SUMMARY
To characterize the role of rare complete human knockouts in autism spectrum disorders (ASD), we identify genes with homozygous or compound heterozygous loss-of-function (LoF) variants (defined as nonsense and essential splice sites) from exome sequencing of 933 cases and 869 controls. We identify a two-fold increase in complete knockouts of autosomal genes with low rates of LoF variation (≤5% frequency) in cases and estimate a 3% contribution to ASD risk by these events, confirming this observation in an independent set of 563 probands and 4,605 controls. Outside the pseudo-autosomal regions on the X-chromosome, we similarly observe a significant 1.5-fold increase in rare hemizygous knockouts in males, contributing to another 2% of ASDs in males. Taken together these results provide compelling evidence that rare autosomal and X-chromosome complete gene knockouts are important inherited risk factors for ASD.
INTRODUCTION
Autism spectrum disorder (ASD) is a highly heritable, common disorder that affects ~1 in 88 individuals (2012). Previous studies have shown a reproducible contribution of de novo copy number variants (CNVs) (Levy et al., 2011; Sanders et al., 2011; Sebat et al., 2007; Weiss et al., 2008) and de novo single nucleotide variants (SNVs) (Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012; Sanders et al., 2012) to ASD risk - though these effects provide little explanation for the widely recognized high heritability (Constantino et al., 2012).
An early segregation analysis on 46 multiplex families (each with multiple affected children) suggested evidence for an autosomal recessive (or ‘2-hit’) model in ASD (Ritvo et al., 1985) with a subsequent study showing that ASD is unlikely to fit a model with a major gene effect (Jorde et al., 1991). Further to this point, the most recent results from de novo CNVs and SNVs point to a model in which hundreds of genes are likely to contribute to autism risk. Building from these observations, as a means of providing insight into the heritable component of ASD risk, we sought to test the hypothesis that 2-hit etiologies exist in ASD and that these events, like the de novo CNVs and SNVs, are most likely to be distributed over many genes. Supporting this hypothesis are historical segregation analyses (Ritvo et al., 1985; Zweier et al., 2009), the successful use of homozygosity mapping in consanguineous populations (Morrow et al., 2008), as well as recent studies showing that ASD probands had a significant excess of homozygous haplotype sharing, suggesting that there are recessive loci in these risk-conferring haplotypes (Casey et al., 2011; Chahrour et al., 2012). Other studies have also implicated the role of a 2-hit or oligogenic model for rare CNVs in ASD (Girirajan et al., 2012).
It has been shown that there are relatively few homozygous or compound heterozygous LoF variants (i.e., complete gene knockouts) in healthy individuals. Most of these complete knockouts found are common (MAF>5%) and are distributed across a very small number (~100–200) of genes, such as the olfactory receptors, that are apparently inessential and do not result in any obvious phenotype or severe medical consequence (MacArthur et al., 2012). We similarly observe in these ASD datasets that an average individual harbors ~5 common complete knockouts (from nonsense and essential splice site variants) distributed across a small subset of genes on the autosomes. In striking contrast, if we consider only LoF variants with frequency ≤5%, fewer than 5% of individuals harbor even a single rare complete knockout (Table 1). While heterozygous LoF mutations are seen in thousands of genes, the very low frequency and paucity of observed complete knockouts suggests a broad pool of genes (including many Mendelian disorders) where 2-hit variants may give rise to severe and reproductively deleterious phenotypes. While genes with common complete knockouts are more likely to be benign (or unlikely to result in severe phenotypes with high penetrance), genes with rare complete knockouts are more likely to be disease-causing (Gorlov et al., 2008) simply because selection prevents deleterious recessive-acting variants from reaching even moderate allele frequencies.
Table 1.
Population Distribution of Rare and Common LoFs
| Average number of homozygous variants |
Number of unique genes with a homozygous variant |
Average number of heterozygous variants |
Number of unique genes with a heterozygous variant |
|
|---|---|---|---|---|
| Rare (≤5%) LoFs | 0.05 variant per individual | 33 genes | 13 variants per individual | 3,409 genes |
| Common (>5%) LoFs | 5 variants per individual | 96 genes | 36 variants per individual | 99 genes |
The average number of rare (≤5%) and common (>5%) homozygous LoF variants, as well as the average number of such variants calculated from the BI case-control dataset.
If a subset of ASD cases were caused by rare 2-hit events with large effects (e.g. odds ratios of >5) distributed across many different genes, then family-based linkage or GWAS would have little power to detect such events, as each locus individually would explain a very small fraction of all cases given the commonness of the outcome and the large number of ASD genes. To evaluate evidence for such 2-hit etiologies in ASD, we studied the distribution and patterns of rare complete knockouts from whole-exome sequence data across two case-control studies comprised of 1,802 European subjects to identify events in which individuals carried 2 LoF autosomal variants in a single gene in trans. In this study, we show that rare complete knockouts on the autosomes (variant allele frequencies of ≤5%) are significantly enriched in cases, suggesting that these events contribute to the genetic etiology of ASD.
A variant with a diploid allele frequency of 5% on the autosomes results in a complete knockout in 0.25% of the individuals. Outside the pseudo-autosomal regions on the X-chromosome in males, a single LoF variant with 0.25% allele frequency also results in a complete knockout in 0.25% of males. Similarly, we found that rare complete knockouts on the X-chromosome (variant allele frequencies of ≤0.25%) are also significantly enriched in male cases, further reinforcing the role of rare complete knockouts as risk factors for ASD.
RESULTS
Exome Capture and Sequencing
To assess the contribution of rare complete knockouts to ASD, we analyzed data from an ethnically-matched case-control population. We selected 933 cases and 869 controls sequenced in this study by matching them with multi-dimensional scaling (MDS) of common variants genotyped on Illumina 1M, Affymetrix 5.0, or 6.0 arrays (Purcell et al., 2007) to reduce potential confounding by population stratification. The exomes were sequenced at two different sequencing centers – the Broad Institute (BI) and the Baylor College of Medicine (BCM). A total of 428 ASD cases selected from the Autism Genetic Resource Exchange (AGRE) and 378 NIMH controls (a total of 806 individuals) were sequenced at BI, and another 505 ASD cases selected from the Autism Simplex Collection (TASC) and 491 NIMH controls (a total of 996 individuals) were sequenced at BCM, resulting in 1,802 individuals across the two case-control datasets. All controls were selected from an NIMH control repository and were ascertained for not having schizophrenia or bipolar mood disorder. Another 563 probands were added into the final analyses (388 trios/quartets from the Simons Simplex Collection (SSC) (Iossifov et al., 2012; Sanders et al., 2012), 175 trios from the Boston Autism Consortium sequenced at BI (104 from (Neale et al., 2012)) and together with 4,605 additional European controls from the NHLBI exome sequencing project and the 1000 Genomes Project, this resulted in a total of >6,000 exomes used in this study (Table S1). The metrics for the case-control datasets are described in Table S2.
Enrichment of Rare Complete Knockouts in ASD
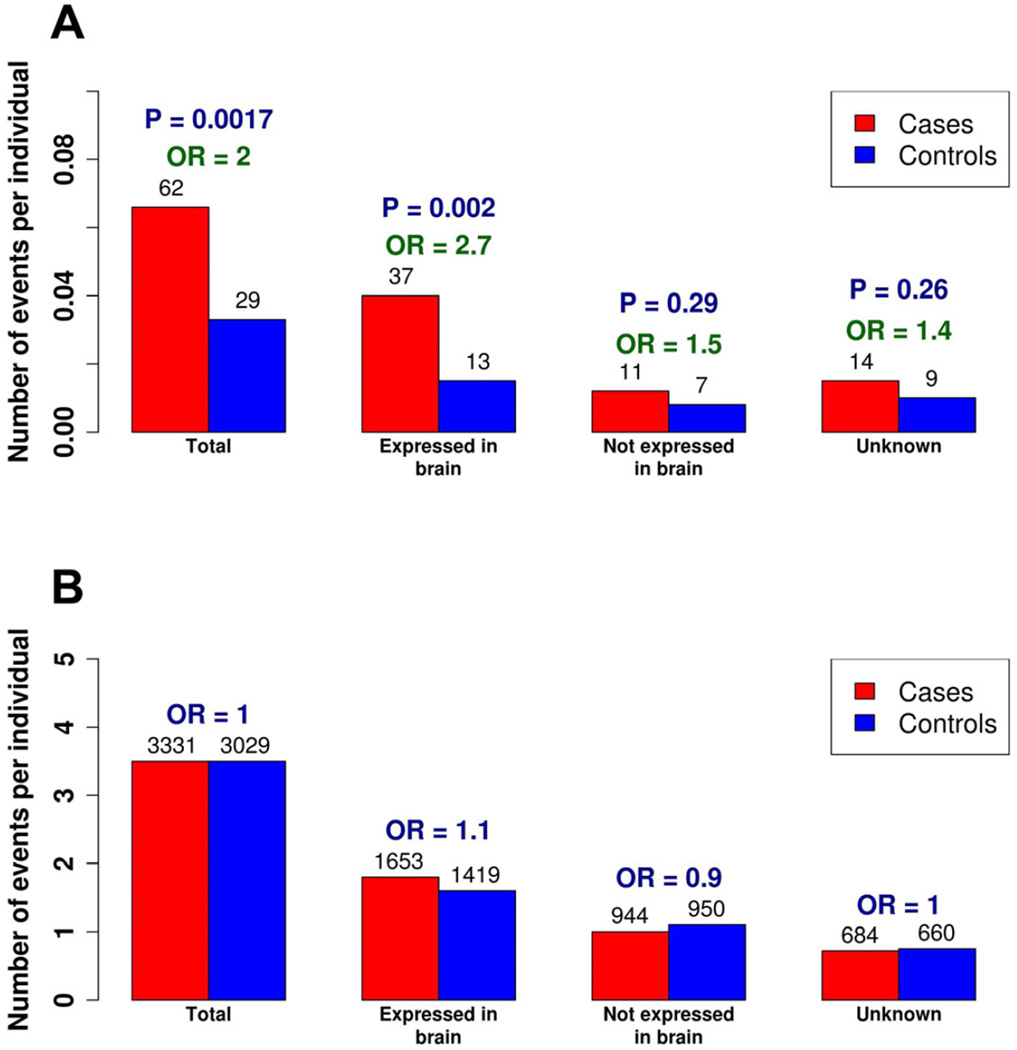
Given that rare complete knockouts consist of both compound heterozygous and homozygous variants on the autosomes, we adapted a statistical phasing approach similar to the four-haplotype test to eliminate instances in which multiple LoF variants may segregate in cis (Figure S1). There are a total of 91 such rare complete knockouts in the case-control datasets, with 62 of these found in the cases compared to 29 in the controls (Table S3), representing a roughly 2-fold enrichment of these events in the cases (odds ratio (OR) = 2.0, 95% CI = [1.5, 2.5], one-sided permutation P = 0.0017). Based on the difference between cases and controls (6% of the cases versus 3.3% of the controls have a rare complete knockout), we estimate a ~3% contribution by rare complete knockouts to ASD. While different capture and sequencing technologies were employed at the two sequencing centers, and different depths of sequencing achieved (Li et al., unpublished), the excess in cases was consistent in the two datasets (ORs = 2.1, 95% CI = [1.5, 2.7] and 1.8, 95% CI = [1.1, 2.5]).
Using the results from a previous study of expression patterns of post-mortem brains (Kang et al., 2011), we observed the enrichment in rare complete knockouts in cases was particularly pronounced in genes found to be expressed in the brain, with 37 events in cases compared to only 13 in the controls (OR = 2.7, 95% CI = [2.1, 3.3], one-sided permutation P = 0.002), although this enrichment in brain-expressed genes was not significantly different from the global enrichment observed (one-sided permutation P = 0.13, Figure 1).
Figure 1. Expression Patterns of the Complete Knockouts.
(A) The enrichment of rare complete knockouts in cases versus controls.
(B) The enrichment observed in rare complete knockouts is not observed in the common complete knockouts.
The x-axis indicates the average number of events per individual in cases and controls and the numbers above the barplots indicate the total number of such events in cases and controls, with the odds ratios (OR) shown above.
To confirm that this excess was not an artifact of any residual uncertainty in statistical phasing, we examined the subset of rare complete knockouts that were homozygous LoF variants alone and found that these events were also significantly enriched by 2-fold (42 in cases and 19 in controls, OR = 2.1, 95% CI = [1.6, 2.6], one-sided permutation P = 0.0059, Table S2). We further ensured that the excess was not driven by inaccuracies in phasing ‘singleton’ variants (variants that were observed only once in a single individual) and found that rare complete knockouts excluding the singleton variants were also significantly enriched (48 in cases and 24 in controls, OR = 1.9, 95% CI = [1.4, 2.4], one-sided permutation P = 0.0081). Since an excess in 2-hit LoF could arise trivially if there was a significant overall difference in rates of LoF variants between cases and controls, we evaluated the total number of single-copy losses (i.e., heterozygous LoF carriers) with variant allele frequencies ≤5% found in cases compared to controls and saw no enrichment (OR = 1.0, 95% CI = [0.9, 1.1], Table S4). Finally, we validated all variants by ensuring that they were either present in dbSNP, the NHLBI Exome Sequencing Project and/or were confirmed using Fluidigm genotyping, Sanger sequencing or Fluidigm PCR with MiSeq sequencing with 94% of these variants validating as true polymorphisms (Table S5, Table S7). Even conservatively assuming all validation failures were false positive SNPs (rather than genotyping assay failures), removing the three events in cases and two in controls from the overall tallies has no impact on the results. As a final check, we used rare homozygous and compound heterozygous (or ‘2-hit’) synonymous events, as well as common complete knockouts, as internal controls and confirmed the enrichment of rare complete knockouts was far greater and significantly different compared to both of these (Table S4).
Knockouts via homozygosity of rare LoF sites could arise from hemizygous LoF variants that were exposed through the deletion of the other copy in the gene region. Using a CNV-calling algorithm for exome sequencing (XHMM) (Fromer et al., 2012), we found that 2 of the homozygous LoFs observed in cases (E201X in KRT83 and E211X in PRAMEF2) were, in fact, LoF variants unmasked by deletions spanning across the regions (11kb and 183kb deletions respectively), although this does not change the fact that they are complete gene knockouts.
To confirm these observations, we examined an independent set of cases (N = 563) from recent trio sequencing efforts (where 2-hit knockout status was certain from the existence of parental sequence data) and compared to a broader population dataset (N = 4,605) from the NHLBI exome sequencing project and 1000 Genomes Project (Table S1). The enrichment (7.6% in cases to 5.5% in controls, hypergeometric test P = 0.016) was replicated in this comparison as well – further confirming the veracity of this observation.
Similar Enrichment of Rare Complete Knockouts Observed on the X-chromosome
Given the gender bias in ASD, with roughly 4 times as many affected males than females (Devlin and Scherer, 2012), we asked analogously whether rare gene knockouts outside the pseudo-autosomal regions on the X-chromosome (arising from hemizygous LoFs in males) were enriched in male cases versus male controls. To further increase the sample sizes, we included the male probands and their unaffected fathers from the trios and quartets. The nucleotide diversity on the X-chromosome is estimated to be between half to three-quarters that of the autosomes and deleterious LoF variants on the X-chromosome are under stronger negative selection given the smaller effective population size and constant exposure in hemizygous males (Gottipati et al., 2011). To match the baseline knockout rate to the autosomes, where we examined variants with ≤5% minor allele frequency (MAF) and therefore ≤0.25% homozygosity, we examined LoF variants with population frequency (assessed in female control samples) of ≤0.25%. On average, we observed less than 1 such rare LoF variant on the X-chromosome in both males and females (Table S6).
Similar to the autosomes, we observed a significant enrichment of rare hemizygous LoFs in male cases (Table 2), with 88 such events observed – 60 of them were found in male cases and 28 of them were found in male controls (OR = 1.5, 95% CI = [1.1, 2.0], one-sided hypergeometric test P = 0.034, Table S7). No enrichment was seen in the internal controls of this comparison - rare hemizygous synonymous variants were not enriched in male cases compared to male controls (OR = 1.0, 95% CI = [0.9, 1.1]), indicating the observed enrichment is specific to rare complete knockouts on the X-chromosome in male ASD cases. Based on the difference between cases and controls, we further estimate another 1.7% contribution by rare complete knockouts on the X-chromosome in male cases. In addition, we found 2 of 170 female cases bearing a rare complete knockout on the X-chromosome and 0 of 452 female controls. As with the autosomes, we attempted validation for 44 of 50 rare X-chromosome LoF variants and all 44 validated.
Table 2.
Number of Rare LoF and Synonymous Variants on the X-chromosome
|
Rare hemizygous / heterozygous LoF variants |
Rare hemizygous / heterozygous synonymous variants |
|
| Hemizygous LoFs in males (N = 2,144) | ||
| Cases (N = 1,245) | 60 events | 2,114 events |
| Controls (N = 899) | 28 events | 1,516 events |
| OR [95% CI] | 1.5 [1.1, 2.0] | 1.0 [0.9, 1.1] |
| Heterozygous LoFs in females (N = 622) | ||
| Cases (N = 170) | 21 events | 641 events |
| Controls (N = 452) | 56 events | 1,256 events |
| OR [95% CI] | 1.0 [0.5, 1.5] | 1.4 [1.2, 1.6] |
|
Rare homozygous LoF variants |
Rare homozygous synonymous variants |
|
| Homozygous LoFs in females (N = 622) | ||
| Cases (N = 170) | 2 events | 5 events |
| Controls (N = 452) | 0 events | 0 events |
| OR [95% CI] | - | - |
The number of rare hemizygous LoF and synonymous variants outside the pseudo-autosomal regions on the X-chromosome in males, as well as the number of rare heterozygous LoF and synonymous variants in females are shown, together with the respective odds ratios.
We screened the list of rare complete knockouts observed on the autosomes and X-chromosome for instances where a knockout was observed only in cases and not in any of the controls (Table 3) and performed a screen for enrichment of pathways and microRNA targets using WebGestalt (Zhang et al., 2005). The top pathway (“Complement and coagulation cascades”) was driven by 2 genes (KNG1 and PLAT; corrected P = 0.0027). Scanning predicted targets of microRNAs, we found one (mir-328) predicted to target 3 genes from the list (HAP1, AFF2 and MECP2; corrected P = 0.0013; Table S8). Additional siblings (affected = 24, unaffected = 11) were available for 22 probands who were genotyped to examine segregation of a proposed recessive model. We observed 18 (expected 14) instances where segregation was consistent with a fully penetrant recessive model, including 4 genes with rare complete knockouts (PTH2R, MECP2, VSIG1 and ZCCHC16) observed in cases only and not in a single control in any wave of our study.
Table 3.
List of Rare Complete Knockouts on Autosomes and X-chromosome Found in Cases Only
| Gene | Chr | # Cases | # Controls | Expressed in the brain? |
|---|---|---|---|---|
| DGAT2L6 | X | 3 | 0 | No |
| SLC22A14 | 3 | 2 | 0 | No |
| LUZP4 | X | 2 | 0 | No |
| MAGEC3 | X | 2 | 0 | Unknown |
| CFHR2 | 1 | 1 | 0 | Yes |
| USH2A* | 1 | 1 | 0 | No |
| PTH2R | 2 | 1 | 0 | Yes |
| KNG1 | 3 | 1 | 0 | No |
| TGM4 | 3 | 1 | 0 | No |
| AGXT2 | 5 | 1 | 0 | No |
| KIAA1919 | 6 | 1 | 0 | Yes |
| MICB | 6 | 1 | 0 | Yes |
| ACTR3C | 7 | 1 | 0 | Unknown |
| LRRC69 | 8 | 1 | 0 | Unknown |
| PLAT | 8 | 1 | 0 | Yes |
| CYP2C18 | 10 | 1 | 0 | Yes |
| C12orf64 | 12 | 1 | 0 | Unknown |
| PZP | 12 | 1 | 0 | Unknown |
| LRRC29 | 16 | 1 | 0 | No |
| DBF4B | 17 | 1 | 0 | Unknown |
| HAP1 | 17 | 1 | 0 | Yes |
| AFF2* | X | 1 | 0 | Yes |
| ARSF | X | 1 | 0 | Yes |
| ARSH | X | 1 | 0 | Unknown |
| ATP1B4 | X | 1 | 0 | No |
| BEND2 | X | 1 | 0 | No |
| CT45A5 | X | 1 | 0 | Yes |
| CXCR3 | X | 1 | 0 | Yes |
| DMD | X | 1 | 0 | Yes |
| DRP2 | X | 1 | 0 | Yes |
| GPR112 | X | 1 | 0 | Unknown |
| GYG2 | X | 1 | 0 | Yes |
| HAUS7 | X | 1 | 0 | Yes |
| ITIH5L | X | 1 | 0 | No |
| KIAA1210 | X | 1 | 0 | Unknown |
| KIAA2022* | X | 1 | 0 | Unknown |
| MCF2 | X | 1 | 0 | Yes |
| MECP2* | X | 1 | 0 | Yes |
| MTMR8 | X | 1 | 0 | Yes |
| PCDH11X+ | X | 1 | 0 | Yes |
| PIR | X | 1 | 0 | Yes |
| PRDX4 | X | 1 | 0 | Yes |
| RNF128 | X | 1 | 0 | Yes |
| SRPX2* | X | 1 | 0 | Yes |
| TMLHE+ | X | 1 | 0 | Yes |
| VSIG1 | X | 1 | 0 | Yes |
| ZCCHC13 | X | 1 | 0 | No |
| ZCCHC16 | X | 1 | 0 | No |
| ZNF157 | X | 1 | 0 | Yes |
A summary of the list of genes with rare complete knockouts observed only in the cases and not in controls - genes found to be involved in known diseases have been marked with “*”, and genes found in CNVs regions previously implicated in ASD risk have been marked with “+”.
Gender and IQ
It has been shown that the male gender bias is stronger in high-functioning ASD cases, and the gender bias is reduced for syndromic cases (Newschaffer et al., 2007). We found that there was a higher rate of rare complete knockouts in females (5.4%) compared to males (4%). Although 16% of the cases sequenced were female, 25% of the cases harboring rare complete knockouts were female (OR = 1.7, 95% CI = [1.3, 2.1], one-sided Fisher’s P = 0.076). While not statistically significant, this trend is similar to previous observations that de novo CNVs and SNVs show a higher fraction of female cases with such events (Iossifov et al., 2012; Levy et al., 2011; Sanders et al., 2011) and consistent with the model that females need a higher dose of genetic risk to manifest a diagnosis of ASD. We also observed a trend in IQ scores from 18 of these cases with rare complete knockouts to another 133 cases (mean Z-score = −0.26 in probands with rare complete knockouts versus 0.035 in other cases), but it was not statistically significant (one-sided Wilcox P = 0.11).
DISCUSSION
As shown previously, de novo copy number variants (CNVs) are extremely rare events in a control population and they occur at 1–2% in controls. Given the rarity of such events, discovery of a global enrichment of these de novo CNVs at a much higher rate of 6–8% in ASD individuals suggested a 6% contribution to ASD by these de novo CNVs (Levy et al., 2011; Sanders et al., 2011; Sebat et al., 2007). This highlighted the significance of such events as risk factors for ASD and subsequent association and replication studies of such events with larger sample sizes pinpointed to specific de novo CNVs that have since been significantly associated with ASD, such as deletions and duplications on chromosome 16p11.2 (Weiss et al., 2008).
Similar to the de novo CNV studies, as well as emerging de novo SNV studies, we observed that rare complete knockouts in the human exome are found in only 3% of a control population, but are present at a 2-fold enrichment in ASD cases. Given that these rare complete knockouts are not found in a single gene but, like the de novo CNVs and SNVs, are distributed across many different genes, these events would have been missed through previous association or linkage studies. As with any genetic screen, population stratification can confound these results. However, the samples selected for sequencing were of European ancestry and individually matched in case-control pairs based on principal component analyses and selected from a much larger pool of potential samples. Owing to occasional sample failure, ultimately 88% of the final samples were matched one-to-one for ancestry and a similar 2-fold enrichment was observed in the subset of matched cases and controls for the rare complete knockouts (49 events in cases versus 25 events in controls, OR = 2, 95% CI = [1.5, 2.5]).
Interestingly, we observed a 1.5-fold enrichment of hemizygous LoF variants on the X-chromosome in male cases compared to male controls, but did not observe a significant global enrichment of heterozygous LoF variants on the X-chromosome in female cases compared to female controls. There are genes on the X-chromosome that can cause ASD-related disorders like Rett Syndrome in an X-linked dominant mode of inheritance such as CDKL5 and MECP2. However, we found that while there is a significant 1.5-fold enrichment in hemizygous LoFs in male cases, we did not observe a significant enrichment in single-copy losses in female cases, consistent with the observation that we did not see an overall difference in single-copy (heterozygous) losses on the autosomes. Given that males have only a single copy of the X-chromosome and would be more susceptible to a complete knockout on the X-chromosome than females, these rare complete knockouts on the X-chromosome can also explain a small part of the male gender bias observed in ASD.
Candidate genes
Among our list of consolidated genes with rare complete knockouts that were observed only in cases (Table 3), we discovered a known autosomal recessive gene in one of the probands from the trios – Usher syndrome 2A protein (USH2A), which has been reported to cause a known autosomal recessive disease Usher Syndrome Type II, characterized by mild to severe hearing loss and sometimes retinitis pigmentosa (Yan and Liu, 2010). We found and confirmed the bilinieal inheritance of two previously unreported compound heterozygous nonsense mutations (W2075X and Y4238X) in USH2A from both parents. Clinical follow-up confirmed an Usher Syndrome Type II diagnosis – a potential confounder in the diagnosis of ASD (Johansson et al., 2010).
When we cross-compared the list of genes harboring rare complete knockouts with previously published literature on de novo SNVs (Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012; Sanders et al., 2012), we found 3 genes that were common between the rare complete knockouts and de novo SNVs – IFIH1 (where a de novo missense variant was found in a proband), ABCC12 (where a de novo silent variant was found in a proband) and PKHD1L1 (where a de novo upstream variant was found in a proband).
We further compared the list of X-chromosome genes with previously published CNVs and found that there are 2 genes that have been previously associated with rare CNVs. We found an affected male with a rare hemizygous splice variant (c.359−2T>C) in the trimethyllysine hydroxylase, epsilon protein – TMLHE, which is involved in the biosynthesis of carnitine (Celestino-Soper et al., 2011). Recently, TMLHE deficiency resulting in dysregulation of carnitine metabolism has also been proposed as a risk factor for ASD (Celestino-Soper et al., 2012; Nava et al., 2012). Another affected male was found to harbor a hemizygous splice variant (c.3034−1G>A) in the protocadherin 11 X-linked protein – PCDH11X. An inherited deletion in PCDH11X, as well as a de novo deletion in PCDH11Y was previously reported in a child with severe language delay, suggesting a potential role for PCDH11X in language development (Speevak and Farrell, 2011).
There were 3 genes with at least 2 male cases harboring rare complete knockouts on the X-chromosome and no controls were found to harbor rare complete knockouts in these genes (SLC22A14, LUZP4, DGAT2L6). In addition, among a list of genes known to be involved in intellectual disability (Neale et al., 2012), we found 4 genes from our list with rare complete knockouts in 4 male cases. One affected male has a nonsense variant Q283X in the Fragile X E mental retardation syndrome protein (AFF2), which causes non-syndromic mental retardation and this nonsense variant results in more than 80% of the protein to be truncated. Another male case has a nonsense variant Q1471X in an uncharacterized protein KIAA2022 and mouse studies revealed that the protein is expressed in the developing brain and plays a role in neurite outgrowth (Ishikawa et al., 2012). A third male case has a splice variant c.961+1G>A in Sushi-repeat containing protein, X-linked 2 (SRPX2), a protein that is found to be expressed in neurons. Mutations in SRPX2 have been reported to be associated with rolandic epilepsy with speech and cognition impairment (Roll et al., 2006) and FOXP2, a gene which is involved in speech and language disorders, has been shown to regulate SRPX2 (Roll et al., 2010). A fourth male with ASD harbored an E495X nonsense variant in methyl CpG binding protein 2 (MECP2). Complete knockouts in MECP2 are lethal in males and heterozygous LoFs in MECP2 cause Rett Syndrome in females. Interestingly, the hemizygous nonsense mutation that was observed in this male case truncates only the last two amino acids of the MECP2 protein and this potentially generates a protein product, which explains why the hemizygous LoF observed in this gene is viable in a male. Late-truncating mutations in MECP2 have been reported to cause the Zappella variant of Rett Syndrome, which is a milder form of Rett Syndrome and autistic behavior is often observed in affected individuals (Renieri et al., 2009).
Total Contribution to ASD From de novo and Inherited Factors
As described previously in various studies, there is an estimated 6% contribution to ASD risk from de novo CNVs (Levy et al., 2011; Sanders et al., 2011; Walsh et al., 2008). Recent studies have estimated another 10% contribution to ASD risk by de novo SNVs (Iossifov et al., 2012; Neale et al., 2012; O'Roak et al., 2012; Sanders et al., 2012). In this study, we estimate a 3% contribution to ASD risk by rare complete knockouts on the autosomes and another 2% contribution by rare complete knockouts on the X-chromosome, resulting in another 5% contribution to ASD risk. Because a comparably reliable and validated set of insertion and deletion variants are not yet available across our entire dataset, we have not fully evaluated the contribution of frameshifts. Given that there is likely a similar number of frameshift mutations as single nucleotide LoF variants (Iossifov et al., 2012; MacArthur et al., 2012), the addition of frameshifts will likely increase this contribution further.
The global enrichment of rare complete knockouts in cases highlights the significance of such events in the overall genetic etiology of ASD. In addition, these events provide further insight into the heritable component of ASD, which have not yet been accounted for by de novo CNVs and SNVs. However, many of these rare complete knockouts are distributed across many different genes. This agrees with our current understanding of ASD genetics to date: that this complex disorder follows a multigenic model where hundreds of genes are involved and that each individual gene accounts for a small fraction of ASD. Together with the ongoing de novo CNV and SNV studies, our study and that of another study in this issue (Yu et al., 2013), demonstrate convincing evidence of a rare recessive contribution to the heritability of ASD.
EXPERIMENTAL PROCEDURES
The institutional review board of all participating institutions approved this study and written informed consent from all subjects was obtained. The datasets and detailed information for the samples have been deposited into dbGAP (accession ID: phs000298.v1.p1).
Data quality control and filtering
BI data was processed with Picard (http://picard.sourceforge.net/), which utilizes base quality score recalibration and local realignment at known indels and BWA for mapping reads to hg19. SNPs were called using GATK (McKenna et al., 2010). BCM data was processed with Picard and reads mapped to hg18 using Bfast (Homer et al., 2009). The quality score recalibration and indel realignment was performed using GATK, followed by SNV identification using AtlasSNP 2 software (Challis et al., 2012). Genotyping data from Affymetrix 5.0 and 6.0 was filtered using an MAF threshold of ≥5% and missing genotypes with ≤2% using PLINK and concordance checks were performed on the variant calls from the sequencing and genotyping arrays. 3 samples with low concordance between the exome sequencing and genotyping arrays (≤90%) were detected in the BI case-control dataset and discarded from further analyses.
The variants used in this study were restricted to sites that passed the standard GATK filters to eliminate SNPs with strand-bias, low quality for the depth of sequencing achieved, homopolymer runs, and SNPs near indels. And variants were required had an average read depth of ≥10× and a quality score of ≥30. Homozygous calls were required to have less than 10% of the alternate allele and heterozygous calls to have an allele balance of between 30% and 70%. A HWE threshold of ≥0.05 was used as well. A set of 160 rare variants was selected for Sequenom validation and the validation rate using these filters was 99.5%.
Annotation and analyses
For the case-control datasets, we annotated each variant according to the longest transcript from the RefSeq database. The trio and quartet datasets were annotated using a custom pipeline that was built on top of the Variant Effect Predictor (McLaren et al., 2010) to allow more stringent filtering of annotation artifacts from the 1000 Genomes Project (MacArthur et al., 2012). The cases and controls in the BI dataset was compared separately from the cases and controls in the BCM dataset before combining the results, to ensure that differences in sequencing technologies and platforms did not affect the results. Variants on the autosomes were filtered using MAF≤5% in the controls from each dataset.
Variants on the X-chromosome were filtered using similar thresholds as the autosomal variants. In addition, variants that were found to be heterozygous in males were removed from the analyses as such inconsistencies were most likely to have resulted from mis-alignment errors. To increase the number of observations for the X-chromosome analyses, male probands from the trios/quartets were added as additional cases to the overall counts from the case-control datasets and their fathers were added as additional controls, since male offspring do not inherit their X-chromosomes from their fathers and the X-chromosomes in their fathers would serve as perfect normal controls. In addition, the MAF for rare variants on the X-chromosome were calculated from a large set of control females from the NHLBI exome sequencing study.
Linkage disequilibrium-based phasing of variant pairs
We adopted a linkage disequilibrium (LD) based method, similar to the four-haplotype test used to detect a recombination event, to phase pairs of variants within the same gene and applied this approach to predict compound heterozygous variants in the case-control datasets. A pair of variants (A and B) was predicted to occur on different chromosomes if:
We observed at least 1 individual who is heterozygous for variant A; and,
we observed at least 1 individual who is heterozygous for variant B; and,
we did not observe any individual who is homozygous at 1 variant and has at least 1 copy of the second variant (Figure S1).
In addition, since we cannot accurately phase singletons, we included all pairs of variants if at least one of them is a singleton.
Statistical analyses for global enrichment
For each variant, we calculated the MAF of the variant in the controls. The MAF of a variant pair is the maximum MAF of either variant in the pair. Multiple variant pairs within the same gene in the same individual were counted as a single complete knockout event. We calculated the normalized enrichment ratio as the (total number of events in cases/total number of events in controls)×(number of controls/number of cases) to handle the imbalance in the number of cases and controls that were sequenced. We assessed the statistical significance of the global enrichment by shuffling the case-control labels for 10,000 permutations. For the enrichment analyses on the X-chromosome, one-sided hypergeometric probabilities were calculated assuming that hemizygous synonymous variants in male cases and controls are largely neutral variants. All the analyses were performed within each case-control dataset separately before combining the results, to ensure that the observations were not driven by a single dataset.
Supplementary Material
HIGHLIGHTS.
Excess of rare complete knockouts provides support for inherited component in ASD.
Estimate a 3% contribution to ASD risk from rare autosomal complete knockouts.
A further 2% contribution to ASD risk in males from X-linked complete knockouts.
Discovered ASD candidate genes from screen of rare human knockouts.
ACKNOWLEDGEMENTS
We are most grateful to the families from all participating studies: Autism Genetic Resource Exchange (AGRE), the Autism Simplex Collection (TASC), National Database for Autism Research (NDAR), Boston Autism Consortium (AC) and Simons Simplex Collection (SSC). This work was directly supported by NIH grants R01MH089208 (MJD), R01 MH089025 (JDB), R01 MH089004 (GS), R01MH089175 (RG) and R01 MH089482 (JSS) and supported in part by NIH grants P50 HD055751 (EHC), RO1 MH057881 (BD), and R01 MH061009 (JSS). We thank Thomas Lehner (NIMH), Adam Felsenfeld (NHGRI), and Patrick Bender (NIMH) for their support and contribution to the project. EB, JDB, BD, MJD (communicating PI), RG, KR, AS, GS, JSS are lead investigators in the ARRA Autism Sequencing Consortium (ASC). We would also like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes supplemental methods, one figure and nine tables.
REFERENCES
- Prevalence of autism spectrum disorders--Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. [PubMed] [Google Scholar]
- Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, Shields DC, Abrahams BS, Almeida J, Bacchelli E, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2011 doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino-Soper PB, Shaw CA, Sanders SJ, Li J, Murtha MT, Ercan-Sencicek AG, Davis L, Thomson S, Gambin T, Chinault AC, et al. Use of array CGH to detect exonic copy number variants throughout the genome in autism families detects a novel deletion in TMLHE. Hum Mol Genet. 2011;20:4360–4370. doi: 10.1093/hmg/ddr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celestino-Soper PB, Violante S, Crawford EL, Luo R, Lionel AC, Delaby E, Cai G, Sadikovic B, Lee K, Lo C, et al. A common X-linked inborn error of carnitine biosynthesis may be a risk factor for nondysmorphic autism. Proc Natl Acad Sci U S A. 2012;109:7974–7981. doi: 10.1073/pnas.1120210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour MH, Yu TW, Lim ET, Ataman B, Coulter ME, Hill RS, Stevens CR, Schubert CR, Greenberg ME, Gabriel SB, et al. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 2012;8:e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA, Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Todorov A, Hilton C, Law P, Zhang Y, Molloy E, Fitzgerald R, Geschwind D. Autism recurrence in half siblings: strong support for genetic mechanisms of transmission in ASD. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.9. [DOI] [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012 doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O'Donovan MC, Owen MJ, et al. Discovery and Statistical Genotyping of Copy-Number Variation from Whole-Exome Sequencing Depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, et al. Phenotypic Heterogeneity of Genomic Disorders and Rare Copy-Number Variants. N Engl J Med. 2012 doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am J Hum Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati S, Arbiza L, Siepel A, Clark AG, Keinan A. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat Genet. 2011;43:741–743. doi: 10.1038/ng.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Miyata S, Koyama Y, Yoshikawa K, Hattori T, Kumamoto N, Shingaki K, Katayama T, Tohyama M. Transient expression of Xpn, an XLMR protein related to neurite extension, during brain development and participation in neurite outgrowth. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Johansson M, Gillberg C, Rastam M. Autism spectrum conditions in individuals with Mobius sequence, CHARGE syndrome and oculo-auriculo-vertebral spectrum: diagnostic aspects. Res Dev Disabil. 2010;31:9–24. doi: 10.1016/j.ridd.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Jorde LB, Hasstedt SJ, Ritvo ER, Mason-Brothers A, Freeman BJ, Pingree C, McMahon WM, Petersen B, Jenson WR, Mo A. Complex segregation analysis of autism. Am J Hum Genet. 1991;49:932–938. [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava C, Lamari F, Heron D, Mignot C, Rastetter A, Keren B, Cohen D, Faudet A, Bouteiller D, Gilleron M, et al. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry. 2012;2:e179. doi: 10.1038/tp.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012 doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newschaffer CJ, Croen LA, Daniels J, Giarelli E, Grether JK, Levy SE, Mandell DS, Miller LA, Pinto-Martin J, Reaven J, et al. The epidemiology of autism spectrum disorders. Annu Rev Public Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renieri A, Mari F, Mencarelli MA, Scala E, Ariani F, Longo I, Meloni I, Cevenini G, Pini G, Hayek G, et al. Diagnostic criteria for the Zappella variant of Rett syndrome (the preserved speech variant) Brain Dev. 2009;31:208–216. doi: 10.1016/j.braindev.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Spence MA, Freeman BJ, Mason-Brothers A, Mo A, Marazita ML. Evidence for autosomal recessive inheritance in 46 families with multiple incidences of autism. Am J Psychiatry. 1985;142:187–192. doi: 10.1176/ajp.142.2.187. [DOI] [PubMed] [Google Scholar]
- Roll P, Rudolf G, Pereira S, Royer B, Scheffer IE, Massacrier A, Valenti MP, Roeckel-Trevisiol N, Jamali S, Beclin C, et al. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15:1195–1207. doi: 10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- Roll P, Vernes SC, Bruneau N, Cillario J, Ponsole-Lenfant M, Massacrier A, Rudolf G, Khalife M, Hirsch E, Fisher SE, et al. Molecular networks implicated in speech-related disorders: FOXP2 regulates the SRPX2/uPAR complex. Hum Mol Genet. 2010;19:4848–4860. doi: 10.1093/hmg/ddq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, Dilullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012 doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speevak MD, Farrell SA. Non-syndromic language delay in a child with disruption in the Protocadherin11X/Y gene pair. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:484–489. doi: 10.1002/ajmg.b.31186. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Yan D, Liu XZ. Genetics and pathological mechanisms of Usher syndrome. J Hum Genet. 2010;55:327–335. doi: 10.1038/jhg.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, Bijlsma EK, Oortveld MA, Ekici AB, Reis A, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.