Abstract
Luteinizing hormone receptor (LHR) undergoes downregulation during preovulatory LH surge through a post-transcriptional mechanism involving an RNA binding protein designated as LRBP. The present study examined the mechanism by which LRBP induces the degradation of LHR mRNA, specifically the role of decapping of LHR mRNA and the translocation of LRBP-bound LHR mRNA to degradative machinery. Immunoprecipitation of the complex with the 5’cap structure antibody followed by real time PCR analysis showed progressive loss of capped LHR mRNA during downregulation suggesting that LHR mRNA undergoes decapping prior to degradation. RNA immunoprecipitation (RIP) analysis confirmed dissociation of eIF4E from the cap structure, a step required for decapping. Furthermore, RIP analysis using antibody against the p body marker protein, DCP1A showed that LHR mRNA was associated with the p bodies, the cytoplasmic foci that contains RNA degradative enzymes and decapping complex. Immunohistochemical studies using antibodies against LRBP and DCP1A followed by confocal analysis showed colocalization of LRBP with DCP1A during downregulation. This was further confirmed by co-immunoprecipitation of LRBP with DCP1A. The association of LRBP and LHR mRNA in the p bodies during downregulation was further confirmed by examining the association of a second p body component, rck/p54, using IP and RIP, respectively. These data suggest that the association of LRBP with LHR mRNA results in the translocation of the mRNP complex to the p bodies leading to decapping and degradation.
Keywords: Luteinizing hormone receptor, mRNA decay, mRNA binding protein, p bodies, decapping
1. Introduction
Luteinizing hormone receptor (LHR) expression undergoes dynamic changes during normal ovarian cycle. More remarkable among these changes is the downregulation of the receptor during preovulatory LH surge [1–4]. This can also be mimicked in vivo by a pharmacological dose of its ligands. Previous studies from our laboratory have shown that downregulation of LHR mRNA occurs through a post-transcriptional mechanism where the mRNA undergoes degradation through its association with an RNA binding protein, designated as LHR mRNA binding protein (LRBP) [5]. Further studies showed that the expression of LRBP follows a reciprocal relationship with LHR mRNA [6]. The RNA binding protein was then identified as being mevalonate kinase [5–7]. In agreement with our studies, Ikeda et al showed that overexpression of MVK abrogated the FSH-induced increase in LHR mRNA expression in rat granulosa cells [8] further supporting the notion that LRBP is a negative regulator of LHR mRNA expression. Furthermore, during downregulation, LRBP translocates to the ribosomes and bind to the coding region of LHR mRNA to form an untranslatable complex, which is then targeted for degradation [9–11].
mRNA is predominantly degraded by processes initiated by shortening of the poly A tail via deadenylation. The deadenylated mRNA then undergoes decapping in the p bodies, where the 5’7mGpppN cap is removed. The decapped mRNA is then hydrolyzed by 5’ to 3’ exonuclease [12, 13]. P bodies, the site of mRNA degradation, are aggregates of RNA and proteins that was found to play an important role in the translational suppression and degradation of mRNAs [14, 15]. Since P-body aggregates contain intermediates of mRNA decay, it has been suggested that they serve as sites of mRNA degradation [14]. They contain decapping enzymes, DCP1 And DCP2 as well as the mammalian 5’-3’ exonuclease XRN1 [15–20]. Since LHR mRNA undergo accelerated degradation during downregulation, we examined the events that lead to the degradative procedures specifically focusing on the role of p body components.
In the present study, we show the association of the untranslatable LRBP-LHR mRNP complex with specific p body markers and subsequent decapping and degradation of LHR mRNA. Thus, the present study unravels the mechanism of accelerated degradation of LHR mRNA during ligand-induced downregulation by identifying the components involved in the process.
2. Materials and Methods
2.1. Materials
Pregnant mare serum gonadotrophin (PMSG) and Anti-2, 2, 7-trimethyl guanosine monoclonal antibody agarose conjugate were purchased from Calbiochem (San Diego, CA). Highly purified human chorionic gonadotrophin (hCG; CR 127) was purchased from Dr. A. F. Parlow (National Hormone and Peptide Program, Torrance, CA). EDTA-free protease inhibitor mixture tablets and RNAse inhibitor (rRNasin) were purchased from Roche Applied Science (Indianapolis, IN) and Promega (Madison, WI) respectively. Real time PCR primers for LH receptor and 18S rRNA (TaqMan Assay-on-Demand Gene Expression Product) as well as Multiscribe reverse transcriptase were from Applied Biosystems (Foster City, CA). Since LRBP was identified as MVK, anti-N-terminal mevalonate kinase IgG was raised against the first 15 N-terminal amino acids of MVK (MLSEVLLVSAPGKVI) and this antibody is referred to as the LRBP antibody in the text. Purified antibodies against β tubulin, DCP1A (Sigma, St. Louis, MO), eIF4E and rck/p54 (Santa Cruz biotech. Santa Cruz, CA) were commercial products. The Super Signal West Femto chemiluminescence kit and antirabbit/anti mouse IgG conjugated to horseradish peroxidase were obtained from Pierce (Rockford, IL). BCA reagent was purchased from GE Healthcare Life Sciences (Piscataway, NJ). ProLong Gold antifade reagent with DAPI, anchored Oligo(dT) primer, oligonucleotides and primers specific for LH receptor, RNase H as well as Alexa fluor 594-labeled goat antimouse IgG and 488-labeled goat anti-rabbit IgGs were from Invitrogen (Grand Island, NY).
2.2. Animals and Tissues
Superovulation was induced in 23 day old Sprague-Dawley rats by subcutaneous injection of 50 IU of PMSG followed by 25 IU of hCG 56 h later. The day of hCG injection was taken as 0. LH receptor downregulation was induced by the injection of 50 IU of hCG on day 5. Ovaries were collected 0, 2, 4 and 6h after hCG injection and were frozen in liquid nitrogen until further use.
2.3. Immunoprecipitation (IP)
Ovaries were homogenized in RIPA buffer and the homogenates were centrifuged at 10, 000×g for 10 min at 4°C. Equal amounts of protein from each sample (S10 fractions) were incubated overnight in primary antibody against the protein of interest at 4°C followed by which they were incubated in Protein A- Agarose beads for two hours at 4°C. The immunoprecipitated proteins were then analyzed by SDS-PAGE and Western Blot.
2.4. RNA Immunoprecipitation (RIP)
Ovaries were homogenized in NET-2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.05% Nonidet P-40) containing RNasin (100 units/0.5 ml of buffer) and protease inhibitors. The homogenates were centrifuged at 10, 000 × g for 10 min at 4°C. The supernatants were collected and 1–1.5 mg of protein/ml from the extracts (S10 fractions) were used for immunoprecipitation of RNP complex as described before [9].
2.5. Real-Time PCR (qPCR) analysis
Total RNAs were reverse-transcribed and subjected to real time PCR quantitation as described before [21]. The fold change in gene expression was calculated using the ΔΔCt method with 18S rRNA as the internal control.
2.6. Western Blot analysis
For this, tissue homogenates (S10 fractions) in RIPA buffer or immunoprecipitates were subjected to 10% SDS-PAGE under reducing conditions followed by Western blot analysis as previously described [6]. The presence of immune complexes was detected by chemiluminescence.
2.7. Immunostaining of the ovaries
Paraffin sections of the ovaries were hydrated, blocked and incubated with anti-DCP1A and MVK antibodies (dilution 1:25 in PBS containing 0.1% BSA) at 4°C overnight. Tissue sections were then double-stained with Alexa Fluor 594-labeled goat anti-mouse IgG (dilution 1:100) and Alexa Fluor 488-labeled goat anti-rabbit IgG (dilution 1:100) at room temperature for 2 h. Sections were mounted using ProLong Gold antifade reagent with DAPI, observed using a confocal photomicroscope (Olympus FluoView FV500) and photographed.
2.8. Statistical Analysis
Statistical analysis was carried out using one-way ANOVA followed by the Tukey multiple comparison test. Values were considered statistically significant for p<0.05. Each experiment was repeated at least three times with similar results. Blots and autoradiograms shown are representative of a minimum of three experiments.
3. Results
3.1. LHR mRNA undergoes decapping prior to downregulation
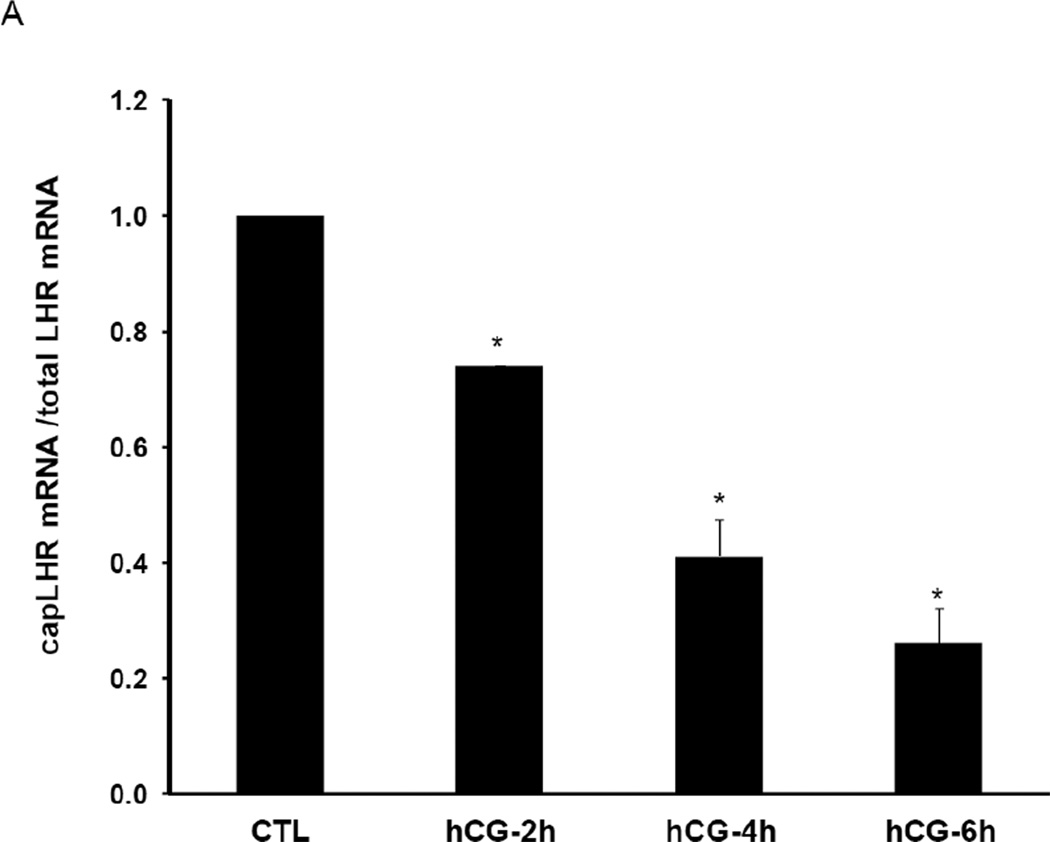
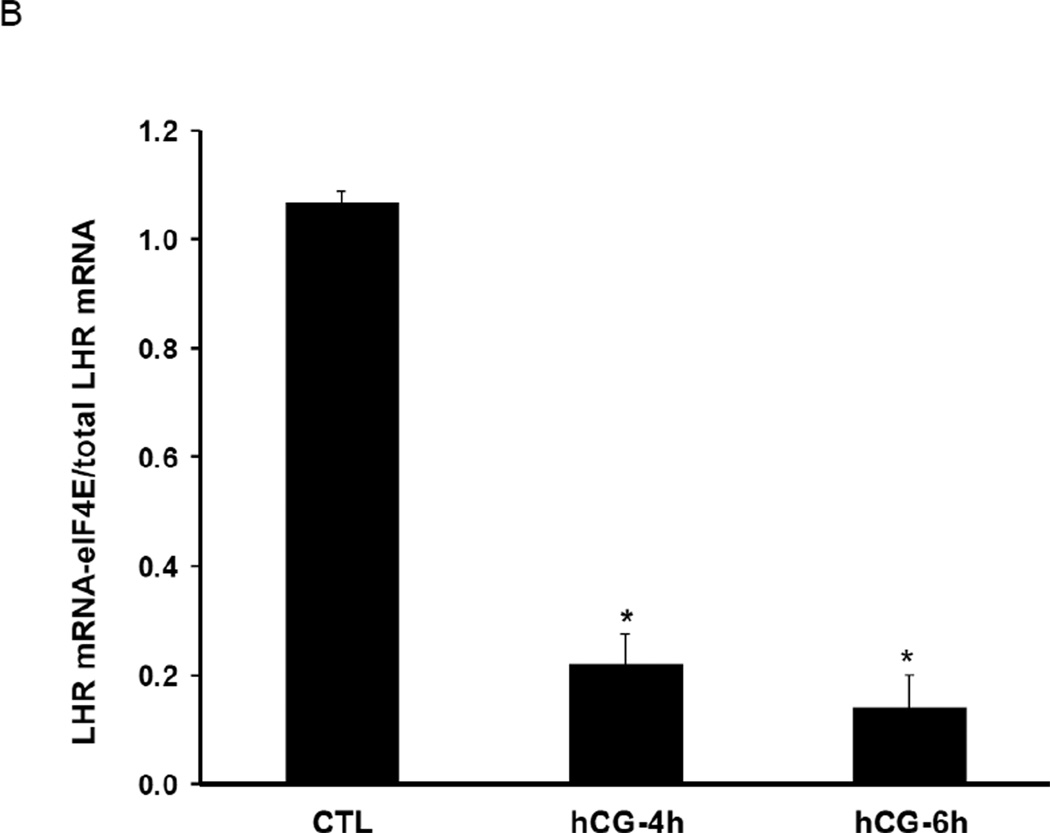
We examined the role of decapping of LHR mRNA during downregulation, since it is believed that decapping precedes 5’-3’ exonuclease mediated mRNA degradation. For this, cytosolic S100 fractions from control and downregulated ovaries were immunoprecipitated with 7-met guanosine (cap structure) antibody. Total RNA was then isolated from the immunoprecipitate which selectively contains capped mRNAs. The amount of capped LHR mRNA present in the immunoprecipitates was then determined by real time PCR using specific primers. The results clearly showed that LHR mRNA undergoes regulated decapping during downregulation and the extent of decapping increased with progression of downregulation. There was a 20% decrease in the levels of capped LHR mRNA by 2h of downregulation (Fig 1A). The decapping showed a further increase at 4 and 6 h, when the capped LHR mRNA in the downregulated samples were reduced to 40% and 25%, respectively of that of the control (Fig 1A). To further confirm this finding, the decreased association of capped LHR mRNA with eukaryotic initiation factor 4E (eIF4E) was examined. eIF4E, which is a component of the cytoplasmic cap-binding complex, stabilizes mRNA by binding to its cap structure [22]. To accomplish this, we performed an RNA-immunoprecipitation in the cytosolic S100 fractions using eIF4E antibody. RNA associated with immunoprecipitates was extracted and the levels of LHR mRNA associated with eIF4E were then determined using real time PCR. We also confirmed the identity of eIF4E by performing Western Blot analysis of the immunoprecipitates (data not shown). As evident from Fig 1B, the levels of LHR mRNA associated with eIF4E were significantly reduced in the downregulated samples when compared to the control. There was an 80% decrease in the association of LHR mRNA with eIF4E by 4h and this association was further reduced at 6h. These results show that during downregulation, eIF4E dissociates from the cap structure that normally stabilizes the complex. Thus, dissociation of eIF4E from the capped LHR mRNA-LRBP complex facilitated decapping to allow LHR mRNA degradation to occur.
Fig. 1. LHR mRNA undergoes decapping prior to downregulation.
(A) Rats were injected with hCG on the fifth day of superovulation, ovaries were collected 0, 2, 4 and 6 h later and cytosolic extracts (S10). Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to immunoprecipitation using agarose conjugated 7-methyl guanosine antibody followed by real time PCR using predesigned primers and probes for rat LH receptor mRNA as described in Materials and Methods. Total LHR mRNA levels present in the S10 fractions were also measured using real time PCR. The graphs represent ratio of LHR mRNA levels in the immunoprecipitate normalized to total LHR mRNA and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=3. (B) Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to RIP using antibodies against eIF4E followed by real time PCR using predesigned primers and probes for rat LH receptor mRNA as described in Materials and Methods. Total LHR mRNA levels present in the S10 fractions were also measured using real time PCR. The graphs represent ratio of LHR mRNA levels in the immunoprecipitate normalized to total LHR mRNA and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=3.
3.2. Translocation of LHR mRNA to the p bodies during hCG-induced downregulation
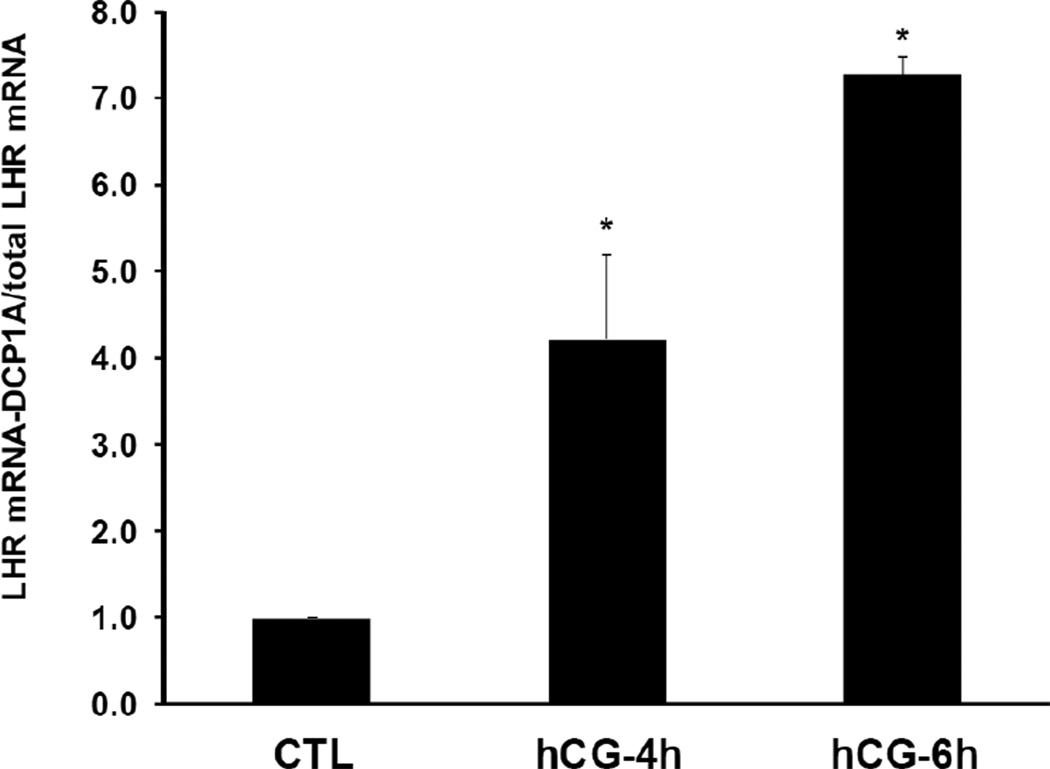
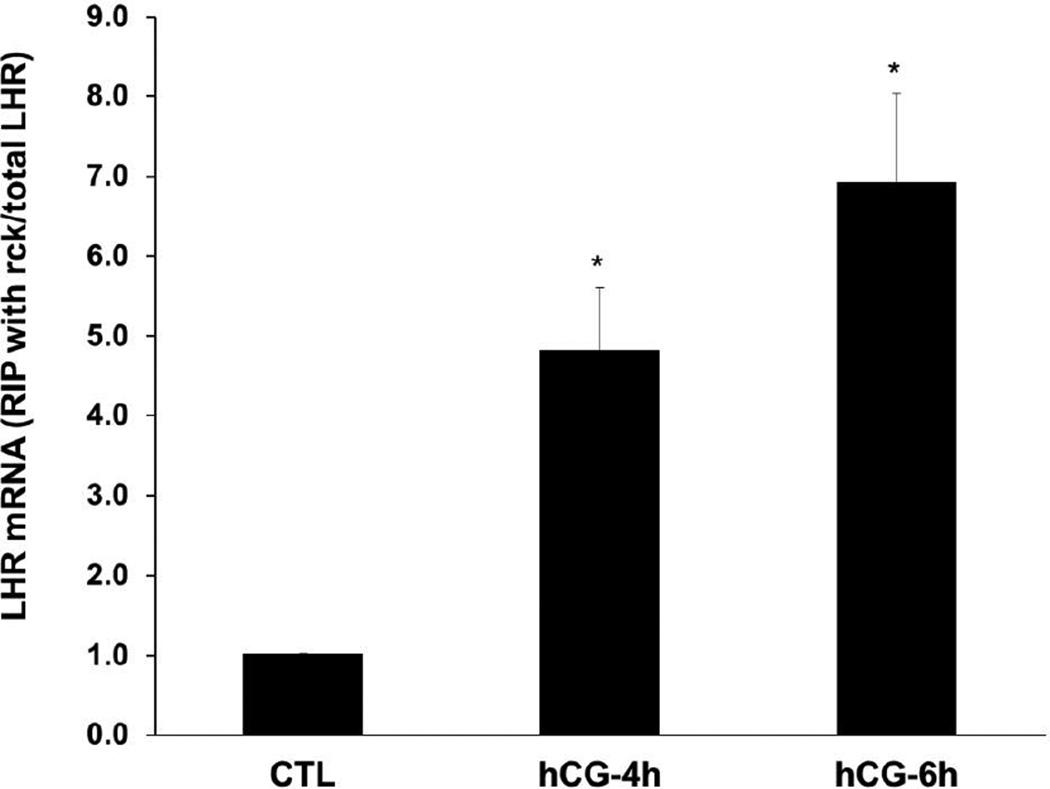
One of the major pathways of mRNA degradation in the eukaryotes involves decapping followed by 5’-3’ degradation in the p bodies. Since our results showed that decapping occurs during downregulation, as a next step we determined if LHR mRNA decay involves translocation of the untranslatable LHR mRNA-LRBP complex from the ribosomes to the p bodies. For this, the interaction of LHR mRNA with one of the p body marker proteins, DCP1A was examined. Immunoprecipitation of the cytosolic S100 fractions with an antibody against DCP1A, followed by real time PCR showed that there was a 4-fold increase in the association of LHR mRNA with the p body component DCP1A in the 4h hCG-treated sample compared to control (Fig 2). This association was further increased to 7 fold at 6h of hCG treatment, showing that the association of LHR mRNA with the p body components increases as downregulation progresses.
Fig. 2. Association of LHR mRNA with p body marker DCP1A during downregulation.
Rats were injected with hCG on the fifth day of superovulation, ovaries were collected 0, 4 and 6 h later and cytosolic extracts (S10). Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to immunoprecipitation using DCP1A antibody followed by real time PCR using predesigned primers and probes for rat LH receptor mRNA as described in Materials and Methods. Total LHR mRNA levels present in the S10 fractions were also measured using real time PCR. The graphs represent ratio of LHR mRNA levels in the immunoprecipitate to total LHR mRNA and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=4.
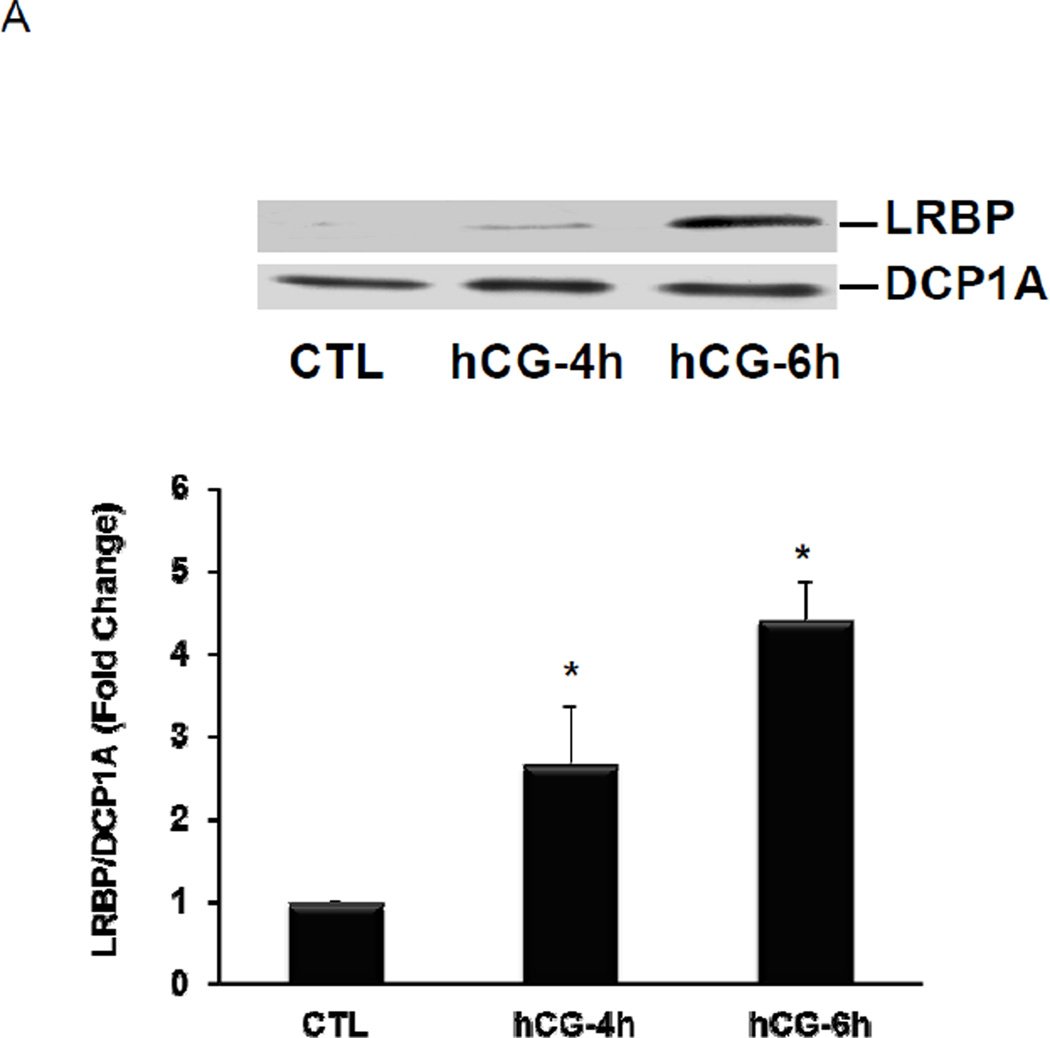
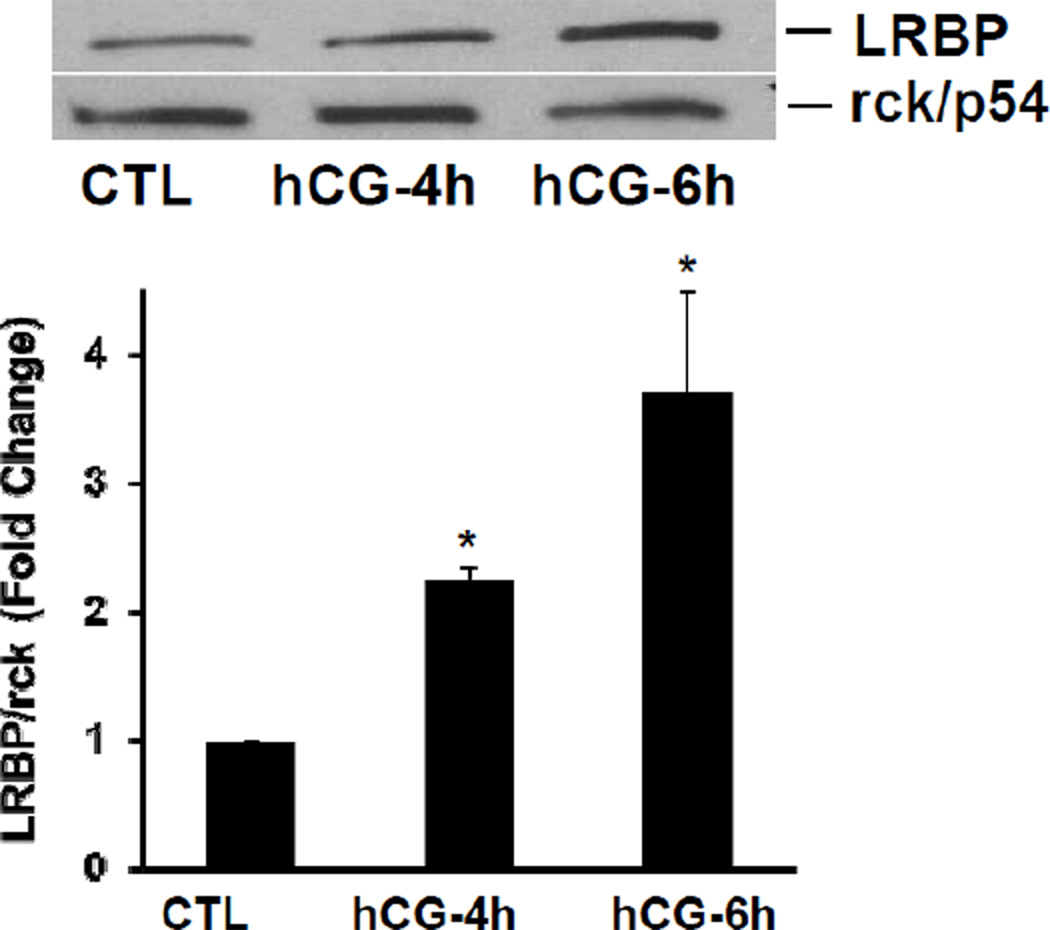
3.3. LRBP associates with p body marker, DCP1A during LHR mRNA downregulation
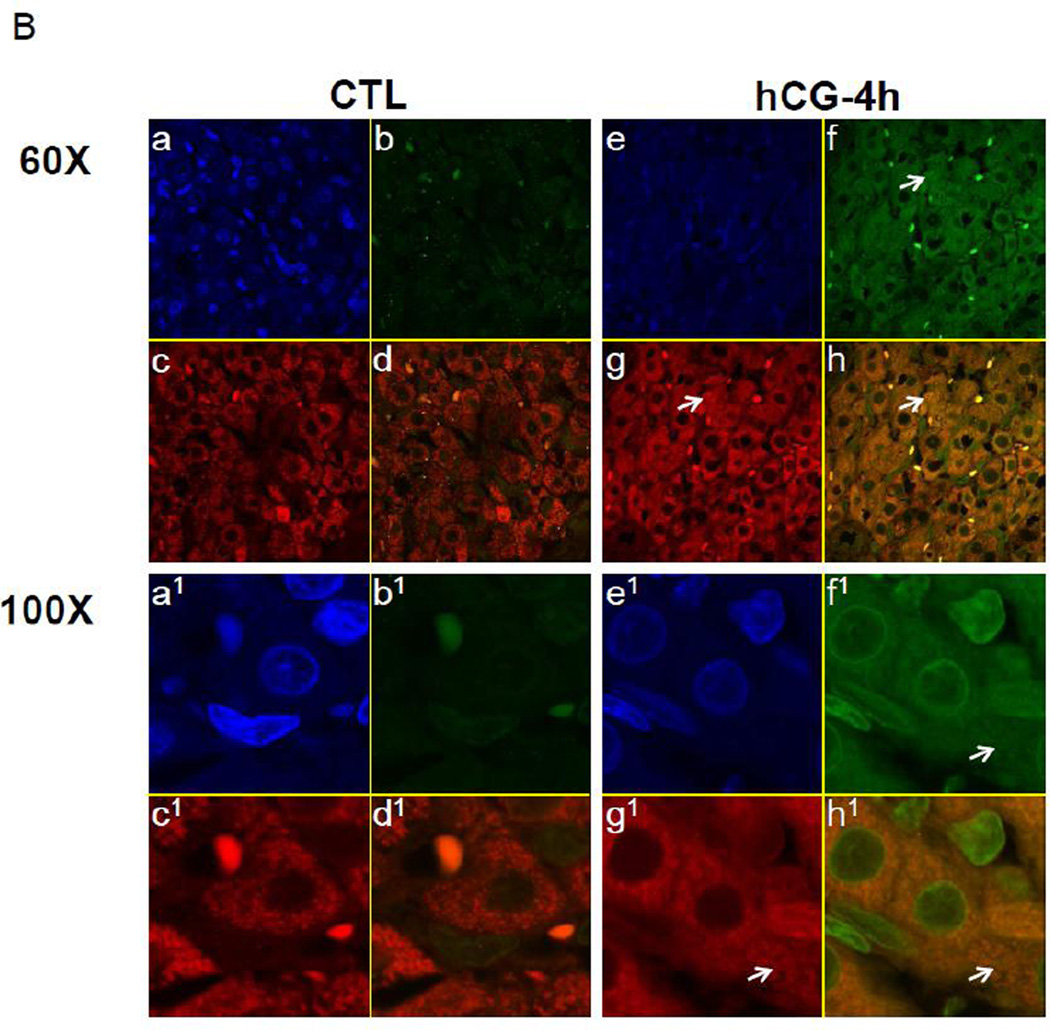
We hypothesized that the association of LHR mRNA with LRBP might be necessary for the translocation of LHR mRNA to the p bodies. To verify this, cytosolic S100 fractions were immunoprecipitated with DCP1A antibody and the immunoprecipitates were subjected to Western blot analysis using LRBP antibody. The results presented in Fig 3 clearly show that direct interaction of LRBP with the p body component, DCP1A, occurs during downregulation and this association was found to be increased significantly after 4 and 6 h of hCG treatment (Fig 3A). The reverse IP experiments using LRBP antibody for immunoprecipitation and DCP1A antibody for Western Blot analysis, also yielded similar results (data not shown). Thus, the association of LRBP with LHR mRNA plays a critical role in the transport of LHR mRNA from the ribosomes to the p bodies for its regulated degradation. To further confirm the association of LRBP with p bodies, immunohistochemistry was performed using primary antibodies against LRBP and DCP1A followed by confocal microscopic analysis. The top left panels in Fig 3B (a, a1 e and e1) show the Dapi images and the right panels (b, b1, f and f1) show LRBP staining in green. The bottom left panels (c, c1, g and g1) show DCP1A staining in red and the bottom right panels (d, d1, h and h1) show the merged image of LRBP and DCP1A staining. The results showed an increase in LRBP staining during downregulation compared to the control. There was no significant change in the intensity of DCP1A staining between control and hCG treated samples. However, the data clearly show that there was a strong association of LRBP with DCP1A as evident from the merged image of the downregulated sample. The yellow stain in the downregulated sample (see arrows in panel h, h1) indicates colocalization of the two proteins, which is not present in the control (panel d, d1). This result further confirmed our notion that during hCG-induced downregulation, LRBP mediates the transport of LHR mRNA to the p bodies for subsequent degradation.
Fig. 3. Association/Colocalization of LRBP with the p body marker during LHR mRNA downregulation.
(A) Rats were injected with hCG on the fifth day of superovulation, ovaries were collected 0, 4 and 6 h later and cytosolic extracts (S10). Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to immunoprecipitation using DCP1A antibody followed by Western Blot analysis using LRBP antibody (upper panel). The membranes were stripped and reprobed for DCP1A (lower panel). The blots shown are representative of three independent experiments. The graphs represent LRBP levels normalized to DCP1A and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=4. [B] Immunofluorescence analysis of control (CTL) and hCG treated (4h) rat ovaries using confocal microscopy. The frozen ovary sections were incubated with antibodies against DCP1A and LRBP, followed by corresponding Alexa Fluor dye conjugated secondary antibodies and then fixed in a DAPI containing antifade reagent. The images are shown in 60X (top) and 100X (bottom) magnifications. The top panels on each section show DAPI (a, a1 & e, e1; blue fluorescence) on the left and LRBP (b, b1 & f, f1; green fluorescence) on the right. Bottom panels show DCP1A (c, c1 & g, g1; red fluorescence) on the left and merged image on the right (d, d1 & h, h1; yellow stain; one representative spot is indicated by an arrow in each panel). Experiment was repeated 4 times with same results.
3.4. Association of LHR mRNA and LRBP with the p body component protein rck/p54 during hCG-induced LHR mRNA downregulation
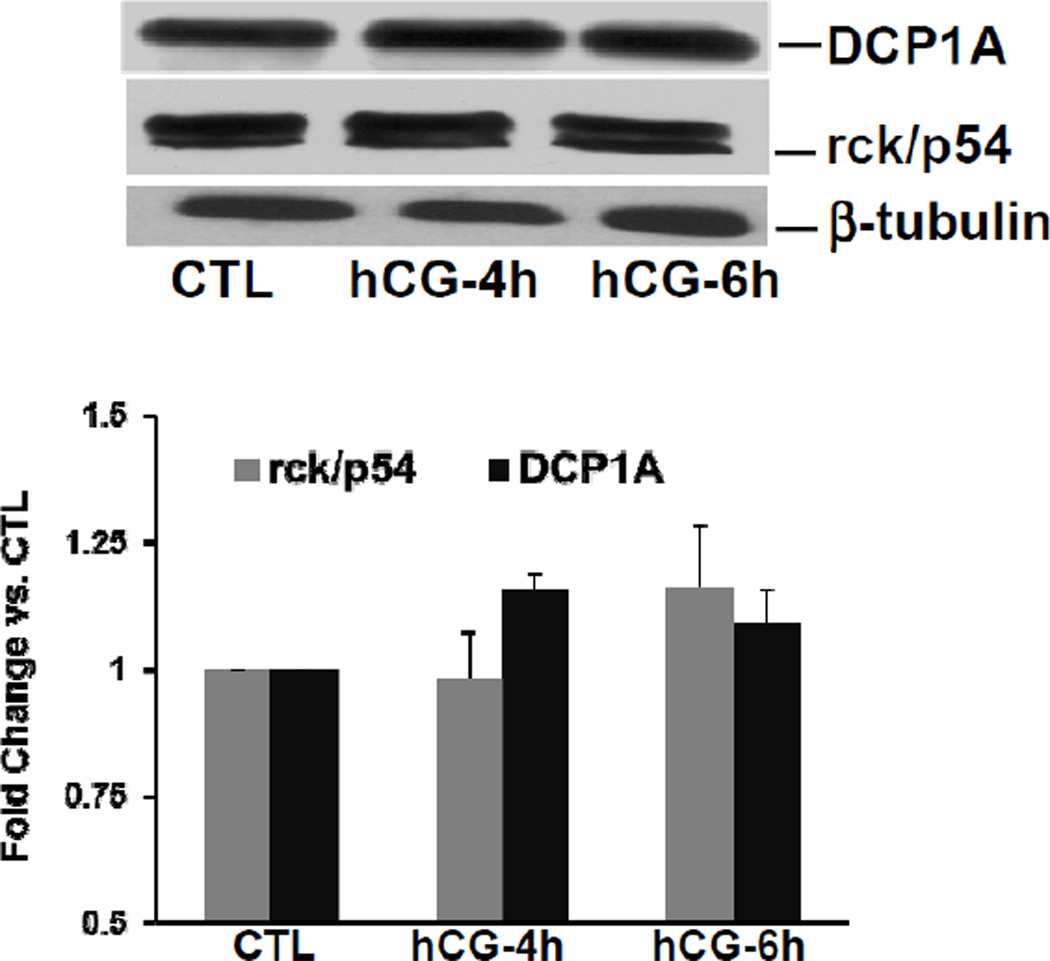
The association of LHR mRNA and LRBP with the p bodies during downregulation was further confirmed by examining their association with another p body marker protein rck/p54. Dead box helicase, DDX6 or DDX6-like proteins such as p54 are known to interact with non-translating mRNA and are present in the p bodies [23]. To examine the association of LRBP-LHR mRNP complex with rck/p54, we immunoprecipitated the cytosolic fractions with an antibody against rck/p54. The immunoprecipitates were either used for total RNA isolation followed by real time PCR to analyze the association of LHR mRNA or subjected to western blot analysis to determine the involvement of LRBP. As shown in Fig 4, there was 5-fold and 7.5-fold increases in the association of LHR mRNA with rck/p54 in the 4 and 6h hCG-treated sample, respectively compared to the control, further confirming the association of LHR mRNA with the p body components during downregulation. Similarly, the results presented in Fig 5 showed that the association of LRBP with rck/p54 increased progressively and significantly as downregulation progressed. In order to determine if there were any changes in the number of p bodies, we also analyzed the changes in the expression of p body marker proteins during downregulation. No change in the expression of either of these proteins was seen during downregulation (Fig 6). This was confirmed by the absence of any changes in the size or number of p bodies in immunohistochemical analysis presented in Fig 3B. There was also no change in the pattern of p bodies with or without hCG treatment (panels d and g). These results suggest that the p body formation occurs at the onset of downregulation and the quantity of p bodies does not appear to change during the time course of downregulation.
Fig. 4. Association of LHR mRNA with p body component protein rck/p54 during downregulation.
Rats were injected with hCG on the fifth day of superovulation, ovaries were collected 0, 4 and 6 h later and cytosolic extracts (S10). Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to immunoprecipitation using rck/p54 antibody followed by real time PCR using predesigned primers and probes for rat LH receptor mRNA as described in Materials and Methods. Total LHR mRNA levels present in the S10 fractions were also measured using real time PCR. The graphs represent ratio of LHR mRNA levels in the immunoprecipitate to total LHR mRNA and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=3.
Fig. 5. Association of LRBP with p body component protein rck/p54 during downregulation.
Rats were injected with hCG on the fifth day of superovulation, ovaries were collected 0, 4 and 6 h later and cytosolic extracts (S10) prepared using RIPA buffer. Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to immunoprecipitation using rck/p54 antibody followed by Western Blot analysis using LRBP antibody (upper panel). The membranes were stripped and reprobed for rck (lower panel). The blots shown are representative of three independent experiments. The graphs represent LRBP levels normalized to rck and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=4.
Fig. 6. Expression of p body markers DCP1A and rck/p54 during LHR mRNA downregulation.
Rats were injected with hCG on the fifth day of superovulation, ovaries were collected 0, 4 and 6 h later and cytosolic extracts (S10) prepared using RIPA buffer. Equal amounts of protein from the control (CTL) or hCG-treated S10 fractions were subjected to Western Blot analysis using DCP1A antibody (upper panel). The membranes were stripped and reprobed first for rck (middle panel) and finally for tubulin (lower panel). The blots shown are representative of three independent experiments. The graphs represent DCP1A and rck levels normalized to tubulin and are shown as fold change vs. control. Error bars represent mean ±SE. *p<0.05, n=4–7.
4. Discussion
Studies employing microarray analyses have revealed that 40–50% of the changes in gene expression pattern in response to cellular signals occur at the level of mRNA stability [24, 25]. These changes are usually induced by alterations in the composition of messenger ribonucleo proteins (mRNPs) that either inhibit or facilitate mRNA decay. A similar mechanism, involving alteration of mRNA stability, is responsible for the changes in LHR mRNA expression during ovarian cycle [6, 26]. Our past studies revealed a crucial role for an mRNA binding protein, designated as LRBP, in LHR downregulation during preovulatory LH surge [5, 6, 26, 27]. We have shown that LRBP associates with LHR mRNA in the ribosomes and the untranslatable complex formed is routed for degradation [9]. Here we show that during ligand-induced downregulation, LHR mRNA accumulates in the p bodies, where it undergoes decapping. Using confocal microscopy and immunoprecipitation experiments, we further show that the LHR mRNA binding protein, LRBP, also associates with the p bodies during LHR mRNA downregulation. These results suggest that the untranslatable mRNP complex consisting of LHR mRNA and LRBP is translocated from the ribosomes to the p bodies, where it undergoes decapping making it a potential target for degradation by 5’→ 3’ exonuclease.
mRNAs are predominantly degraded by two possible pathways in eukaryotic cells that are initiated by shortening of the poly A tail (deadenylation). In one pathway, the deadenylated mRNAs undergo decapping to remove the 5’7mGpppN cap prior to degradation by 5’→ 3’ exonucleases in the p bodies. In the second pathway, deadenylated mRNAs are degraded by 3’→ 5’ exonucleases in the cytoplasmic exosomes [12, 13]. Since LHR mRNA undergoes decapping prior to degradation, it points to the strong possibility that LHR mRNA is degraded by the 5’→ 3’ exonuclease in the p bodies. This is consistent with the notion that translationally suppressed mRNAs accumulate in the p bodies to undergo decapping [28]. P bodies are dynamic structures that are formed to degrade translationally suppressed mRNAs [14, 15]. Using two different markers of p bodies, we were able to demonstrate that LHR mRNA associates with p body components during downregulation. However, the number or size of the p bodies showed no significant changes during downregulation. This was not surprising since LHR mRNA accounts for only a small proportion of the total mRNAs in the ovary. Furthermore, stimulation of the cAMP/PKA pathway, in response to LH/hCG treatment, instituted to induce LHR mRNA downregulation, causes the activation of transcription of a large number of unrelated genes that might further dilute the relative abundance of LHR mRNA. The results presented in this study demonstrate that LRBP is also colocalized with LHR mRNA in the p bodies during downregulation. Thus, during downregulation, LRBP remains associated with actively translating LHR mRNA from the outset until it is degraded in the p bodies. Since LRBP directly binds to both the p body marker proteins (DCP1A and rck) during downregulation, it is likely that the p bodies might interact with LHR mRNA that has been bound to LRBP.
There are several other examples where mRNA binding proteins have been shown to deliver mRNAs to p bodies by interacting with p body components [29–32]. In particular, the cytoplasmic polyadenylation element-binding protein (CPEB-1), an RNA-binding protein that plays a role in translational control in oocytes and localized in p bodies, has been shown to interact with a number of p body components such as rck/p54 [33]. It is also possible that LRBP might interact with other yet unidentified factors/proteins for transporting the complex to the p bodies. In this context, our previous studies [34] have shown that LRBP can interact with other proteins such as ubiquitin conjugating enzyme 2i (UBCE2i), a SUMO conjugating enzyme and eIF5A, a ubiquitous protein believed to be involved in translational suppression and decay in other systems. Thus it is possible that the interaction of LRBP with one or more of these proteins might be involved in the translocation of LRBP-LHR mRNP complex to the p bodies.
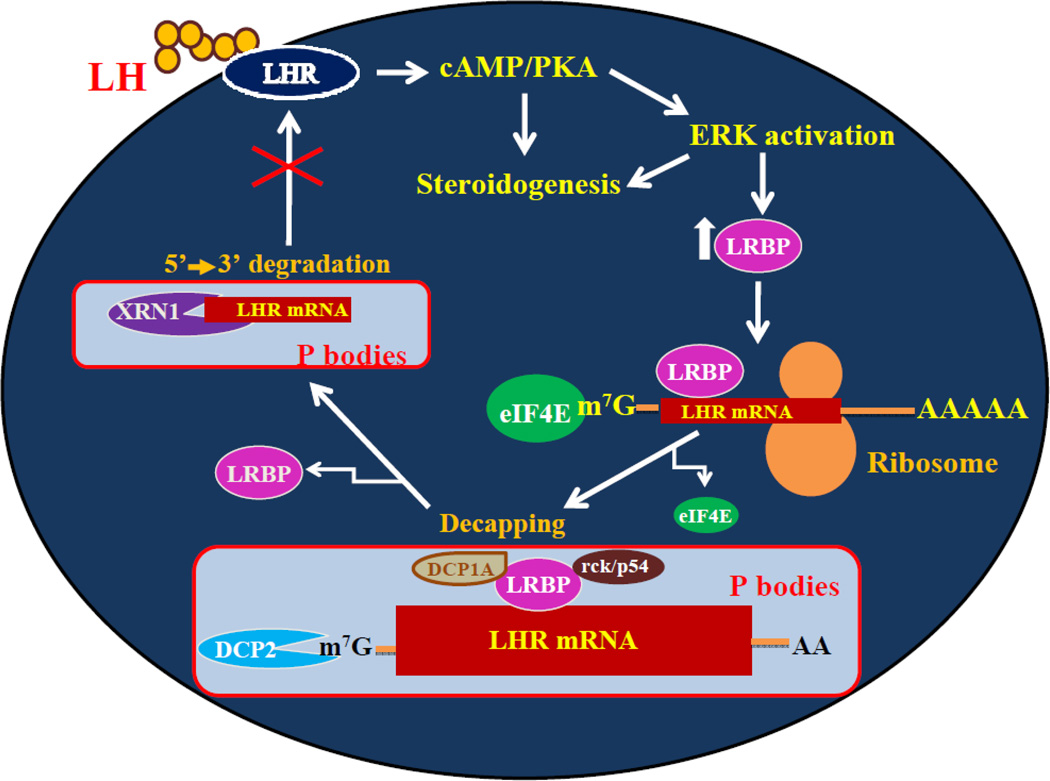
In summary, a model for LRBP-mediated LHR mRNA decay is proposed, which is illustrated in Fig 7. LH activates cAMP/PKA/ERK pathway leading to an increase in the expression of LRBP [35, 36]. LRBP binds to the coding region of LHR mRNA in the ribosomes to form an untranslatable complex [9–11]. LRBP then associates with the p body components such as DCP1A and rck/p54 thereby inducing the p body localization of LRBPLHR mRNP complex. This is accompanied by the dissociation of eIF4E from the cap structure and decapping. Exposure of the uncapped, nascent 5’ end of LHR mRNA facilitates its degradation by the 5’-3’ exonuclease, XRN1, culminating in the downregulation of LHR.
Fig. 7. Proposed mechanism of LH/hCG stimulated and LRBP mediated degradation of LHR mRNA in the p bodies.
LH signaling through its receptor activates cAMP/PKA followed by the ERK pathway, leading to increased levels of LRBP. LRBP binds to the coding region of LHR mRNA in the ribosomes, to form an untranslatable ribonucleoprotein complex. LRBP also associates with the p body components such as DCP1A and rck/p54 resulting in the p body localization of LRBP-LHR mRNP complex. This is accompanied by the dissociation of eIF4E from the cap structure. In the next step, LHR mRNA undergoes decapping in the p bodies initiated by the enzyme DCP2. This leads to the 5’-3’ exonuclease (XRN1) mediated degradation of LHR mRNA, in the final step.
Highlights.
Accumulation of LHR mRNA in p bodies during downregulation.
Role of LRBP in LHR mRNA accumulation in p bodies.
Decapping of LHR mRNA in p bodies leads to mRNA degradation.
Acknowledgement
This work was supported by National Institutes of Health Grant R37 HD06656. We thank Meghan Franzo-Romain for technical assistance. We are grateful to Helle Peegel and other members of the laboratory for critical reading of the manuscript and many helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lu DL, Menon KM. 3' untranslated region-mediated regulation of luteinizing hormone/human chorionic gonadotropin receptor expression. Biochemistry. 1996;35:12347–12353. doi: 10.1021/bi961019a. [DOI] [PubMed] [Google Scholar]
- 2.LaPolt PS, Oikawa M, Jia XC, Dargan C, Hsueh AJ. Gonadotropin-induced up- and down-regulation of rat ovarian LH receptor message levels during follicular growth, ovulation and luteinization. Endocrinology. 1990;126:3277–3279. doi: 10.1210/endo-126-6-3277. [DOI] [PubMed] [Google Scholar]
- 3.Segaloff DL, Wang HY, Richards JS. Hormonal regulation of luteinizing hormone/chorionic gonadotropin receptor mRNA in rat ovarian cells during follicular development and luteinization. Mol Endocrinol. 1990;4:1856–1865. doi: 10.1210/mend-4-12-1856. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman YM, Peegel H, Sprock MJ, Zhang QY, Menon KM. Evidence that human chorionic gonadotropin/luteinizing hormone receptor down-regulation involves decreased levels of receptor messenger ribonucleic acid. Endocrinology. 1991;128:388–393. doi: 10.1210/endo-128-1-388. [DOI] [PubMed] [Google Scholar]
- 5.Kash JC, Menon KM. Identification of a hormonally regulated luteinizing hormone/human chorionic gonadotropin receptor mRNA binding protein. Increased mrna binding during receptor down-regulation. J Biol Chem. 1998;273:10658–10664. doi: 10.1074/jbc.273.17.10658. [DOI] [PubMed] [Google Scholar]
- 6.Nair AK, Kash JC, Peegel H, Menon KM. Post-transcriptional regulation of luteinizing hormone receptor mRNA in the ovary by a novel mRNA-binding protein. J Biol Chem. 2002;277:21468–21473. doi: 10.1074/jbc.M111653200. [DOI] [PubMed] [Google Scholar]
- 7.Nair AK, Menon KM. Isolation and characterization of a novel trans-factor for luteinizing hormone receptor mRNA from ovary. J Biol Chem. 2004;279:14937–14944. doi: 10.1074/jbc.M309484200. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda S, Nakamura K, Kogure K, Omori Y, Yamashita S, Kubota K, Mizutani T, Miyamoto K, Minegishi T. Effect of estrogen on the expression of luteinizing hormonehuman chorionic gonadotropin receptor messenger ribonucleic acid in cultured rat granulosa cells. Endocrinology. 2008;149:1524–1533. doi: 10.1210/en.2007-1163. [DOI] [PubMed] [Google Scholar]
- 9.Menon B, Peegel H, Menon KM. Evidence for the association of luteinizing hormone receptor mRNA-binding protein with the translating ribosomes during receptor down-regulation. Biochim Biophys Acta. 2009;1793:1787–1794. doi: 10.1016/j.bbamcr.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair AK, Menon KM. Regulation of luteinizing hormone receptor expression: evidence of translational suppression in vitro by a hormonally regulated mRNA-binding protein and its endogenous association with luteinizing hormone receptor mRNA in the ovary. J Biol Chem. 2005;280:42809–42816. doi: 10.1074/jbc.M503154200. [DOI] [PubMed] [Google Scholar]
- 11.Kash JC, Menon KM. Sequence-specific binding of a hormonally regulated mRNA binding protein to cytidine-rich sequences in the lutropin receptor open reading frame. Biochemistry. 1999;38:16889–16897. doi: 10.1021/bi9915770. [DOI] [PubMed] [Google Scholar]
- 12.Garneau NL, Wilusz J, Wilusz CJ. he highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 13.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 17.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of posttranscriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Nair AK, Menon KM. Ribonucleic acid binding protein-mediated regulation of luteinizing hormone receptor expression in granulosa cells: relationship to sterol metabolism. Mol Endocrinol. 2007;21:2233–2241. doi: 10.1210/me.2007-0102. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, Yang X, Wang W, Wood WH, 3rd, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci U S A. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menon KM, Nair AK, Wang L. A novel post-transcriptional mechanism of regulation of luteinizing hormone receptor expression by an RNA binding protein from the ovary. Mol Cell Endocrinol. 2006;246:135–141. doi: 10.1016/j.mce.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 27.Menon KM, Nair AK, Wang L, Peegel H. Regulation of luteinizing hormone receptor mRNA expression by a specific RNA binding protein in the ovary. Mol Cell Endocrinol. 2007;260–262:109–116. doi: 10.1016/j.mce.2006.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 29.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoecklin G, Anderson P. In a tight spot: ARE-mRNAs at processing bodies. Genes Dev. 2007;21:627–631. doi: 10.1101/gad.1538807. [DOI] [PubMed] [Google Scholar]
- 32.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 33.Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Gulappa T, Menon KM. Identification and characterization of proteins that selectively interact with the LHR mRNA binding protein (LRBP) in rat ovaries. Biochim Biophys Acta. 2010;1803:591–597. doi: 10.1016/j.bbamcr.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menon B, Franzo-Romain M, Damanpour S, Menon KM. Luteinizing hormone receptor mRNA down-regulation is mediated through ERK-dependent induction of RNA binding protein. Mol Endocrinol. 2011;25:282–290. doi: 10.1210/me.2010-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peegel H, Towns R, Nair A, Menon KM. A novel mechanism for the modulation of luteinizing hormone receptor mRNA expression in the rat ovary. Mol Cell Endocrinol. 2005;233:65–72. doi: 10.1016/j.mce.2004.12.009. [DOI] [PubMed] [Google Scholar]