Abstract
We determined the effects of severe hypoxia (∼0.1% O2) on acute myeloid leukemia cells expressing the AML1/ETO oncogene. Incubation of Kasumi-1 cells in hypoxia induced growth arrest, apoptosis and reduction of AML1/ETO protein expression. The conditional expression of AML1/ETO in U937-A/E cells showed that hypoxia induces marked apoptosis in AML1/ETO-expressing cells only, pointing to AML1/ETO as a factor predisposing cells to hypoxia-induced apoptosis. In AML1/ETO-expressing cells, hypoxia enhanced TRAIL expression and its proapoptotic effects. AML1/ETO was found to bind TRAIL promoter and induce TRAIL transcription, although TRAIL expression was restrained by a concomitant relative transcription block. In hypoxia, such a TRAIL repression was removed and an increase of TRAIL expression was induced. Finally, blocking anti-TRAIL antibodies markedly reduced (Kasumi-1 cells) or completely inhibited (U937-A/E cells) hypoxia-induced apoptosis. Taken together, these results indicated that hypoxia induces apoptosis in AML1/ETO-expressing cells via a TRAIL/caspase 8-dependent autocrine loop and that TRAIL is a key regulator of hypoxia-induced apoptosis in these cells.
Keywords: AML, AML1/ETO, apoptosis, hypoxia, TRAIL
Acute myeloid leukemia (AML) is a hematopoietic neoplasia characterized by an impaired control of proliferation and differentiation. In 15–20% of AML cases, defined core-binding factor AML (CBF-AML), the disease is caused by either of two chromosomal aberrations, inv(16) or t(8;21). The latter results in the expression of the AML1/ETO fusion protein, which is responsible for a constitutive recruitment of a protein complex, which includes histone-deacetylases (HDAC) and DNA-methyltransferase (DNMT), resulting in the transcriptional repression of genes involved in myeloid maturation.1, 2 Although a number of proteins and biological functions has been shown to be targeted by AML1/ETO, the molecular mechanisms linking AML1/ETO to apoptosis are still to be clarified.3, 4
The regulative role of hypoxia on normal and leukemic hematopoiesis has been long addressed in our laboratory.5, 6, 7, 8 We found that the behavior of leukemia cell populations is strongly influenced by the response to hypoxia, which is a physiological feature of bone marrow, a preferential site of leukemia cell maintenance and growth. Hypoxia activates intracellular mediators of either apoptosis or survival depending on cell type and function.9, 10, 11, 12 Thus, the characterization of the response to hypoxia of a specific type of leukemia is helpful to understand the natural history of disease. In this respect, we focused on AML1/ETO-expressing AML cells.
The study reported here, after showing that hypoxia induces apoptosis in AML1/ETO-expressing cells, was directed to deepen the mechanism involved in this phenomenon. In these cells, the pro-apoptotic effect of hypoxia was found to be mediated by tumor necrosis factor-α-related apoptosis-inducing ligand (TRAIL; APO2L), and TRAIL expression to be critically regulated by AML1/ETO. This finding appears of particular interest as TRAIL has been shown to induce apoptosis in a number of tumor cell lines as well as in some primary tumors, whereas cells from most normal tissues are highly resistant to TRAIL-induced apoptosis.13, 14 This fact makes of TRAIL a tumor-selective apoptosis-inducing cytokine.15 The results presented here provide the first evidence linking AML1/ETO expression, hypoxia and TRAIL and point to TRAIL as a key target in AML1/ETO-expressing cells, which may reside in hypoxic tissue areas.
Results
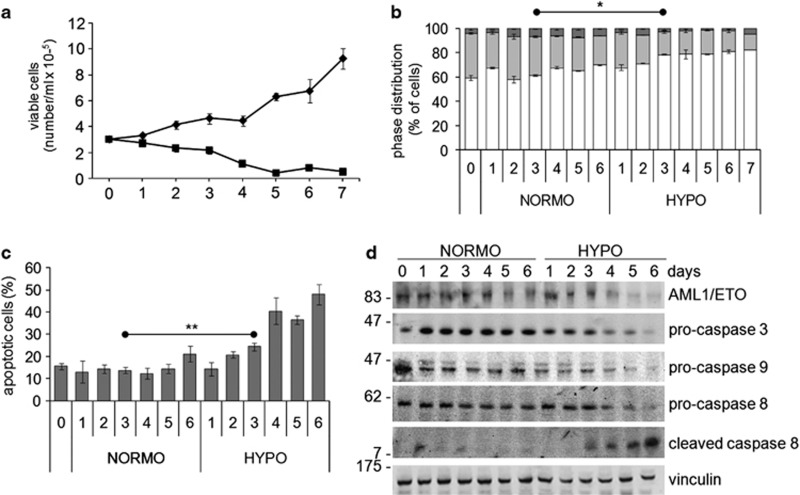
The expansion of Kasumi-1 cell population occurring in routine culture conditions (i.e., in normoxia) was suppressed in hypoxia, where the number of viable cells actually decreased with respect to time 0 (Figure 1a). The effect of incubation in hypoxia on cell number was related to a reduced proliferation, as indicated by the significantly (from day 3 on) decreased percentages of cells in the S phase of mitotic cycle and the increased percentage of G0/G1 cells (Figure 1b). Cell number reduction was paralleled by a significant increase of apoptotic rate from day 3 on, as determined by annexin V test (Figure 1c), as well as by the activation and consequent consumption of pro-caspases 3, 8, 9 and the appearance of cleaved caspase 8 (Figure 1d). Hypoxia also reduced, from day 4 on, the expression of the oncogenic AML1/ETO fusion protein (Figure 1d).
Figure 1.
Effects of hypoxia on viability, proliferation, cell-cycle distribution and apoptosis of Kasumi-1 cells. Cells were incubated in normoxia (diamond, NORMO) or hypoxia (square, HYPO) for the indicated times (days). (a) Viable cells were subjected to the trypan blue exclusion test and counted in a hemocytometer. (b) Mitotic cycle phase distribution of cells was measured by flow cytometry following propidium iodide staining; the percentages of cells in the G0/G1 (white), S (light gray) or G2/M (dark gray) phases are reported. (c) The percentage of apoptotic cells was measured by the Annexin V test and flow cytometry. (a–c) Values represent the average±S.E.M. of data from three (a, b) or four (c) independent experiments. (b, c) Differences, as determined by the Student's t-test for paired samples, were significant at day 3 (indicated in the graphs) and at all later incubation times (*P<0.05; **P<0.01); significance in (b) refers to either G0/G1 or S values. (d) Cell lysates were subjected to SDS-PAGE and immunoblotting with antibodies raised against ETO or the indicated proteins. Equalization of protein loading was verified on the same membrane. Representative results from one out of three independent experiments are shown
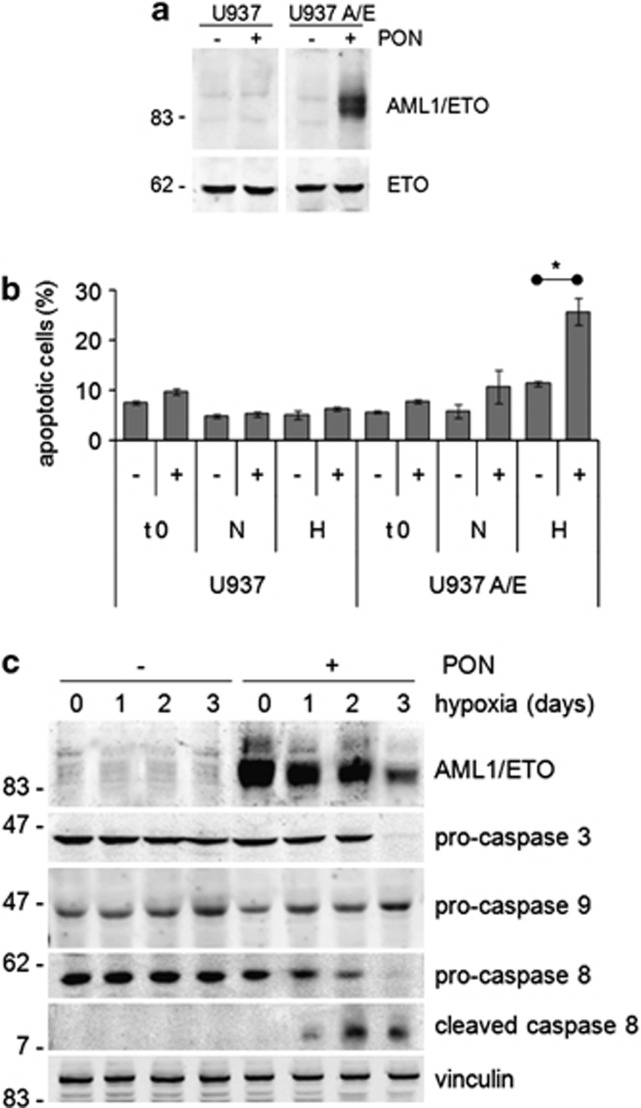
The relationship of AML1/ETO expression to hypoxia-induced apoptosis was addressed using U937 cells where ectopic AML1/ETO could be conditionally expressed (U937-A/E cells) following treatment with ponasterone.16, 17 In U937-A/E cells, ponasterone induced, as expected, the expression of AML1/ETO protein (Figure 2a). In ponasterone-treated, but not -untreated, U937-A/E cells, apoptosis increased significantly after 3 days of incubation in hypoxia, but not normoxia (Figure 2b), indicating a pro-apoptotic effect of AML1/ETO expression in hypoxia. It is worth pointing out that this effect was not due to ponasterone administration in hypoxia (compare histograms 1–2 to 6–12). Accordingly, the activation of caspase 3 (suppression of procaspase 3 at day 3) and caspase 8 (cleavage at days 1–3), but not caspase 9, was induced in AML1/ETO-expressing (but not control) U937-A/E cells in hypoxia, but not normoxia (Figure 2c). Hypoxia also reduced, from day 3 on, AML1/ETO expression in keeping with the results obtained with Kasumi-1 cells.
Figure 2.
Effects of AML1/ETO expression on hypoxia-induced apoptosis. U937 or U937-A/E cells were incubated for 2 days (from day −2 to time 0, t 0) in normoxia, in the absence (–) or the presence (+) of 5 μM ponasterone (PON) added daily (a–c), and then in normoxia or hypoxia (H, N) for 3 days (b) or in hypoxia for the indicated times (days; c). (a, c) Cell lysates were subjected to SDS-PAGE and immunoblotting with antibodies raised against ETO or the indicated proteins. Equalization of protein loading was verified on the same membrane. Representative results from one out of three independent experiments of each type are shown. (b) The percentages of apoptotic cells, as determined by Annexin V test and flow cytometry, are reported as average±S.E.M. of data from four independent experiments. Significance of differences was determined by the Student's t-test (*P<0.05)
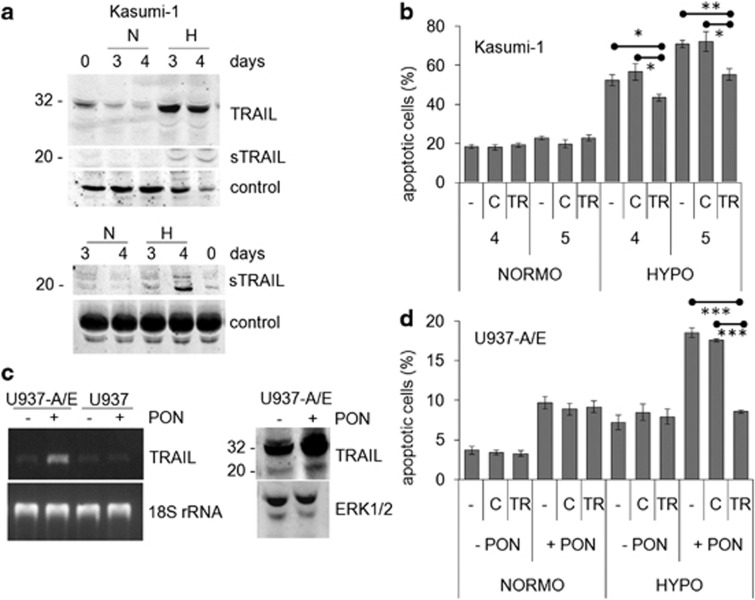
The involvement of the extrinsic pro-apoptotic pathway, indicated by caspase 8 activation, in AML1/ETO-related hypoxia-induced apoptosis was novel and investigated further. The expression of FAS or FAS-L, mediators of the extrinsic pathway, which are constitutively expressed in Kasumi-1 cells,18 was not increased in these cells in hypoxia (Supplementary Figure S1). Thus, we focused on TRAIL, the involvement of which in hypoxia-induced apoptosis of cancer cells had been reported.19, 20 Kasumi-1 cells express both the full-length and soluble forms of TRAIL protein (Figure 3a, time 0); their expression was markedly increased in hypoxia in either cell lysates (upper panel) or culture supernatants (lower panel). Figure 3b shows that the treatment of Kasumi-1 cells with blocking anti-TRAIL antibodies reduced significantly, although not completely, the increase of apoptosis determined by incubation in hypoxia. Control IgG antibodies determined negligible effects. This indicates that hypoxia-induced apoptosis in Kasumi-1 cells was at least in part TRAIL-dependent. Whether the effects of hypoxia on TRAIL expression and apoptosis were specifically related to AML1/ETO was addressed using its conditional expression in U937-A/E cells. Ponasterone stimulation of U937-A/E, but not wild-type U937, cells increased TRAIL mRNA (Figure 3c, left panel). Furthermore, the expression of both the full-length and soluble forms of TRAIL protein was increased by incubation in hypoxia for 2 days (Figure 3c, right panel). This increase is consistent with the timing of the induction of apoptosis shown in Figure 2b. The treatment of AML1/ETO-expressing U937-A/E cells with blocking anti-TRAIL antibodies, but not control IgG, abolished apoptosis induced by 2 days of incubation in hypoxia (Figure 3d). These results, well in keeping with those obtained with Kasumi-1 cells, indicated that AML1/ETO sensitizes, via TRAIL, leukemia cells to the pro-apoptotic effects of hypoxia.
Figure 3.
Effects of AML1/ETO and hypoxia on TRAIL expression and apoptosis. (a) Kasumi-1 cells were cultured for the indicated times in normoxia (N) or hypoxia (H) and cell lysates (top) or culture supernatants (bottom) were subjected to SDS-PAGE and immunoblotting with the anti-TRAIL antibody; sTRAIL: soluble form of TRAIL. Equalization of protein loading was verified on the same membrane (control). Representative results from one out of three independent experiments are shown. (b) Kasumi-1 cells were incubated in normoxia (NORMO) or hypoxia (HYPO) for the indicated times (days) in the absence (–) or the presence of a blocking anti-TRAIL antibody (TR) or of control rabbit IgG (c), and then the percentages of apoptotic cells were measured by Annexin V test and flow cytometry and reported as average±S.E.M. of data from three independent experiments. (c) (Left) U937-A/E or U937 cells were incubated for 2 days (from day −2 to time 0) in normoxia, in the absence (–) or the presence (+) of 5 μM ponasterone (PON) added daily, and then cells were lysed and total RNA extracted. RT-PCR for TRAIL or 18S rRNA was then performed. Representative results from one out of three independent experiments are shown. (Right) U937-A/E cells treated as above were incubated for further 2 days in hypoxia. Cell lysates were subjected to SDS-PAGE and immunoblotting with the indicated antibodies. (d) U937-A/E cells were incubated for 2 days (from day −2 to time 0) in normoxia, in the absence (–PON) or the presence (+PON) of 5 μM ponasterone added daily, and then incubated and treated for 2 days as described for (b). Data, reported as described for (b), are from three independent experiments. (b, d) Significance of differences was determined by the Student's t-test for paired samples (*P<0.05; **P<0.01; ***P<0.005)
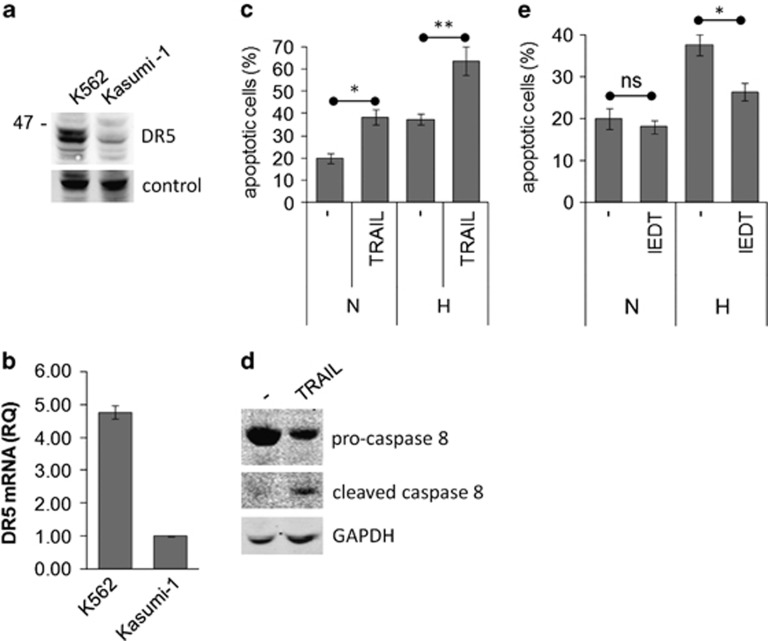
The responsiveness of Kasumi-1 cells to TRAIL was investigated further. The expression of the TRAIL receptor DR5 was assessed by western blotting (Figure 4a) and quantitative polymerase chain reaction (Q-PCR) (Figure 4b), verifying that DR5 is expressed in Kasumi-1 cells, although at a lower level than in K562 cells used as positive control. The sensitivity of Kasumi-1 cells to TRAIL-induced apoptosis was confirmed by adding exogenous TRAIL to cultures (Figure 4c). The robust induction of apoptosis observed in hypoxia was paralleled by a marked caspase 8 activation (Figure 4d). The involvement of caspase 8 activation was further confirmed by the significant prevention of apoptosis in hypoxia obtained by the caspase 8 inhibitor II Z-IEDT-FMK (Figure 4e). These results suggest that, in hypoxia, Kasumi-1 cells undergo induction of TRAIL expression/secretion and thereby of autocrine/paracrine apoptosis via the extrinsic pathway.
Figure 4.
Involvement of the extrinsic pathway in hypoxia-induced apoptosis. (a, b) Routinely cultured Kasumi-1 or K562 (positive control) cells were lysed and proteins subjected to SDS-PAGE and immunoblotting with the anti-DR5 antibody and equalization of protein loading verified on the same membrane (a), or total mRNA was recovered and the relative expression of DR5 calculated by Q-PCR using GAPDH for normalization and Kasumi-1 cells as calibrator (b). (c–e) Kasumi-1 cells were incubated in hypoxia for 4 days in the absence or the presence of 100 ng/ml human recombinant TRAIL (c, d) or 50 μM z-IEDT-FMK caspase 8 inhibitor II (IEDT; e). (c, e) The percentage of apoptotic cells was measured by Annexin V test and flow cytometry. Values represent the average±S.E.M. of data from two independent experiments. The statistical significance of differences was determined by the Student's t-test for paired samples (*P<0.05; **P<0.01; ns: not significant). (d) Cells were lysed and proteins subjected to SDS-PAGE and immunoblotting with anti-caspase 8 antibody. Equalization of protein loading was verified on the same membrane
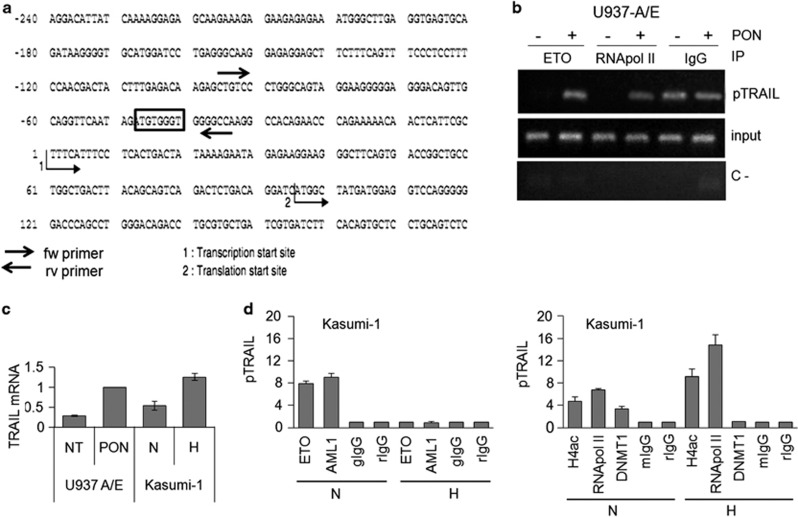
The above observations prompted us to determine whether AML1/ETO protein interact with TRAIL promoter directly. We identified a possible consensus sequence for the binding of the physiological AML1 transcription factor within the TRAIL promoter. This sequence (TGTGGT) is placed at −47 bp from the site of transcription start (Figure 5A). The binding of AML1/ETO to TRAIL promoter was evaluated by chromatin immunoprecipitation (ChIP) using primers designed to amplify a 61-bp fragment of TRAIL promoter including the above possible consensus sequence. Figure 5b shows the results of ChIP carried out using anti-ETO antibodies and reverse transcriptase PCR (RT-PCR) for TRAIL promoter with U937-A/E cells incubated for 2 days in the absence or the presence of ponasterone. When its expression was induced, AML1/ETO itself was recruited, together with RNApol II, to TRAIL promoter. AML1/ETO binding to TRAIL promoter was paralleled by the increase of TRAIL transcription, as determined by either RT-PCR (Figure 3c) or Q-PCR (Figure 5c). The fact that in the absence of ponasterone, no amplification of TRAIL promoter occurred following ETO immunoprecipitation (Figure 5c) confirmed ChIP reliability. These results represent the first demonstration that AML1/ETO binds and recruits the transcriptional complex (i.e., RNApol II) to TRAIL promoter (possibly at the AML1 consensus sequence identified) and that this results in the increase of TRAIL transcription. Figure 5c also shows that hypoxia induced TRAIL transcription in Kasumi-1 cells.
Figure 5.
Effects of AML1/ETO and hypoxia on TRAIL promoter. (a) Portion of TRAIL promoter (180 bp, exon 1) containing the possible consensus sequence (box) for AML1. (b) U937-A/E cells were incubated for 2 days (from day −2 to time 0) in normoxia, in the absence (–) or the presence (+) of 5 μM ponasterone added daily (PON) and lysed. Lysates were subjected to ChIP using antibodies against ETO, RNApol II or control IgG, and then to RT-PCR for the TRAIL promoter (pTRAIL). (c) U937-A/E cells treated (PON) or not (NT) with ponasterone like in (b), or Kasumi-1 cells incubated for 3 days in normoxia (N) or hypoxia (H) were lysed and Q-PCR for TRAIL mRNA was then performed. Histograms report the relative quantification of TRAIL mRNA (normalized for GAPDH) with respect to ponasterone-treated U937-A/E cells (used as calibrator). (d) Kasumi-1 cells were incubated in normoxia (N) or hypoxia (H) for 3 days, lysed, lysates subjected to ChIP using the indicated antibodies and Q-PCR for TRAIL promoter (pTRAIL) then performed. (c, d) Histograms represent the average±S.E.M. of data from three independent experiments
The binding of AML1/ETO to TRAIL promoter in relation to the effects of hypoxia was then evaluated in Kasumi-1 cells by ChIP using antibodies against either AML1 or ETO. Under routine culture conditions, that is in normoxia, AML1/ETO was bound to TRAIL promoter (Figure 5d, left graph) as expected following the results obtained in U937-A/E in Figure 5B. AML1/ETO binding in normoxia apparently occurred while TRAIL promoter was acetylated in H4 and RNApol II recruited (Figure 5d, right graph), indicating that transcription at that site was active. However, the presence of DNMT1 at TRAIL promoter hints to a concomitant relative TRAIL transcription block. This is in keeping with the constitutive, but relatively low, TRAIL expression shown in Figures 3a and 5c. In hypoxia, the relative TRAIL repression was removed, as AML1/ETO (Figure 5d, left) and DNMT1 (Figure 5d, right) abandoned TRAIL promoter, while RNApol II was recruited and H4 acetylation increased (Figure 5d, right). The release of TRAIL repression in hypoxia is in keeping with the maximal expression of TRAIL protein (Figure 3a) and mRNA (Figure 5c) observed in hypoxia with respect to normoxia.
Discussion
This study addressed the relationship of the expression of a leukemogenic oncogene, AML1/ETO, to the induction of apoptosis in AML cells in response to hypoxia and the involvement of TRAIL in this process. In the Kasumi-1 stabilized cell line, AML1/ETO expression decreased in the course of incubation in hypoxia, pointing to AML1/ETO suppression as a signal eliciting apoptosis. A rather different scenario emerged when the pro-apoptotic effects of hypoxia were tested on U937-A/E cells induced to express ectopic AML1/ETO. In these cells, hypoxia determined an increase of apoptosis only when AML1/ETO was induced, indicating that it is the expression of AML1/ETO, rather than its suppression, to make AML cells sensitive to hypoxia-induced apoptosis.
Our results pointed to the activation of extrinsic pathway/caspase 8 as the main mechanism driving hypoxia-induced apoptosis in AML1/ETO-expressing cells. When this mechanism was investigated further, we obtained the novel findings that TRAIL was involved and that the role of AML1/ETO in this phenomenon was critical. It emerged that: (a) TRAIL expression was linked to that of AML1/ETO and was therefore either constitutive (Kasumi-1 cells) or inducible (U937-A/E cells); (b) in AML1/ETO-positive cells, hypoxia enhanced TRAIL expression and its proapoptotic effects. That TRAIL expression is determinant for the induction of apoptosis in hypoxia was demonstrated via the addition of blocking anti-TRAIL antibodies to cultures, which completely suppressed hypoxia-induced apoptosis in ponasterone-treated U937-A/E cells. In Kasumi-1 cells, on the other hand, this impairment was marked, although incomplete, when driven by either a blocking anti-TRAIL antibody or a caspase 8 inhibitor, indicating that pro-apoptotic mediators other than TRAIL/caspase 8 are activated in these cells in response to hypoxia. One of these is certainly centered on caspase 9, which we showed involved in the hypoxia-induced apoptosis of Kasumi-1 cells, in keeping with the results obtained for other cell types.9
Contradicting results about the role of TRAIL in hypoxia-induced apoptosis had been reported previously. First, it has been shown that hypoxia induces a tumor cell phenotype highly sensitive to the cytotoxic effects of TRAIL19, 20 via HIF1α and PKCɛ20 and enhances TRAIL-induced PARP cleavage as well as the activation of caspase 8 and caspase 3. With respect to caspase 9, it has been reported that it is either involved21 or not,22 in keeping with our Kasumi-1 or U937-A/E cells data, respectively. By contrast, several studies showed that hypoxia inhibits TRAIL-mediated apoptosis.23, 24, 25, 26 The current hypothesis to explain this contradiction is centered on p53 activity: when p53 is wild-type, like in Kasumi-1 cells,27 sensitivity to TRAIL-induced apoptosis in hypoxia is high.28 Furthermore, when a long (like in our experiments) exposure to hypoxia stabilizes p53, the possibilities that TRAIL-dependent apoptosis is induced are higher.28
The basal TRAIL expression in Kasumi-1 cells could not explain the relatively high apoptotic rate we described previously17, 18 and show here for routine maintenance cultures, as the percentage of apoptotic cells in normoxia did not change following treatment with anti-TRAIL antibodies or caspase 8 inhibitor. Thus, the high basal apoptosis of Kasumi-1 cells may depend on the expression of AML1/ETO itself but not on its effects on TRAIL. Such a hypothesis would imply that in normoxia the growth-promoting oncogenetic effects of AML1/ETO override its pro-apoptotic signaling. Likewise, the increase of apoptosis induced by ponasterone in normoxic cultures of U937-A/E cells was insensitive to the treatment with anti-TRAIL antibodies, although ponasterone per se induced TRAIL. All above indicates that incubation in hypoxia is necessary to reveal the pro-apoptotic effects of autocrine TRAIL. Autocrine TRAIL was indeed capable to sensitize leukemia cells to the pro-apoptotic effects of hypoxia, but not to trigger apoptosis in normoxia. On the other hand, AML1/ETO expression appeared to steer the effects of hypoxia towards the induction of TRAIL-mediated apoptosis. Hypoxia, in turn, made AML1/ETO behave as a transcription inducer, rather than repressor, at least as far as TRAIL is concerned.
The question of how, via TRAIL, AML1/ETO sensitizes leukemia cells to the pro-apoptotic effect of hypoxia prompted us to determine by ChIP whether AML1/ETO protein interacted with TRAIL promoter directly. By way of ponasterone-treated U937-A/E cells, we demonstrated that AML1/ETO binds, probably through the AML1 consensus sequence identified, and recruits RNApol II to, TRAIL promoter, thereby inducing TRAIL transcription. ChIP studies directed to establish the interference of hypoxia with AML1/ETO binding to TRAIL promoter were carried out using Kasumi-1 cells. In routine cultures of these cells (i.e., in normoxia), TRAIL promoter was bound to AML1/ETO itself and was accessible to RNApol II, and TRAIL was expressed. However, the presence of an appreciable level of the DNMT1 repressor at TRAIL promoter hinted to a certain degree of concomitant TRAIL repression. Thus, a complex interplay involving AML1/ETO, TRAIL and hypoxia emerged, where AML1/ETO induced TRAIL expression constitutively, but at levels below the threshold of induction of TRAIL-dependent apoptosis. Hypoxia forces AML1/ETO to abandon TRAIL promoter, thereby resulting in the enrichment of TRAIL transcriptional machinery (increase of H4 acetylation and RNApol II and decrease of DNMT1) and the consequent induction of apoptosis. Finally, at delayed times, hypoxia drives AML1/ETO degradation, thereby suppressing its oncogenetic growth-promoting signaling. The demonstration that AML1/ETO is a positive regulator of TRAIL transcription in Kasumi-1 and U937-A/E cells is novel and in agreement with recent observations.3, 29 It is not known whether the removal of TRAIL repression following the detachment of AML1/ETO from TRAIL promoter is due to the fact that hypoxia enable other hypoxia-induced factors to bind the promoter and activate TRAIL transcription. It is to note that there is no contradiction within the fact that hypoxia drives an increase of TRAIL expression via AML1/ETO as well as a decrease of AML1/ETO expression. The reduction of viable cells exposed to hypoxia, as shown in Figure 1, appears actually to result from the combination of these factors. Hypoxia is likely indeed to play a dual role in the control of apoptosis in our system, inducing first the pro-apoptotic signaling operated by AML1/ETO via autocrine TRAIL and then suppressing the anti-apoptotic signals that AML1/ETO oncogene most likely directly generates.
The suppression of oncogenetic proteins, thereby preventing clonal expansion, has been shown in our laboratory to parallel adaptation to hypoxia of several different leukemia cell populations.6, 7, 30, 31 In the case of AML1/ETO-expressing AML, an additional reason for its suppression in hypoxia may be represented by the capacity of AML1/ETO to bind TRAIL promoter, induce TRAIL expression and thereby decrease the capacity of leukemia cells to stand hypoxia.
Materials and Methods
Cells and culture conditions
The Kasumi-1 human AML1/ETO-positive cell line,32 a kind gift of Dr. C Chomienne (Université Denis Diderot, Institut Universitaire d'Hématologie, Paris, France) in 1998, was routinely (every 2–3 months) checked for the expression of AML1/ETO by Q-PCR and/or western blotting. The U937 human promonocytic leukemia cell line33 and U937-A/E cells, where AML1/ETO expression is inducible34 following treatment with 5 μM ponasterone-A (Sigma-Aldrich, St Louis, MD, USA), were a kind gift of Dr. M Lübbert (Freiburg Universität, Department of Medicine, Freiburg, Germany). Cells were routinely cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 4 mM glutamine, 50 U/ml penicillin and 50 mg/ml streptomycin and incubated at 37 °C in a water-saturated atmosphere containing 5% CO2. Experiments were performed with exponentially growing cells plated at 3 × 105/ml and incubated as above (normoxia; 21% O2) or in a Concept 400 anaerobic incubator (Ruskinn Technology Ltd., Bridgend, UK) flushed with a water-saturated preformed gas mixture containing 0.1% O2, 5% CO2 and ≈95% N2 (hypoxia). In selected experiments, cells were treated with the cell-permeable irreversible caspase 8 inhibitor II Z-IEDT-FMK (BioVision Inc., Milpitas, CA, USA) or human recombinant TRAIL (ImmunoTools, Friesoythe, Germany).
Measure of cell viability, apoptosis and cell-cycle phases
Cell viability was measured by the trypan blue exclusion test, by counting trypan-negative cells in a hemocytometer. To quantify apoptosis, cells were centrifuged, resuspended in antibody-binding buffer (HEPES-buffered saline solution with 2.5 mM CaCl2) and incubated with FITC-labeled Annexin-V (Roche Diagnostics, Basel, Switzerland) and propidium iodide (PI) for 15 min at room temperature (RT) in the dark. Flow cytometry was performed using a FACSCanto (Beckton & Dickinson, San Josè, CA, USA). Annexin-V+/PI- cells were considered early apoptotic and annexin-V+/PI+ cells late apoptotic, whereas Annexin-V+ cells defined total apoptosis. In some experiments, cells were incubated in the presence of a blocking anti-TRAIL antibody (0.5 μg/ml; Abcam, Cambridge, UK, 9959-100) or control rabbit IgG (0.5 μg/ml; Sigma-Aldrich). To determine cell-cycle phase distribution, cells were centrifuged and pellet resuspended in 1 ml of PI solution (50 μg/ml PI, 0.1% trisodium citrate, 0.1% NP40). After 30 min of incubation in the dark, nuclei were analyzed by flow cytometry.
Cell lysis and western blotting
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) containing 100 μM orthovanadate and lysed by incubation in Laemmli buffer (Tris/HCl 62.5 mM pH 6.8, 10% glycerol, 0.005% blue bromophenol, 2% SDS) at 95 °C for 10 min in the presence of 100 mM 2-mercaptoethanol. Lysates were then clarified by centrifugation (20 000 × g, 10 min, RT). In some experiments, cell culture supernatants (50 μg) were subjected to western blotting. Protein concentration was determined by the BCA method and 15 μg protein/sample were separated by SDS-PAGE in 9–15% polyacrylamide gel and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA) by electroblotting. Membranes were incubated for 1 h at RT in Odyssey Blocking Buffer diluted 1 : 1 with PBS, and then for 16–18 h at 4 °C in PBS containing 0.1% Tween-20 and the primary antibody. Primary antibodies used were: rabbit polyclonal anti-procaspase 3 (#sc-7272), rabbit polyclonal anti-procaspase 8 (#sc-7890), goat polyclonal anti-GAPDH (#sc-20357), rabbit polyclonal anti-procaspase 9 (#sc-7885), goat polyclonal anti-ETO (#sc-9737), rabbit polyclonal anti-FAS (#sc-7886), rabbit polyclonal anti-FAS-L (#sc-834), rabbit polyclonal anti-ERK1 (#sc-93) from Santa Cruz Biotechnology Inc., Dellas, TX, USA; rabbit polyclonal anti-TRAIL (#9959-100) from Abcam, mouse monoclonal anti-vinculin (#v9131) from Sigma-Aldrich, mouse monoclonal anti-cleaved caspase 8 (#9748) from Cell Signaling Technology, Inc., Danvers, MA, USA. After extensive washing with 0.1% Tween-20 in PBS, membranes were incubated for 1 h at 4 °C in Odyssey Blocking Buffer diluted 1 : 1 with PBS containing an IRDye800CW- or IRDye680-conjugated secondary antibody. Bands were visualized by infrared imaging using an Odyssey detector (Licor, Lincoln, NE, USA), images were recorded as TIFF files and band intensity was measured with the Adobe Systems Incorporated Photoshop software (San Jose, CA, USA).
Chromatin immunoprecipitation
Aliquots of 106 Kasumi-1 cells were treated with 1% formaldehyde for 10 min at 37 °C and cross-linking stopped by adding glycine at 0.125 M final concentration. Cells were centrifuged at 1300 r.p.m. for 5 min at 4 °C and washed twice using ice-cold PBS containing protease inhibitors: 1 mM phenyl-methyl-sulphonyl-fluoride (PMSF), 1 μg/ml aprotinin and 1 μg/ml pepstatin-A. Cell pellet was lysed in 200 μl of lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS, 1 mM PMSF, 1 μg/ml aprotinin and 1 μg/ml pepstatin-A) for 10 min on ice and then sonicated (165 pulses, 12 s on and 28 s off each, at maximum power in a Sonoplus apparatus; Bandelin GM3200) to generate DNA fragments of 100–500 bp. After centrifugation (13 000 r.p.m., 10 min, 4 °C), the supernatant was 10-fold diluted with ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100) and 4% of this sample recovered and used as an indicator of chromatin content in each sample (input). In a new tube, 2 μg antibody was incubated with 14 μl Protein G- Dynabeads (Invitrogen, Carlsbad, CA, USA, #100.03D) and 200 μl PBS/Tween-20 0.02%, for 30 min at RT under constant agitation. The bead–antibody complexes were recovered with a Dynal magnet (Invitrogen), resuspended in 100 μl PBS/BSA 100 mg/ml and added to each sample to be incubated for 16–18 h at 4 °C under constant agitation. In parallel, each sample was subjected to the same procedure without antibody (negative control). Antibody-protein–DNA complexes were transferred on the magnet and supernatant was removed. After extensive washing, complexes were eluted from beads with 500 μl elution buffer (0.1 M NaHCO3, 1% SDS). Following addition of 0.2 M NaCl, all samples, including input, were incubated for 4 h at 65 °C to revert cross-linking. After treatment with 10 μM RNAase and digestion with 40 μM proteinase-K, DNA was extracted using QIAcquick PCR purification, according to the manufacturer's recommendations (Qiagen, German-Town, MD, USA, #28106) and DNA eluted in 50 μl of water. ChIP-grade antibodies used were: rabbit polyclonal anti-pan-acetylated-H4 (Millipore, #06-598), anti-RNApol II (Abcam, #ab5408), anti-DNMT1 (Abcam, #ab5208); goat polyclonal anti-ETO (Santa Cruz Biotechnology, #sc-9737). Other antibodies used were: goat polyclonal anti-AML1 (#sc-8563) and goat IgG (#sc-20357) from Santa Cruz Biotechnology; rabbit IgG (#G5518) and mouse IgG (#M7023) from Sigma-Aldrich.
Reverse Transcriptase Polymerase Chain Reaction
After total RNA extraction by RNeasy Plus Mini as recommended by the manufacturer (Qiagen, #28106), 1 μg/sample of total RNA was subjected to reverse transcription with SuperScriptVILO-Reverse Transcriptase (Invitrogen) for 10 min at 25 °C, 1 h at 42 °C and 5 min at 85 °C, utilizing 50 pmol hexameric random primers. RT-PCR was performed on a MasterCycler (Eppendorf, Milano, Italy) using GoTaq DNA Polymerase (Promega, Madison, WI, USA). TRAIL mRNA expression was evaluated using the following primers: for 5′-GAGAGTAGCAGCTCACATAAACT-3′ rev 5′-TCATGGATGACCAGTTCACCATT-3′. 18 S rRNA was used as control with the following primes: for 5′-CGGCTACCACATCCAAGGAA-3′ rev 5′-GCTGGAATTACCGCGGCT-3′. All primers were used at 0.2 μM. RT-PCR parameters were: 2 min at 94 °C, 35 cycles at 94 °C for 20 s, 60 °C for 20 s and 72 °C for 20 s; final extension at 72 °C for 5 min. RT-PCR products were visualized by a run in a 3% agarose gel and image acquisition (Kodak EDAS 299 camera, Rochester, NY, USA).
Quantitative Real-Time PCR
Q-PCR (2 min at 50 °C, 5 min at 95 °C, 40 cycles at 95 °C for 15 s and 60 °C for 1 min) was performed with the ABI Prism 7500 Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA) using Power SYBR Green PCR master mix (Applied Biosystems). A melting curve analysis was performed to discriminate between specific and non-specific PCR products. The relative expression of TRAIL mRNA (using the same primers indicated above) was calculated by using a comparative threshold cycle method and the formula 2−(ΔΔCt).35 The GAPDH mRNA was used for normalization with the following primers: for 5′-AACAGCCTCAAGATCATCAGCAA-3′ rev 5′-CAGTCTGGGTGGCAGTGAT-3. The relative amount of immunoprecipitated TRAIL promoter DNA was determined using the following primers: for 5'-CTGTCCCTGGGCAGTAGGAA-3' rev 5′-GGCCCCACCCACATCTATT-3′. Data were normalized by input DNA and expressed with respect to those of control IgG (used as calibrator). The amount of immunoprecipitated TRAIL promoter DNA was also monitored by RT-PCR using the same primers indicated for Q-PCR. The amount of TRAIL mRNA was determined also by Q-PCR using the same primers indicated for RT-PCR. DR5 mRNA was determined using the following primers: for 5'-GCCTCATGGACAATGAGATAAAGGT-3' rev 5′- TCAATCTTCTGCTTGGCAAGTCTCT-3'.
Acknowledgments
This work was supported by Istituto Toscano Tumori, Associazione Italiana per la Ricerca sul Cancro, Ministero della Salute (Ricerca Finalizzata, grant #RF-TOS-2008-1163728), Regione Toscana (Programma per la Ricerca in Materia di Salute), Associazione Italiana per la Lotta contro le Leucemie e i Linfomi (sezione di Prato), Fondazione Cassa di Risparmio di Volterra, Fondazione Oretta Bartolomei-Corsi.
Author contributions
ER and VB conceived, designed and performed the experiments, analyzed the data and wrote the manuscript. PDS contributed to the interpretation of data and wrote the manuscript. IT and MGC performed the experiments. All authors read and approved the final version of manuscript.
Glossary
- AML
acute myeloid leukemia
- CBF-AML
core-binding factor AML
- HDAC
histone-deacetylases
- DNMT
DNA-methyltransferase
- TRAIL
tumor necrosis factor-α-related apoptosis-inducing ligand
- FBS
fetal bovine serum
- PI
propidium iodide
- RT
room temperature
- PBS
phosphate-buffered saline
- ChIP
chromatin immunoprecipitation
- PMSF
phenyl-methyl-sulphonyl-fluoride
- Q-PCR
quantitative real-time PCR
- RT-PCR
reverse transcriptase polymerase chain reaction
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by H-U Simon
Supplementary Material
References
- Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. Protein ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR. The AML1-ETO chimaeric transcription factor in acute myeloid leukaemia: biology and clinical significance. Br J Haematol. 1999;106:296–308. doi: 10.1046/j.1365-2141.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, et al. M. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet. 2008;4:e1000275. doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–5679. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- Cipolleschi MG, Dello Sbarba P, Olivotto M. The role of hypoxia in the maintenance of hematopoietic stem cells. Blood. 1993;82:2031–2037. [PubMed] [Google Scholar]
- Giuntoli S, Rovida E, Barbetti V, Cipolleschi MG, Olivotto M, Dello Sbarba P. Hypoxia suppresses BCR/Abl and selects imatinib-insensitive progenitors within clonal CML populations. Leukemia. 2006;20:1291–1293. doi: 10.1038/sj.leu.2404224. [DOI] [PubMed] [Google Scholar]
- Giuntoli S, Rovida E, Gozzini A, Barbetti V, Cipolleschi MG, Olivotto M, et al. P. Severe hypoxia defines heterogeneity and selects highly immature progenitors within clonal erythroleukemia cells. Stem Cells. 2007;25:1119–1125. doi: 10.1634/stemcells.2006-0637. [DOI] [PubMed] [Google Scholar]
- Cipolleschi MG, Rovida E, Ivanovic Z, Praloran V, Olivotto M, Dello Sbarba P. The expansion of murine bone marrow cells preincubated in hypoxia as an in vitro indicator of their marrow-repopulating ability. Leukemia. 2000;14:735–739. doi: 10.1038/sj.leu.2401744. [DOI] [PubMed] [Google Scholar]
- Shroff EH, Snyder C, Chandel NS. Role of Bcl-2 family members in anoxia-induced cell death. Cell Cycle. 2007;6:807–809. doi: 10.4161/cc.6.7.4044. [DOI] [PubMed] [Google Scholar]
- Han SH, Kim M, Park K, Kim TH, Seol DW. Blockade of processing/activation of caspase3 by hypoxia. Biochem Biophys Res Commun. 2008;375:684–688. doi: 10.1016/j.bbrc.2008.08.091. [DOI] [PubMed] [Google Scholar]
- Kim M, Park SY, Pai HS, Kim TH, Billiar TR, Seol DW. Hypoxia inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by blocking Bax translocation. Cancer Res. 2004;64:4078–4081. doi: 10.1158/0008-5472.CAN-04-0284. [DOI] [PubMed] [Google Scholar]
- Greljer AE, van der Wall E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J Clin Pathol. 2004;57:1009–1014. doi: 10.1136/jcp.2003.015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchiero P, Zauli G. Tumor-necrosis-factor-related apoptosis inducing ligand and the regulation of hematopoiesis. Curr Opin Hematol. 2008;15:42–48. doi: 10.1097/MOH.0b013e3282f15fa6. [DOI] [PubMed] [Google Scholar]
- Manzo F, Nebbioso A, Miceli M, Conte M, De Bellis F, Carafa V, et al. L. TNF-related apoptosis-inducing ligand: signalling of a 'smart' molecule. Int J Biochem Cell Biol. 2009;41:460–466. doi: 10.1016/j.biocel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Noteborn MH. Proteins selectively killing tumor cells. Eur J Pharmacol. 2009;625:165–173. doi: 10.1016/j.ejphar.2009.06.068. [DOI] [PubMed] [Google Scholar]
- Lu Y, Xu YB, Yuan TT, Song MG, Lübbert M, Fliegauf M, et al. GQ. Inducible expression of AML1-ETO fusion protein endows leukemic cells with susceptibility to extrinsic and intrinsic apoptosis. Leukemia. 2006;20:987–993. doi: 10.1038/sj.leu.2404218. [DOI] [PubMed] [Google Scholar]
- Barbetti V, Gozzini A, Rovida E, Morandi A, Spinelli E, Fossati G, et al. V. Selective anti-leukaemic activity of low-dose histone deacetylase inhibitor ITF2357 on AML1/ETO-positive cells. Oncogene. 2008;27:1767–1778. doi: 10.1038/sj.onc.1210820. [DOI] [PubMed] [Google Scholar]
- Rovida E, Gozzini A, Barbetti V, Giuntoli S, Santini V, Dello Sbarba P. The c-Jun-N-terminal-Kinase inhibitor SP600125 enhances the butyrate derivative D1-induced apoptosis via caspase 8 activation in Kasumi-1 t(8;21) acute myeloid leukaemia cells. Br J Haematol. 2006;135:653–659. doi: 10.1111/j.1365-2141.2006.06365.x. [DOI] [PubMed] [Google Scholar]
- Weinmann M, Marini P, Jendrossek V, Betsch A, Goecke B, Budach W, et al. Influence of hypoxia on TRAIL-induced apoptosis in tumor cells. Int J Radiat Oncol Biol Phys. 2004;58:386–396. doi: 10.1016/j.ijrobp.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Gobbi G, Masselli E, Micheloni C, Nouvenne A, Russo D, Santi P, et al. P. Hypoxia-induced down-modulation of PKCepsilon promotes trail-mediated apoptosis of tumor cells. Int J Oncol. 2010;37:719–729. doi: 10.3892/ijo_00000721. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Lee KH, Kim HR, Jessup JM, Seol DW, Kim TH, et al. YK. Sodium nitroprusside enhances TRAIL-induced apoptosis via a mitochondria-dependent pathway in human colorectal carcinoma CX-1 cells. Oncogene. 2001;20:1476–1485. doi: 10.1038/sj.onc.1204225. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Moon MS, Kwon SJ, Rhee JG. Hypoxia and low glucose differentially augments TRAIL-induced apoptotic death. Mol Cell Biochem. 2005;270:89–97. doi: 10.1007/s11010-005-5261-8. [DOI] [PubMed] [Google Scholar]
- Park SY, Billiar TR, Seol DW. Hypoxia inhibition of apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) Biochem Biophys Res Commun. 2002;291:150–153. doi: 10.1006/bbrc.2002.6421. [DOI] [PubMed] [Google Scholar]
- Kim M, Park SY, Pai HS, Kim TH, Billiar TR, Seol DW. Hypoxia inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by blocking Bax translocation. Cancer Res. 2004;64:4078–4081. doi: 10.1158/0008-5472.CAN-04-0284. [DOI] [PubMed] [Google Scholar]
- Nagaraj NS, Vigneswaran N, Zacharias W. Hypoxia inhibits TRAIL-induced tumor cell apoptosis: involvement of lysosomal cathepsins. Apoptosis. 2007;12:125–139. doi: 10.1007/s10495-006-0490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LD, Bailey CK, Brandt SJ, Thompson ME. BRCA1 accumulates in the nucleus in response to hypoxia and TRAIL and enhances TRAIL-induced apoptosis in breast cancer cells. FEBS J. 2007;274:5137–5146. doi: 10.1111/j.1742-4658.2007.06033.x. [DOI] [PubMed] [Google Scholar]
- Nijhuis EH, Poot AA, Feijen J, Vermes I. Hsp70- and p53-reponses after heat treatment and/or X-irradiation mediate the susceptibility of hematopoietic cells to undergo apoptosis. Int J Radiat Biol. 2008;84:99–105. doi: 10.1080/09553000701817084. [DOI] [PubMed] [Google Scholar]
- Mayes PA, Campbell L, Ricci MS, Plastaras JP, Dicker DT, El-Deiry WS.Modulation of TRAIL-induced tumor cell apoptosis in a hypoxic environment Cancer Biol Ther 200541068–1074.(ex-23). [DOI] [PubMed] [Google Scholar]
- Dunne J, Gascoyne DM, Lister TA, Brady HJ, Heidenreich O, Young BD. AML1/ETO proteins control POU4F1/BRN3A expression and function in t(8;21) acute myeloid leukemia. Cancer Res. 2010;70:3985–3995. doi: 10.1158/0008-5472.CAN-09-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli S, Tanturli M, Di Gesualdo F, Barbetti V, Rovida E, Dello Sbarba P. Glucose availability in hypoxia regulates the selection of chronic myeloid leukemia progenitor subsets with different resistance to imatinib-mesylate. Haematologica. 2011;96:204–212. doi: 10.3324/haematol.2010.029082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanturli M, Giuntoli S, Barbetti V, Rovida E, Dello Sbarba P. Hypoxia selects bortezomib-resistant stem cells of chronic myeloid leukemia. PLoS One. 2011;6:e17008. doi: 10.1371/journal.pone.0017008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asou H, Tashiro S, Hamamoto K, Osuji A, Kita K, Kamada N. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
- Larrick JW, Fischer DG, Anderson SJ, Koren HS. Characterization of a human macrophage-like cell line stimulated in vitro: a model of macrophage functions. J Immunol. 1980;125:6–12. [PubMed] [Google Scholar]
- Fliegauf M, Stock M, Berg T, Lubbert M. Williams-Beuren syndrome critical region-5/non-T-cell activation linker: a novel target gene of AML1/ETO. Oncogene. 2004;23:9070–9081. doi: 10.1038/sj.onc.1208042. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.