Abstract
MicroRNAs (miRNA) have tumor suppressive and oncogenic potential in human cancer, but whether and how miRNAs control cell cycle progression is not understood. To address this question, we carried out a comprehensive analysis of miRNA expression during serum stimulation of quiescent human cells. Time course analyses revealed that four miRNAs are up-regulated and >100 miRNAs are down-regulated, as cells progress beyond the G1-S phase transition. We analyzed the function of two up-regulated miRNAs (miR-221 and miR-222) that are both predicted to target the cell growth suppressive cyclin-dependent kinase inhibitors p27 and p57. Our results show that miR-221 and miR-222 both directly target the 3′ untranslated regions of p27 and p57 mRNAs to reduce reporter gene expression, as well as diminish p27 and p57 protein levels. Functional studies show that miR-221 and miR-222 prevent quiescence when elevated during growth factor deprivation and induce precocious S-phase entry, thereby triggering cell death. Thus, the physiologic upregulation of miR-221 and miR-222 is tightly linked to a cell cycle checkpoint that ensures cell survival by coordinating competency for initiation of S phase with growth factor signaling pathways that stimulate cell proliferation.
Introduction
MicroRNAs (miRNA) are fundamental gene regulators that control proliferation, differentiation, and apoptosis during development (1–5). These broad functions suggest that miRNAs may contribute to a range of diseases, including cancer, and recent evidence indicates that miRNAs can function as tumor suppressors and oncogenes (reviewed in refs. 1, 6–9). The first report linking miRNAs and cancer showed preferential expression of miR-15 and miR-16 in patients with B-cell chronic lymphocytic leukemia (CLL; ref. 10). The genes for both of these miRNAs are located at chromosome 13q14, a locus frequently deleted in abnormalities, including CLL and prostate cancers. In addition, other miRNA expression signatures have been associated with the initiation and progression of human cancers (reviewed in refs. 1, 6–9). Although tumorigenesis is accompanied by loss of cell cycle control, how differences in miRNA expression influence the complex regulatory machinery that mediates cell cycle progression and cell survival remains to be investigated (11).
miRNAs are a class of small (~22 nt), conserved, noncoding RNAs that play important regulatory roles in posttranscriptional repression (reviewed in refs. 1, 2, 12). They target a large number of mRNAs (predicted to be up to 30% of the human genome) and induce mRNA degradation or inhibition of translation by targeting the 3′ untranslated regions (UTR) through specific, albeit imperfect, base-pairing (2). Several methods for predicting target mRNAs rely on conserved base-pairing in the “seed” miRNA region (13–17). miRNAs are synthesized by RNA polymerase II as long primary transcripts (pri-miRNAs) that form stem-loop structures (18). These transcripts are capped, polyadenylated, and processed in the nucleus by a complex consisting of at least two proteins: the RNase III enzyme Drosha and the double-stranded RNA-binding protein Pasha/DGCR8 (19–24). These imperfect hairpin products are then exported into the cytoplasm by exportin-5, wherein the 65 to 75 nt pre-miRNAs are further cleaved by Dicer to generate double-stranded miRNAs of ~22 nt (25, 26). The duplex is subsequently incorporated into a functional miRISC complex, wherein the mature miRNA strand is preferentially retained to identify mRNA targets (reviewed in refs. 27, 28). Thus, cells have developed sophisticated mechanisms to produce miRNAs that can target selected groups of mRNAs. Investigating the cellular pathways that are regulated by miRNAs represents the next frontier in advancing our knowledge of growth control and differentiation and will provide insights into miRNA-dependent deregulation of cell proliferation and survival in cancer.
In the present study, we examined the emerging concept that expression of miRNAs influences the activity of cell cycle regulatory proteins that control competency for cell proliferation. We tested this idea by performing expression profiling of miRNAs that are regulated when quiescent cells are stimulated to proliferate. Our results show that miR-221 and miR-222 are preferentially expressed during serum-stimulated entry into the cell cycle and target the cyclin-dependent kinase (CDK) inhibitors p27 and p57 to promote cell cycle progression. Transfection of cells with either miRNA during serum deprivation prevents quiescence and causes premature S-phase entry with subsequent cell death. We propose that the normal up-regulation of miR-221 and miR-222 during serum stimulation is conducive for cell cycle progression but must be coupled with a cell cycle checkpoint that monitors appropriate activation of growth factor-dependent cell signaling pathways.
Materials and Methods
Cell culture
Human T98G glioblastoma cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mmol/L l-glutamine, 100 units/mL penicillin G, and 100 µg/mL streptomycin. Human leukemic K562 cells were maintained in RPMI (Invitrogen) supplemented with 10% FBS, 2 mmol/L l-glutamine, 100 units/mL penicillin G, and 100 µg/mL streptomycin.
Cell cycle and cell proliferation analyses
T98G cells were synchronized by serum starvation for 72 h, followed by stimulation from quiescence with media containing 20% FBS. Cell cycle distribution was monitored by propidium iodide staining of cells and fluorescence-activated cell sorting (FACS; UMASS Medical School Flow Cytometry Core Facility) at different time points after release. For proliferation/quiescence studies, T98G cells were transfected with miRNA oligonucleotides using Oligofectamine (Invitrogen) for 24 h before serum deprivation. Cells were photographed and harvested for analysis at different times after serum withdrawal.
miRNA expression profiling
Microarray and bioinformatic analyses, as well as significance analysis of microarrays, were done as described before (29). Results from three independent experiments are expressed as log2 of fluorescence and as a hierarchical clustering of the average values by dCHIP software (30).
Protein preparation and Western blotting
Total protein extracts from mammalian cell lines were prepared in direct lysis buffer [50 mmol/L Tris-HCl (pH 6.8), 100 mmol/L DTT, 2% SDS, 10% glycerol, 1× Complete (Roche), and 0.2% bromophenol blue]. Cells were boiled for 5 min and centrifuged at 16,100 × g for 5 min. Total proteins were loaded onto 10% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride Immobilon-P membrane (Millipore) for 30 min at 10 V in a semidry transfer apparatus (model HEP-1, Owl Separation Systems). Immunodetection was done using an appropriate dilution of specific antibodies with the western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences). The following dilutions of primary antibodies used were p57 C-20 rabbit polyclonal 1:1,000, cyclin A C-19 rabbit polyclonal 1:2,000, and CDK2 M-2 rabbit polyclonal 1:5,000 from Santa Cruz Biotechnology, Inc.; p27 mouse monoclonal IgG1 1:1,000 from BD Biosciences; and caspase-3 Asp175 rabbit polyclonal 1:1,000, caspase-8 1C12 mouse monoclonal 1:1,000, and caspase-9 rabbit polyclonal 1:1,000 from Cell Signaling.
Northern blot analysis of miRNAs
Total cellular RNA from synchronized T98G cells was prepared using TRIzol reagent following the manufacturer’s instructions (Invitrogen). Total RNA (15 µg) was separated by electrophoresis in 0.5× Tris-borate EDTA [TBE; 44.5 mmol/L Tris, 44.5 mmol/L sodium borate, and 1 mmol/L EDTA (pH 8.3)] through denaturing 12% urea-polyacrylamide (0.75 mm × 16 cm × 16 cm) gels at 15 to 20 W. RNA was electrotransferred to a Zeta-Probe GT membrane (Bio-Rad) in a semidry apparatus in 0.5× TBE for 1 h at 300 mA. RNA was fixed to the membrane by UV irradiation (optimal cross-linking setting, Spectrolinker XL-1000 UV cross-linker, Spectronics Corporation) and baked at 80°C for 30 min under vacuum. Membranes were prehybridized at 42°C for 2 h in buffer (7% SDS/0.2 mol/L sodium phosphate, pH 7.2) and then hybridized with oligonucleotide DNA probes radiolabeled with T4 polynucleotide kinase overnight at 42°C in the same buffer. Membranes were washed for 30 min at 42°C twice each time in wash buffer 1 (5% SDS/ 20 mmol/L sodium phosphate, pH 7.2) and wash buffer 2 (1% SDS/ 20 mmol/L sodium phosphate, pH 7.2), respectively. Band intensities were quantified with a PhosphoImager using ImageQuant 5.0 software (Storm 840, Molecular Dynamics), followed by exposure to X-ray film (Kodak), and expressed as fold-increase of the specific miRNA relative to U6 RNA, which was used as loading control. Probes used were as follows (in 5′ to 3′ direction): hsa-miR-221 GAA ACC CAG CAG ACA ATG TAG CT, hsa-miR-222 GAG ACC CAG TAG CCA GAT GTA GCT, and U6 ATA TGG AAC GCT TCA CGA ATT.
Sequence alignment
Alignments of miR-221 and miR-222 to p57 and p27 3′ UTR sequences were taken from TargetScan (14, 17).
Luciferase constructs and reporter gene assays
Genomic DNA from human WI-38 normal diploid fibroblasts was isolated following a standard protocol (31). p57-Luc and p27-Luc plasmids were constructed by cloning segments of the 3′ UTRs spanning the miRNA recognition sites into pMIR-REPORT miRNA Expression Reporter Vector (Ambion) by PCR from WI-38 genomic DNA using the following primers (in 5′ to 3′ direction): for p57-Luc, GGG TAC TAG TTT TAG AGC CCA AAG AGC C (the SpeI site is underlined) and GGT AAG CTT TAC ACC TTG GGA CCA GT (the HindIII site is underlined); for p27-Luc, GGG TAC TAG TTC CTT GTT TAT CAG ATA CAT CAC T (the SpeI site is underlined) and GGT AAG CTT TTT AAA TGA AGT ATC AGC TGT CTC (the HindIII site is underlined).
Transient transfection of K562 cells (5 × 104 cells per well) was carried out in 24-well plates with Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Cells were transfected with 800 ng of the luciferase constructs p57 3′ UTR-Luc or p27 3′ UTR-Luc and 5 ng of Renilla luciferase plasmid and cotransfected with human pre-miR-221 miRNA precursor or pre-miR-222 miRNA precursor (30 nmol/L) obtained from Ambion. After 48 h, cells were harvested and lysates were measured for luciferase activity and normalized for transfection efficiency to Renilla luciferase (phRL-null) activity (dual-luciferase reporter assay system, Promega). The relative reporter activity was obtained by comparison to a control oligonucleotide arbitrarily taken as 100 (pre-miR miRNA precursor negative control 2, Ambion). Significant differences were determined in three independent experiments by Student’s t test (**, P < 0.01).
miRNA transfections
K562 or T98G cells were transfected with human miR-221, miR-222, or negative control (miR-NC) oligonucleotides (Ambion or Dharmacon) using Oligofectamine following the manufacturer’s instructions. The miRNA oligonucleotide concentrations used ranged from 30 to 150 nmol/L, with or without a 5-fold excess of anti-miRNA inhibitor. Protein extracts were prepared and analyzed by Western blot as described above. Cell cycle distribution was monitored by FACS analysis.
BrdUrd incorporation and immunofluorescence microscopy
Incorporation of BrdUrd into actively proliferating T98G cells was determined using 5-bromo-2′-deoxy-uridine labeling and detection kit I (Roche Applied Science) following the manufacturer’s instructions. Cells were grown in six-well plates with coverslips (Fisher Scientific), seeded at a density of 50,000 cells per well, and transfected after 24 h with miRNA oligonucleotides for 24 h, as described above. Cells were allowed to incorporate BrdUrd for 30 min at 37°C, washed, and then fixed with an ethanol/glycine (pH 2.0) solution for 20 min at −20°C. BrdUrd incorporation into DNA was detected in whole-cell preparations by incubation with a mouse monoclonal antibody against BrdUrd at a 1:10 dilution for 30 min at 37°C. The secondary antibody (antimouse-immunoglobulin-fluorescein) was used at a 1:10 dilution for 30 min at 37°C. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at 4°C. Immunostaining of cell preparations was captured by an epifluorescence microscope (Zeiss Axioplan II) equipped with a charge-coupled device camera. Digital images were acquired and analyzed with MetaMorph software (Molecular Devices).
Results
miR-221 and miR-222 levels are up-regulated after stimulation of quiescent T98G cells to proliferate
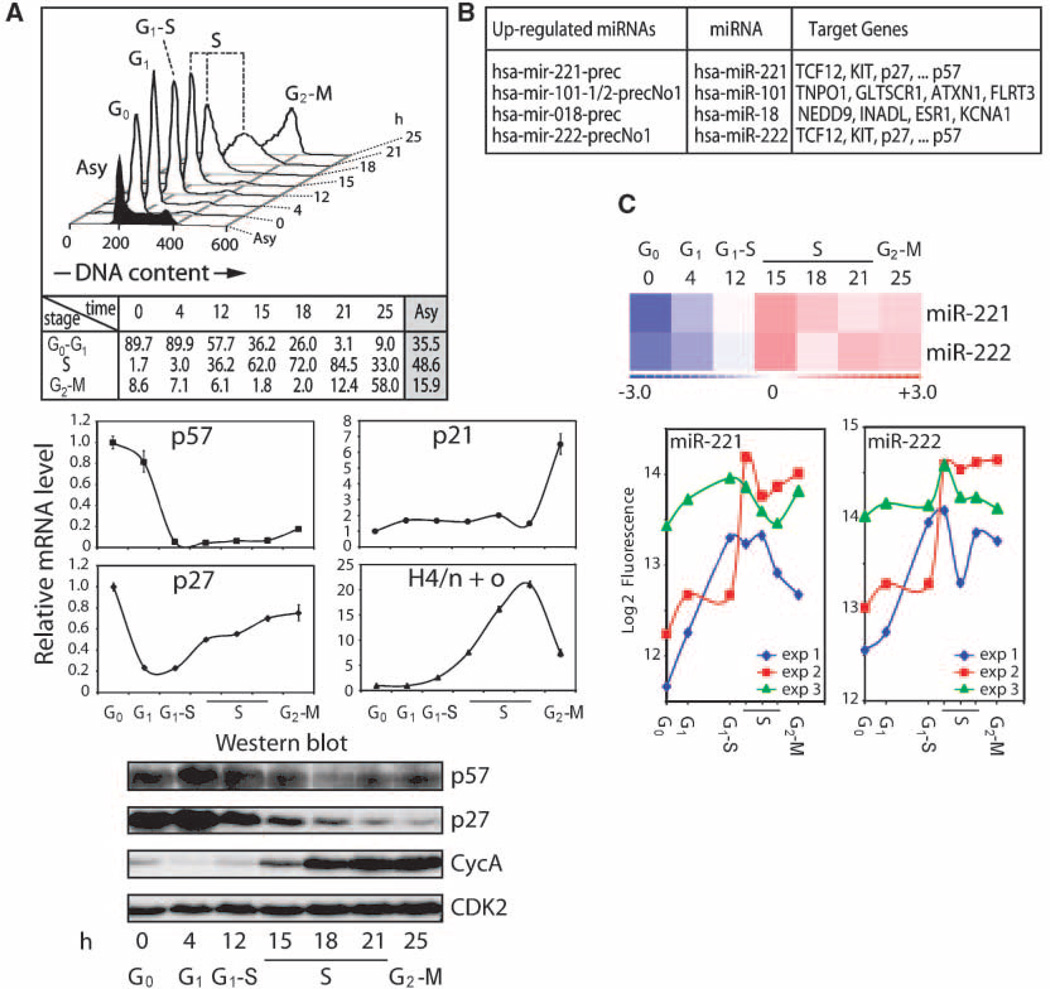
To identify miRNAs that support cell proliferation, we examined miRNA expression during cell cycle entry upon growth factor stimulation of quiescent human T98G cells (32). Cells were synchronized in G0 by serum deprivation and released from the arrest with complete media (Fig. 1A, top). As expected, histone H4 mRNA levels are transiently up-regulated at the G1-S phase transition (12 hour), whereas p57 mRNA levels are significantly down-regulated (Fig. 1A, middle). In contrast, p27 mRNA levels decrease immediately when cells enter G1 (4 hour) and are slightly up-regulated during S- and G2-M phases. Levels of p57, p27, and cyclin A proteins exhibit characteristic cell cycle stage–specific oscillations: p57 and p27 decrease at the G1-S phase transition, whereas cyclin A increases as cells enter S phase (Fig. 1A, bottom).
Figure 1.
Up-regulation of miR-221 and miR-222 in quiescent T98G cells stimulated to proliferate. A, synchronization of T98G cells was achieved by serum deprivation, and cells were released from the arrest with serum-containing media. Cell cycle distribution determined by FACS analysis with cell cycle stages after release from the block along with the percentage in each cell cycle phase (h; top). mRNA (middle) and protein (bottom) profiles for the different cell cycle stages. Characteristic cell cycle stage–specific oscillations of mRNA and proteins are depicted (see text for details). B, up-regulated miRNAs identified by microchip analysis as a function of cell cycle and selected putative targets for each miRNA. C, three independent experiments are displayed as hierarchical clustering (average; top) and log2 of fluorescence of microchip data (bottom). In the top panel, red shades represent expression above the mean across all samples and blue shades represent expression below the mean. Both miR-221 and miR-222 are up-regulated when quiescent T98G cells are stimulated to proliferate.
We analyzed miRNA profiles at G0 (t = 0), G1 (4 hours), G1-S transition (12 hours), S phase (15, 18, and 21 hours), and G2-M phase (25 hours) using microarrays containing 316 human and 318 mouse precursor miRNAs (29). Strikingly, using this panel of 634 miRNAs, we found that four miRNAs are up-regulated and >100 miRNAs are down-regulated when cells reenter the cell cycle upon serum stimulation (Fig. 1B and data not shown). The four up-regulated miRNAs are miR-18, miR-101, miR-221, and miR- 222; miR-18 was first discovered in HeLa cells (33), miR-101was detected during megakaryocytopoiesis (34), whereas expression of miR-221/222 was found to be enhanced in glioblastoma (35). We focused on miR-221 and miR-222 because several of their predicted targets are cell cycle–related factors. Statistical analyses of the temporal expression profiles revealed that both miR-221 and miR-222 are robustly up-regulated in serum-stimulated T98G cells (Fig. 1C). Northern blot analysis confirmed the cell cycle–specific increase in the expression of miR-221 and miR-222, as well as of their respective precursors when T98G cells are stimulated to proliferate (Fig. 2). This finding suggests that miR-221 and miR-222 may promote competency for cell proliferation.
Figure 2.
miRNA-221 and miRNA-222 levels during cell proliferation. miR-221 and miR-222 levels were analyzed in T98G cells released from quiescence by serum stimulation. Total RNA was isolated from samples taken at the indicated time points after release and analyzed by Northern blot using specific probes (left). Bottom, cell cycle stages. Right, band quantification of mature miRNAs expressed as fold change compared with U6 RNA. Maximum value was arbitrarily considered as 1.
miR-221 and miR-222 reduce protein levels of the Kip/Cip family members p57 and p27
We searched for potential mRNA targets of miR-221 and miR-222 using Web-based resources, including TargetScan, TargetCombo, and PicTar (14, 16, 17, 36). Our target analysis revealed that two members of the Kip/Cip family of CDK inhibitors, p57 and p27, are putative target mRNAs. Both p57 and p27 mRNAs contain seed sequences and base-pairing potential in the 3′ UTR for miR-221 and miR-222: the 3′UTR of p57 harbors one site expected to be recognized by both miR-221 and miR-222, whereas the 3′UTR of p27 contains two sites for both miRNAs (Fig. 3A).
Figure 3.
p57 3′UTR and p27 3′UTR are targets of miR-221 and miR-222. A, p57 and p27 mRNA 3′ UTRs depicting putative target sites for miR-221 and miR-222. The 3′ UTRs of p57 and p27 contain one and two sites expected to be recognized by both miR-221 and miR-222, respectively (arrowheads). Numbers represent the position of the seed match within the UTR sequences. Hs, human; Mm, mouse; Rn, rat; Cf, dog; Gg, chicken. B, repression of activity of the p57 3′ UTR-luciferase and p27 3′ UTR-luciferase constructs by ectopic expression of miR-221 and miR-222 oligonucleotides in K562 cells. Transfection of a nonfunctional miRNA (miR-NC2) was used as a negative control. Firefly luciferase activity was determined 48 h after transfection and normalized to Renilla luciferase activity from a promoter-less vector. Statistical significance was determined by the Student’s t test for three independent experiments (**, P < 0.01). C, K562 cells were either mock-transfected or transfected with miR-NC2, miR-221, miR-222, or a combination of miRNA and a 5-fold excess of the corresponding anti-miRNA inhibitor for 48 h. Total protein extracts were used to analyze p57 and p27 levels by Western blot with the specific antibodies (right). Transfection with miR-221 and miR-222 significantly reduced the levels of endogenous p57 and p27 proteins (lanes 3 and 7) when compared with mock (lanes 1 and 5) or miR-NC2 transfection (lanes 2 and 6). This decrease was reverted by the corresponding miRNA inhibitors (lanes 4 and 8). CDK2 is shown as loading control.
We validated p57 and p27 mRNAs as bona fide targets for both miR-221 and miR-222. The 3′ UTR regions of both genes encompassing the highly conserved putative miR-221/miR-222 binding sites were fused downstream of the coding sequences for a luciferase reporter. Cotransfection of these reporter vectors in K562 cells, which do not express miR-221 or miR-222 (37), with either miR-221 or miR-222 oligonucleotides significantly reduced p57 3′ UTR-luciferase and p27 3′ UTR-luciferase reporter activities by ~60% and ~77%, respectively (Fig. 3B). Relative reporter activities were not altered by a negative control miRNA (miR-NC2). Thus, both miRNAs functionally inhibit expression of the luciferase reporter vectors. We also examined the effect of exogenous miR-221 and miR-222 by oligonucleotide transfection on the expression of endogenous levels of p57 and p27 proteins in K562 cells. Western blot analysis shows that both miR-221 and miR-222 greatly reduce the levels of p57 and p27 proteins (Fig. 3C). Cotransfection of miR-221 and miR-222 with the corresponding anti-miRNA oligonucleotides reverses the effect on the protein levels (compare lanes 4 and 8 with lanes 3 and 7, respectively). Our findings show that p57 and p27 mRNAs are both direct targets of miR-221/miR-222.
miR-221 and miR-222 abrogate cell survival upon growth factor deprivation
To understand the functional role of miR-221 and miR-222 in regulating the quiescence/proliferation transition, we investigated the effects of ectopic expression of miR-221/ miR-222 in serum-deprived T98G cells. Cells were transfected with miRNAs and then induced to enter G0 by removal of serum. We anticipated that the premature presence of miR-221 and miR-222 would prevent the expected elevated levels of p57 and p27 that are normally present in quiescent T98G cells (see Fig. 1A) and perhaps would permit continuous proliferation. Strikingly, cells pretreated for 24 h with either miR-221 or miR-222 undergo massive cell death upon serum deprivation for 72 h (Fig. 4A). Mock-transfected cells or cells pretreated with either a negative control (miR-NC2) or anti–miR-222 oligonucleotides were morphologically indistinguishable from untreated control cells. Cell cycle analysis by flow cytometry revealed that T98G cells transfected with miR-221 or miR-222 exhibited highly reduced cell numbers and distinctive sub-2N populations (Fig. 4B), apart from changes in the distribution of cells in S phase (Fig. 4B; see below). Untreated and mocktransfected miR-NC2–transfected, and anti–miR-222–transfected T98G cells showed a characteristic G0 peak indicative of arrested cells.
Figure 4.
Ectopic expression of miR-221 and miR-222 causes cell death during serum withdrawal. A, T98G cells were transfected with miRNAs at 150 nmol/L for 24 h and then forced to enter G0 by serum deprivation. Photographs and samples for FACS analysis (see below) were taken after incubating cells in serum-free media for 72 h. Massive cell death occurs in samples transfected with miR-221 and miR-222 compared with the morphologically normal mock-transfected, NC2-transfected, and anti–miR-222–transfected cells. B, cell cycle distribution of samples taken as in A. Black lines, profiles of transfected cells. For comparison, the asynchronous T98G population is represented as gray histograms. Sub-2N (arrowhead) and S-phase accumulation (asterisk) are observed in miR-221–transfected and miR-222–transfected T98G cells. C, time course of miRNA-transfected T98G cells during serum withdrawal. Cells were transfected with miRNA oligonucleotides at 75 nmol/L for 24 h and photographed after 24 and 36 h of serum deprivation. The appearance of floating cells indicative of cell death was first observed at 24 h, which was more pronounced at 36 h and later times of serum-starvation. Increased cell death was observed only with miRNA transfection and not in anti–miR inhibitor-transfected cells. UT, untreated.
We also analyzed the effects of miR-222 pretreatment in T98G cells during a time course of serum starvation for 24, 36, 48, 60, and 72 hours (Fig. 4C and data not shown). We observed a slight increase in cell death by 24 hours, which became very pronounced as early as 36 hours after serum deprivation in cells transfected with miR-222 oligonucleotide (Fig. 4C). For comparison, cells pretreated with either a negative control (miR-NC2) or anti–miR-222 oligonucleotides were morphologically indistinguishable from mock-transfected or untreated controls at all time points (Fig. 4C and data not shown). Taken together, our results indicate that high exogenous levels of miR-221 and miR-222 during serum withdrawal compromise cell survival and suggest that low physiologic levels of the two miRNAs in quiescent T98G cells may protect against unscheduled cell death.
High levels of miR-221/222 permit precocious S-phase entry in the absence of serum growth factors
T98G cells transfected with miR-222 were analyzed for cell cycle distribution as a time course after serum withdrawal for 24, 48, and 72 h. Within 24 h of serum deprivation, there is a clear accumulation of S-phase cells concomitant with the appearance of a sub-2N peak in cells treated with miR-222 oligonucleotide (Fig. 5A). Western blot analysis confirmed the decrease in p27 protein level in miR-222–treated T98G cells, as well as an increase of the S-phase marker cyclin A as expected (Fig. 5B). Untreated, mock-transfected, miR-NC2– transfected, and anti-miR-222–transfected cells showed a characteristic G0 peak. Thus, serum deprivation in the presence of miR-222 seems to be linked to both cell death and S-phase entry.
Figure 5.
Elevated levels of miR-221/miR-222 drive cells into S phase in the absence of serum growth factors. A, cell cycle distribution of T98G cells transfected with 75 nmol/L miRNAs for 24 h and then serum-deprived for 24 h. Cell cycle distribution was analyzed by FACS sorting, and profiles along with the percentage in each cell cycle stage are shown underneath. Cells transfected with miR-222 show accumulation of cells in S phase (asterisk), as well as a sub-2N peak (arrowhead). B, western blot analysis of samples taken from A confirmed the decrease in p27 protein level in miR-222–treated T98G cells; an increase in cyclin A protein is also observed, consistent with increased S-phase accumulation. Right, antibodies used. CDK2 was used as loading control. Asy, asynchronous.
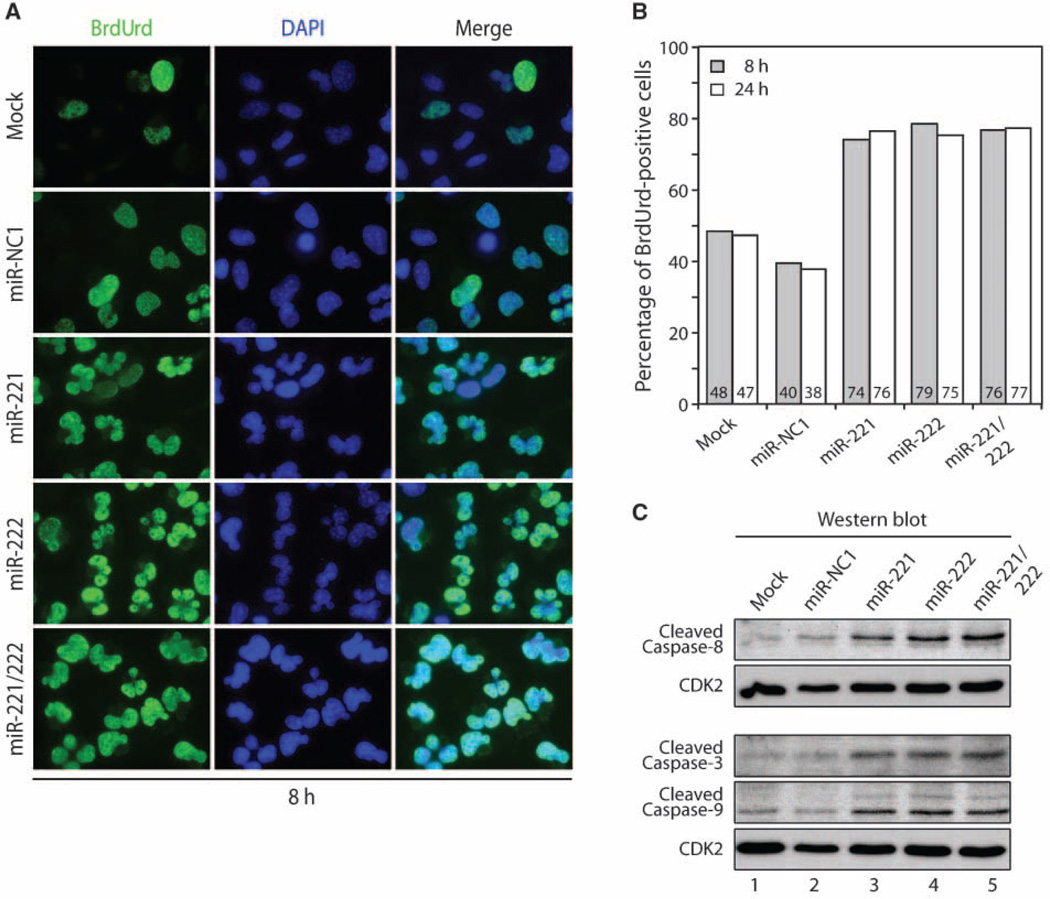
To examine whether miR-221/miR-222–containing cells are capable of entering S phase during growth factor withdrawal, we monitored DNA synthesis by BrdUrd incorporation after serum starvation for 8 and 24 hours (Fig. 6A and B and data not shown). Transfection of miR-221, miR-222, or a combination of both miRNA (miR-221/miR-222) oligonucleotides renders ~76% of cells labeled with BrdUrd compared with ~39% of an miRNA-negative control (miR-NC1) at both 8 and 24 hours (Fig. 6 and data not shown). Cell cycle distribution by FACS analysis confirmed an S-phase accumulation of 76% in miR-222–transfected T98G cells after serum deprivation for 24 h (data not shown). Furthermore, both DAPI-staining and BrdUrd labeling show that nuclei exhibit a lobulated shape suggestive of apoptosis (Fig. 6A). Therefore, we monitored the levels of activated caspase-3, caspase-8, and caspase-9 by Western blot analysis (Fig. 6C). We find that the proteolytically cleaved activated forms of all three caspases are modestly elevated by 24 hours when we first observed the presence of dead cells (see Fig. 4C). We conclude that miR-221 and miR-222 promote entry into S phase under conditions that are not conducive for normal mitotic division and result in apoptotic cell death.
Figure 6.
miR-221 and miR-222 induce apoptosis in the absence of growth factors. A, BrdUrd incorporation was monitored by immunofluorescence microscopy after transfection of T98G cells with miRNA oligonucleotides (75 nmol/L) for 24 h (left) and serum-deprived for 8 h. Note the presence of lobulated nuclei indicative of apoptosis. B, quantitative analysis of BrdUrd-positive cells in A (data not shown). Transfection of miR-221, miR-222, and the combination miR-221/miR-222 oligonucleotides shows ~76% of BrdUrd-positive cells compared with ~39% in miRNA negative control (miR-NC1) at both 8 and 24 h (>400 cells were counted). C, analysis of caspase activation. Samples treated as in A were resuspended in loading buffer and analyzed by Western blot for caspase cleavage products. Left, antibodies used. CDK2 was used as loading control.
Discussion
In this study, we have systematically characterized miRNAs that are selectively regulated during serum stimulation of growth factor-dependent T98G cells. We detect only four miRNAs that exhibit enhanced expression when cells initiate proliferation from quiescence. Two of these, miR-221 and miR-222, target mRNAs encoding the CDK inhibitory proteins p57 and p27, thus increasing competency for progression beyond the G1-S transition. Importantly, our results show that high exogenous levels of miR-221 and miR-222 during serum withdrawal compromise cell survival. Our findings suggest that, whereas miR-221 and miR-222 coordinately suppress the expression of p57 and p27 proteins, this reduction is not sufficient to sustain active cell proliferation in the absence of growth factors. We propose that low physiologic levels of the two miRNAs in quiescent T98G cells may protect against unscheduled proliferation that will trigger apoptosis.
While our investigation was in progress, several studies have explored the relationship between p27 and miR-221/miR-222. For example, Visone and colleagues (38) have shown that miR-221/ miR-222 elevation suppresses p27 expression and promotes cell cycle progression. Other studies showed that miR-221 and miR-222 are up-regulated in aggressive prostate cancer cell lines or glioblastoma cells that contain low levels of p27 (39, 40). In addition, the Agami laboratory (41) identified miR-221/miR-222 during a screen for miRNAs capable of reducing p27 levels and increasing cell proliferative capacity of several tumor cell lines. Our results extend the findings of these studies by showing that miR-221 and miR-222 are up-regulated upon exit from quiescence and that these miRNAs not only target p27/Kip1 but also the closely related protein p57/Kip2. Thus, these miRNAs are growth regulatory mediators that coordinately modulate the levels of two critical inhibitors of CDK2/cyclin complexes in late G1 when competency for cell cycle progression is monitored.
miR-221 and miR-222 are both up-regulated in several tumor-derived cell lines and in cancer patients (35, 42–44). Both miRNAs are clustered within a 1-kb genomic interval and are transcribed from the same strand. Thus, both miRNAs may be generated from the same primary transcript, which could account for simultaneous enhancement of their expression in tumor cells observed in previous studies (35, 42–44). Up-regulation of miR-221/miR-222 in tumor cells may be directly related to our findings that show proliferation-specific expression of these miRNAs, reflected by their increased levels when cells enter the cell cycle from quiescence.
It has been proposed that miR-221/miR-222 are oncogenic based on their up-regulation in tumor cells and the suppressive effect on p27 expression, which permits increased cell proliferation (38–41). Our data show that up-regulation of these miRNAs promotes S-phase entry but does not suffice for uncontrolled cell proliferation. We find that under conditions of growth factor deprivation, elevation of miR-221 and miR-222 causes unwarranted onset of S phase when cells have apparently not fulfilled the requisite cellular conditions for completion of genomic replication and mitotic division. Instead, forced increase of miR-221/miR-222 levels bypasses the restriction point and induces S-phase entry in the absence of serum. This precocious induction of S phase triggers an intra-S cell cycle checkpoint that activates apoptosis and prevents completion of cell division. Our studies have at least one important ramification for the pathologic roles of miR-221/miR-222 in the molecular etiology of cancer. Whereas increased expression of miR-221 and miR-222 clearly has oncogenic potential, cooperating events must occur to sustain cell proliferation in tumors when growth factor concentrations are limiting.
Acknowledgments
Grant support: NIH grants GM32010 and CA082834 (G.S. Stein). Core facilities support was provided by DK32520.
We thank Judy Rask for expert assistance in the preparation of this manuscript; the members of our research group, especially Jitesh Pratap and Zhaoyong Li for stimulating discussions; Richard Konz for support; and the staff of UMASS Flow Cytometry Core Facility for sample analysis.
References
- 1.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 17.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X, Hagedorn CH, Cullen BR. Human micro-RNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 21.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 22.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of micro-RNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 27.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 29.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 32.Stein GH. T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro . J Cell Physiol. 1979;99:43–54. doi: 10.1002/jcp.1040990107. [DOI] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 34.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 36.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 37.Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 39.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 40.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 41.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 44.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]