased vimentin expression. Interestingly, the main features of this phenotype were independent of p53 and p21 status as cells lacking these genes and treated with shPttg2 also adopted EMT features including spindle-like cell morphology (Figure 4d and data not shown), loss of the epithelial cellular marker E-cadherin (data not shown) and increased survival (Figures 4e and g).
Discussion
The pttg2 gene shares high-sequence homology with pttg1/securin. However, to date, no studies have been conducted to elucidate the biological role of this poorly understood securin family member. Here, we partially characterize Pttg2 and found that it has other activities different from Pttg1, among which we highlight a novel role in cellular adherence and induction of epithelial–mesenchymal transition, presumably through the regulation of E-cadherin levels.
pttg2 is a single exonic gene. Initially, the loss of introns was associated with the origin of processed pseudogenes. However, more recently, it has been demonstrated that some of them have acquired upstream regulatory elements and become functional (retrogenes).23, 24 It has been suggested that introns may contain signals to stabilize transcription or retard regulatory responses. We found that Pttg2 is minimally expressed in normal cells. This is in agreement with the fact that intronless genes are tipically expressed at significantly lower levels and in a narrower range of tissues than spliced genes with the exception of histones.25
In this study, we have compared the biochemical properties of PTTG1 and PTTG2. We first tested whether PTTG2 could also bind to separase. Under our experimental conditions, PTTG2 was unable to bind separase, a prerequisite to function as a securin. The binding site of PTTG1 to separase has been mapped to the N- and C-terminal regions. Importantly, these motifs are not conserved in PTTG2, explaining why PTTG2 failed to interact with separase. Similarly, PTTG2 did not exhibit detectable transactivation function. These results revealed that most likely PTTG1 and PTTG2 play distinct cellular roles.
Through our characterization of HCT116 cells depleted of Pttg2, we demonstrate that the absence of Pttg2 results in morphologic changes consistent with loss of adherence and cell death by apoptosis. Although we cannot unequivocally attribute this effect to the lack of Pttg2 alone, several evidences indicate that Pttg2 depletion specifically induces cell death by loss of anchorage. First, a clear induction of the apoptotic response has been observed in shPttg2-treated HEK293T cells showing a more specific depletion of Pttg2 than HCT116 cells treated in the same way. Furthermore, HCT116 depleted of Pttg2 but expressing Pttg1 at similar levels as untreated cells, showed comparable levels of apoptosis with respect to cells displaying a concomitant reduction of Pttg2 and Pttg1 levels. Moreover, our data indicate that, under the experimental conditions used, Pttg1 deficiency alone does not affect cell survival. This has been further supported by a recent study showing unaltered cell survival following siRNA Pttg1 treatment in HCT116 wild-type cells.26 In addition, homozygous loss of pttg1 is not lethal to human cells.4 Given that the lack of Pttg1 results in no obvious lack of adhesion phenotype, it appears more likely, that these changes are, in fact, the result of decreased PTTG2 level. In fact, recent evidences suggest a role for PTTG1 in the induction of EMT.27, 28 Overexpression of PTTG1 results in decreased E-cadherin and increased vimentin and integrin α-V levels as well as impaired integrin-mediated focal adhesion complexes and actin cytoskeletal reorganization. Interestingly, the lack of Pttg2 result in morphological changes resembling those appeared in the presence of elevated PTTG1 levels. Although pull down experiments suggest that Pttg1 and Pttg2 do not interact (unpublished results), whether or not the two proteins account for the observed morphological changes leading to EMT in our cells remains to be determined.
Although a large number of shPttg2-treated cells presented a mesenchymal morphology and proliferated at a normal rate, 20–30% of cells underwent apoptosis 3 days following shPttg2 treatment. Most importantly, the number of dead cells following Pttg2 depletion was significantly reduced in cells harboring a homozygous deletion of the p53 or p21 gene, indicating the absolute requirement of these genes during shPttg2-induced cell death. Several studies have suggested that p21 may promote apoptosis under certain cellular stresses through both p53-dependent and p53-independent mechanisms although exactly how p21 participates in the apoptotic process is not clear.20 Therefore, we conclude that low Pttg2 levels sensitize cells to anoikis and we attribute this response to the lack of proper adhesion and the activation of wild-type p53.
Pttg2-depleted cells displayed loss of cell–cell contacts, which was accompanied by cell rounding and many prominent elongated spindle-shaped cells. In addition, a decrease of the epithelial marker E-cadherin and increased vimentin levels supported EMT induction. Moreover, the array data indicated that cells with decreased Pttg2 levels also exhibit higher levels of the E-cadherin repressors, Snail and Zeb1 (1.33 and 1.39-fold-induction in the arrays experiments, respectively) (Table 1; Supplementary Table S2) although it is not clear whether PTTG2 may regulate E-cadherin through the induction of these transcription factors. In summary, even under EMT inhibitory conditions such as Pttg1 downregulation28 and p5310, 29, 30, 31, 32 and p21 induction,10 we found that cells with low Pttg2 levels displayed EMT markers. Altogether, these results suggest that Pttg2 is involved in the repression of EMT.
To our knowledge, no studies have been previously conducted to characterize Pttg2. Our results establish for the first time an unexpected role for Pttg2 in EMT induction. Further, epithelial cells, which undergo EMT are known to acquire enhanced invasiveness. This raises the possibility of a potential implication of Pttg2 in cell invasion. Further studies are needed to determine whether the lack of this protein may contribute to cancer cell motility under specific genetic backgrounds that permit the survival of cells with defective adhesion properties.
Materials and Methods
Cell culture and transfections
The HCT116 human colon carcinoma cell line and the derivative cell lines, HCT116 Pttg1−/−,4 HCT116 p53−/−33 and HCT116 p21−/−,34 were kindly provided by B Vogelstein (The John Hopkins Oncology Center). HCT116 cells stably expressing H2B-mCherry were obtained by transfection of these cells with the pcDNA3-H2B-mCherry (Addgene plasmid 20972)35 expression plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and antibiotic selection of G418-resistant clones (1 mg/ml). The HCT116 cell lines were grown in McCoy's medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum (Sigma) and 2 mM ℓ-glutamine, 100 U of penicillin/ml and 100 μg of streptomycin/ml (Sigma) in a 5% CO2 humidified atmosphere at 37 °C. HEK293T cells were grown in Dulbecco's Modified Eagle medium (Sigma) under the same conditions as HCT116 cells.
Cell adhesion assays
For cell-adhesion assays, six-well plates were coated twice with 12 mg/ml poly-HEMA (poly(2-hydroxyethyl methacrylate) (Sigma) in 95% ethanol and dried overnight at 37 °C. Poly-HEMA-coated plates were washed twice with PBS before seeding cells. HCT116 cells infected with control or shPttg2lentivirus for 72 h were seeded onto poly-HEMA-coated or not coated six-well plates at a density of 6 × 104 cells/ml (final volume: 3 ml/per well) for 48 h. As a control, cells were suspended in complete growth medium containing 2% Methylcellulose (Sigma) and plated at the same density onto poly-HEMA-coated six-well plates for 48 h.
Lentiviral production and infection
For the generation of lentiviral particles expressing a shRNA against Pttg1, the HIV packaging (pCMVDR8.91) and VSVG (pMDG) plasmids and the lentiviral vector pHRSIN-DUAL-GFP (also known as pHRSIN-CSGWdINotI_pUb_Em)36 were kindly provided by Mary K Collins (Windeyer Institute, London). For Pttg2 silencing experiments, a set of five MISSION TRC shRNA clones were purchased from Sigma. The different clones were assessed for Pttg2 silencing efficiency by qPCR analysis and one clone (TRCN0000107158) selected for further experiments based on highest Pttg2 silencing efficiency. This clone was modified to express GFP instead of the original puromycin resistance gene. For lentivirus production, 3 × 106 293T cells were seeded onto a 10-cm Petri dish and transfected after 24 h with CalPhos Mammalian Transfection kit (BD Biosciences Clontech, Palo Alto, CA, USA) using 15 μg of the functional construct, 10 μg of pCMVDR8.91 and 5 μg of pMDG. Lentivirus were harvested 72 h post-transfection, passed through a 0.45-μm filter, and concentrated by ultracentrifugation at 22 000 r.p.m. for 90 min. Virus particles were resuspended in cell growth medium and stored at − 80 °C. Lentiviral particles titers were determined by FACS analysis of GFP-positive cells. For infection with the lentiviral stock, HCT116 cells were seeded 24 h prior to infection and then the lentiviral particles were added (MOI=10 for Pttg1 and MOI=5 for Pttg2).
Production and purification of recombinant proteins
To produce a glutathione S-transferase (GST)-PTTG2 fusion protein, the Pttg2 ORF containing BamHI and EcoRI flanking sites was subcloned into the BamHI and EcoRI sites of the pRSET vector (Invitrogen) or into the pGEX4-T (Amersham, Piscataway, NJ, USA) for the production of His-tagged or GST-tagged proteins, respectively. The GST fusion proteins were expressed in Escherichia coli BL21 codon plus (Stratagene, La Jolla, CA, USA) and purified as described.37 For the purification of His-tagged proteins, cells were induced with 1-mℳ IPTG for 3 h at 37 °C and harvested. The cell pellet was resuspended in 1/25 of the bacterial culture volume in buffer (50 mM sodium phosphate pH 7.5, 300 mℳNaCl, 0.5% Sarkosyl, 1 mM PMSF and 1 × protease inhibitor cocktail) and incubated for 15 min on ice. Just before sonication 20% Triton was added. Cell lysates were cleared by centrifugation at 15 000 r.p.m. for 30 min and incubated with TALON metal affinity resin (BD Biosciences Clontech) beads (GE Healthcare, Madrid, Spain) overnight at 4 °C. Beads were washed three times with 10 volumes of lysis buffer lacking the protease inhibitor cocktail. Yield and purity of all recombinant proteins were assessed by 10% SDS-PAGE and Coomassie staining. Protein concentrations were determined using a BSA standard.
Anti-pttg2 antibody production
To generate anti-PTTG2 antibodies, recombinant GST-fused or His-tagged PTTG2 cDNA (ORF) was produced in E. coli and purified on glutathione-agarose beads (Sigma) and TALON His-Tag purification resin (BD Biosciences Clontech), respectively. Alternatively, antibodies were made against a synthetic peptide located at the C-terminus of PTTG2 protein (amino-acid sequence CNLFAVSFKHSVDPG). Rabbit polyclonal antibodies against PTTG2 were affinity-purified by absorption of the sera to His or GST-fused PTTG2 protein immobilized onto CNBr-activated Sepharose (GE Healthcare) and elution at high pH or using the SulfolinkImmobilization Kit for Peptides (Pierce, Rockford, IL, USA).
Western blot analysis
Protein expression was determined by immunoblotting using the following specific antibodies. The human polyclonal rabbit Pttg1 antibody was generated as described8 and used at 1 : 2000 dilution. Monoclonal antibodies against α-tubulin, acetylated-α-tubulin, FLAG and β-actin were purchased from Sigma and anti-p53 (DO1), anti-myc and anti-GAPDH from Santa Cruz, CA, USA. The polyclonal antibodies against active, cleaved caspase-3 were obtained from Cell Signaling (Boston, MA, USA) and against p21 and E-cadherin from Santa Cruz and Beckton Dickinson, respectively. Protein levels were normalized to β-actin or GAPDH expression. Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (150 mmol/l NaCl, 50 mmol/l Tris (pH 8.0), 0.5% deoxycholic acid, 1% NP40, 0.1% SDS and 1 × protease cocktail inhibitor (Roche, Basel, Switzerland). Protein content was quantified using Bradford (Bio-Rad, Madrid, Spain). Fold change in specific protein expression was determined on the basis of β-actin or GAPDH expression as a loading control.
Quantitative RT-PCR (qRT-PCR)
Cells were seeded and infected with control or shPttg2lentivirus for 72 h. Total RNA was prepared using TRIZOL reagent (Invitrogen) followed by treatment with TURBO DNase (Ambion, Austin, TX, USA). RT-PCR experiments were carried out using 1 μl of cDNAs generated from 1 μg of total RNA using Supersript III (Invitrogen). Expression of human Pttg1, Pttg2 and Pttg3 expression was determined using the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). To determine human Pttg1 mRNA levels, we used two Pttg1 sequence-specific PCR primers and a FAM-labeled probe obtained from Operon Biotechnologies (Ebersberg, Germany) (Supplementary Table S1). Expression of Pttg2 and Pttg3 was measured using the Applied BiosystemsTaqMan Gene Expression Assays Hs00747713_sH and Hs00607485_s1. Reactions were carried out at 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The levels of E-cadherin and Vimentin were analyzed using the primers indicated in Supplementary Table S138 and SYBR green master mix (Applied Biosystems). The expression of human hprt1 gene was used to normalize variations in cDNA quantities from different samples.
Immunocytochemistry
Cells grown on coverslips were fixed with methanol at −20 °C for 6 min or 4% paraformaldehyde/phosphate-buffered saline at room temperature for 15 min. After fixation, the coverslips were incubated with primary antibodies as indicated. Species-specific secondary antibodies conjugated with AlexaFluor488, AlexaFluor546 and AlexaFluor633 (Invitrogen) were used. Nuclei were labeled with 4′-6-diamidino-2-phenylindole (DAPI) (200 ng/ml). Wide-field fluorescence imaging was performed using a LeicaDM6000B microscope (Leica, Wetzlar, Germany). Confocal images were captured using a Leica TCS SP5 microscope and HCX PL APO Lambda blue 63 × 1.4 oil objective. Images were analyzed using LAS AF (Leica) and Adobe Photoshop software (Adobe Systems Iberica, Barcelona, Spain).
Immunoprecipitations
For immunoprecipitations, 3 × 106 HEK293T cells were seeded onto a 60-mm Petri dish and transfected with Lipofectamine 2000 (Invitrogen) with plasmids as indicated. After 24 h, cells were lysed in NP40 lysis buffer (100 mℳNaCl, 20 mM Tris-HCl pH 7.5, 5% glycerol, 1mM EDTA pH 8, 1 mM MgCl2 and 0.1–0.5% NP40 with 1 mM PMSF and 1 × protease cocktail inhibitor from Roche). Cellular lysated were precleared with magnetic Dynabeads protein A for 1 h at 4 °C. After magnetic separation, beads were discarded and supernatants incubated overnight at 4 °C with monoclonal antibodies as indicated or protein A followed by beads for 2 h. Beads were washed five times with lysis buffer and bound proteins were dissolved into SDS sample buffer at 95 °C for 5 min and subjected to SDS-PAGE and analyzed by western blot.
Luciferase-based transactivation assays
Cells were seeded in 24-well plates at a density of 105 cells/well and allowed to reach 60–70% confluence and transfected with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. After 24 h, cells were lysed and the luciferase activity was assayed in the lysates using Promega's Dual-luciferase assay system according to the manufacturer's protocol (Promega, Madrid, Spain). Values are normalized to the ratio of Renilla to firefly luciferase activity in the mock-transfected cells.
Cell-cycle analysis
Cell-cycle distribution was determined by propidium iodide staining. Cells were cultured under experimental conditions and harvested by first detaching adherent cells with trypsin and washing the cells with PBS. Cells were fixed by resuspension in 1 ml 70% ethanol and incubated at 4 °C overnight. Fixed cells were stained in 100 μl PBS containing 50 μg/ml PI (Sigma) and 250 μg/ml RNAse. Samples were incubated for at least 30 min at 37 °C before being analyzed by flow cytometry.
BrdU staining
Cells were pulsed with 30 μℳ BrdU (Sigma) for 30 min, fixed in 70% ethanol, and incubated with PE-conjugated anti-BrdU antibody (BD Biosciences). DNA was then stained using 7-AAD (BD Biosciences) and cellular fluorescence was determined using FACS calibur flow cytometer (Becton Dickinson, Madrid, Spain). Data were analyzed using CellQuest-Pro 5.2.1 software (Becton Dickinson).
Mitotic index
To increase the number of cells in mitosis, HCT116 cells were treated with Nocodazole (Sigma) at 1 μM for 24 h, fixed in 70% ethanol and incubated with primary anti-phospho-histone H3 antibody (Millipore, Bedford, MA, USA) at 1 : 200 dilution at room temperature for 2 h, and then with Cy3-conjugated goat anti-rabbit secondary antibody at 1 : 200 dilution for 1 h at room temperature in the dark. DNA was then stained using 7-AAD (BD Biosciences) and cellular fluorescence was determined using FACS calibur flow cytometer (Becton Dickinson). Data were analyzed using CellQuest-Pro 5.2.1 software.
Microarray-based gene expression analysis
Cells were seeded and infected with control or shPttg2lentivirus for 72 h. Total RNA was extracted from both groups of cells using the guanidiniumisothiocynate method (TRIzol reagent; Invitrogen) followed by TURBODNasetreatment (Ambion). The isolated RNA was quantified by a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA). RNA quality was assessed by using the Agilent Model 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). An aliquot (300 ng) of the total RNA was subjected to an AffymetrixGeneChip Human Gene 1.0 ST Array (genome-wide expression profiling chip, 28.869 genes, Affymetrix, Santa Clara, CA, USA) according to the GeneChip Whole Transcript (WT) Sense Target Labeling Assay manual as previously reported.39, 40 After the hybridization of the labeled single-strand cDNAs produced from the total RNAs to the GeneChips, the chips were scanned using a Fluidics 450 station and GeneChip Operating Software (Affymetrix). The quality control of the cDNA synthesis, labeling and hybridization process was verified to be optimal. Statistical analysis was based in data normalization and ANOVA (including t-tests) and was used to identify differentially expressed genes.
Gene ontology analysis
The differently expressed probe sets were analyzed with GO terms, which describe the functional attributes of a gene product in each of these three categories: molecular function, biological process and cellular component. The DAVID version 6.7 software provides statistical analyses in each of the three categories. One-sample two-sided binomial test was used to identify which GO terms of a particular population of probe sets had a significantly different proportion from the overall population (P<0.05). The resulting significant GO terms were manually organized and simplified usually with the help of the hierarchy annotated at the GO database (www.geneontology.org).
Acknowledgments
We thank Dr. Isabel Fabregat (IBIDELL, Barcelona, Spain) and Dr. Sally Wheatley (University of Nottingham, UK) for helpful discussions and Dr. Ana Losada (CNIO, Madrid, Spain) for her assistance to conduct PTTG2/Separase interaction experiments. We are also grateful to Dr. Robert Benezra (Memorial Sloan-Kettering Center) for sharing the pcDNA3-H2B-mCherry plasmid. JAP-T was supported by grants from the Ministerio de Educación y Cultura of Spain and the Dirección General de Universidades e Investigación of Junta de Andalucía. CM-V and MAM-M were recipients of a postdoctoral contract from the Spanish National Research Council (JAE-DOC) and Junta de Andalucía, respectively.
Glossary
- EMT
epithelial–mesenchymal transition
- FACS
fluorescence-activated cell sorter
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- H2B
histone H2B
- ORF
open reading frame
- Pttg
pituitary tumor-transforming gene
- qPCR
quantitative polymerase chain reaction
- S.E.M.
standard error of the mean
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- Salehi F, Kovacs K, Scheithauer BW, Lloyd RV, Cusimano M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer. 2008;15:721–743. doi: 10.1677/ERC-08-0012. [DOI] [PubMed] [Google Scholar]
- Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev. 2007;28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, et al. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Prezant TR, Kadioglu P, Melmed S. An intronless homolog of human proto-oncogene hPTTG is expressed in pituitary tumors: evidence for hPTTG family. J Clin Endocrinol Metab. 1999;84:1149–1152. doi: 10.1210/jcem.84.3.5658. [DOI] [PubMed] [Google Scholar]
- Chen L, Puri R, Lefkowitz EJ, Kakar SS. Identification of the human pituitary tumor transforming gene (hPTTG) family: molecular structure, expression, and chromosomal localization. Gene. 2000;248:41–50. doi: 10.1016/s0378-1119(00)00096-2. [DOI] [PubMed] [Google Scholar]
- Zur A, Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20:792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez A, Ramos-Morales F, Romero F, Rios RM, Dreyfus F, Tortolero M, et al. hpttg, a human homologue of rat pttg, is overexpressed in hematopoietic neoplasms. Evidence for a transcriptional activation function of hPTTG. Oncogene. 1998;17:2187–2193. doi: 10.1038/sj.onc.1202140. [DOI] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansieau S, Courtois-Cox S, Morel AP, Puisieux A. Failsafe program escape and EMT: a deleterious partnership. Semin Cancer Biol. 2011;21:392–396. doi: 10.1016/j.semcancer.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger I, Gimenez-Abian JF, Wernic D, Peters JM. Regulation of human separase by securin binding and autocleavage. Curr Biol. 2002;12:1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- LaRue KE, Bradbury EM, Freyer JP. Differential regulation of cyclin-dependent kinase inhibitors in monolayer and spheroid cultures of tumorigenic and nontumorigenic fibroblasts. Cancer Res. 1998;58:1305–1314. [PubMed] [Google Scholar]
- Wu RC, Schonthal AH. Activation of p53-p21waf1 pathway in response to disruption of cell-matrix interactions. J Biol Chem. 1997;272:29091–29098. doi: 10.1074/jbc.272.46.29091. [DOI] [PubMed] [Google Scholar]
- O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem L, Steinman R. A proapoptotic function of p21 in differentiating granulocytes. Leuk Res. 2005;29:1315–1323. doi: 10.1016/j.leukres.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Mueller S, Cadenas E, Schonthal AH. p21WAF1 regulates anchorage-independent growth of HCT116 colon carcinoma cells via E-cadherin expression. Cancer Res. 2000;60:156–163. [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol. 2010;27:1745–1749. doi: 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SH, Yang PM, Peng CW, Yu YC, Chiu SJ. Securin depletion sensitizes human colon cancer cells to fisetin-induced apoptosis. Cancer Lett. 2011;300:96–104. doi: 10.1016/j.canlet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Shah PP, Fong MY, Kakar SS. PTTG induces EMT through integrin alpha(V)beta(3)-focal adhesion kinase signaling in lung cancer cells. Oncogene. 2011;31:3124–3135. doi: 10.1038/onc.2011.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Kakar SS. Pituitary tumor transforming gene induces epithelial to mesenchymal transition by regulation of Twist, Snail, Slug, and E-cadherin. Cancer Lett. 2011;311:66–76. doi: 10.1016/j.canlet.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert J, Brabletz T. p53 Spreads out further: suppression of EMT and stemness by activating miR-200c expression. Cell Res. 2011;21:705–707. doi: 10.1038/cr.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- Mendez-Vidal C, Wilhelm MT, Hellborg F, Qian W, Wiman KG. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res. 2002;30:1991–1996. doi: 10.1093/nar/30.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend PA, Villanova I, Uhlmann E, Peyman A, Knolle J, Baron R, et al. An antisense oligonucleotide targeting the alphaV integrin gene inhibits adhesion and induces apoptosis in breast cancer cells. Eur J Cancer. 2000;36:397–409. doi: 10.1016/s0959-8049(99)00275-0. [DOI] [PubMed] [Google Scholar]
- Hwang SH, Choi YG, Jeong MY, Hong YM, Lee JH, Lim S. Microarray analysis of gene expression profile by treatment of Cinnamomi Ramulus in lipopolysaccharide-stimulated BV-2 cells. Gene. 2009;443:83–90. doi: 10.1016/j.gene.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Lin HQ, Choi R, Chan KL, Ip D, Tsim KW, Wan DC. Differential gene expression profiling on the muscle of acetylcholinesterase knockout mice: a preliminary analysis. Chem Biol Interact. 2010;187:120–123. doi: 10.1016/j.cbi.2010.03.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.