Abstract
Objective
Changes in metabolic homeostasis in pregnant diabetic women are potential determinants of increased adiposity of the fetus. The aim of this study was to characterize diabetes-induced changes in genes for fetal-placental energy metabolism in relation to fetal adiposity.
Research Design and Methods
Placentas of women with type 1 diabetes (T1DM), gestational diabetes (GDM) or no complications were analyzed using microarray profiling. Pattern of gene expression were assessed in primary placental cell cultures.
Results
Diabetes was associated with 49 alterations in gene expression at key steps in placental energy metabolism with 67 % related to lipid and 9 % related to glucose pathways. Preferential activation of lipid genes was observed in pregnancy with GDM. T1DM induced fewer lipid modifications but an enhancement of glycosylation and acylation pathways. Oleate enhanced expression of genes for fatty acid esterification and the formation of lipid droplets 3 times as much as glucose in cultured placental cells.
Conclusions
These results point to fatty acids as preferential lipogenic substrates for placental cells and, suggest that genes for fetal-placental lipid metabolism are selectively enhanced in GDM. The recruited genes may be instrumental in increasing transplacental lipid fluxes hence delivery of lipid substrates for fetal use.
Keywords: diabetes mellitus, fetus, human, metabolism, obesity, placenta, pregnancy
Background and Objectives
Diabetic macrosomia results from modification of body composition with increased adiposity at birth rather than changes in lean mass (1). How maternal diabetes translates into enhanced lipid deposition in adipose tissue of the fetus is not currently understood. The association between high maternal glucose levels and increased birth weight has long been documented in diabetic pregnancy (2). However fetal macrosomia is also observed in diabetic pregnancy with satisfactory glycemic control (3–5) as well as in offspring of obese women with normal glucose tolerance and only mild or no hyperglycemia (6–8). This suggests that either modest variation in fetal glycemia or in energy substrates other than glucose contribute to enhance fat deposition in fetal adipose tissue. Based on the dogma of adipocyte biology, enhanced adipogenesis relies on plethora of adipogenic substrates in association with a hyperinsulinic milieu. The availability of fetal energy substrates is regulated in the first place by their maternal circulating concentrations and to the extent they are transported across the placenta. Hence the hyperglycemia and hypertriglyceridemia facilitated by the insulin resistance of the diabetic mothers are potential factors to enhance substrate availability to the fetus (9–11). The structural and functional characteristics of the trophoblast cells, allow flexibility in glucose uptake, metabolism and transfer to adapt to concentration changes (12). Maternal FFA are also taken up by placental cells for direct fetal transfer to the fetus (13). However, complex lipids such as triglycerides can only be transported after hydrolysis by specific trophoblast lipases (14). The subsequent re-esterification of FFA suggests that interim storage takes place within the placenta as suggested in rodent model of diabetes (15). However there is only scant information to assess the regulation of placental-fetal lipid fluxes in pregnancy with abnormal lipid homeostasis (16).
In a first step towards the characterization of the mechanisms regulating fetal adipogenic capacity we have characterized the changes in placental glucose and lipid pathways induced by diabetes. In order to encompass an array of diabetic pathologies and neonatal outcomes, we have included pregnancies with type 1 diabetes (T1DM) and gestational diabetes (GDM), a milder diabetic pathology similar in many respects to type 2 diabetes (17). In this report we show that diabetes induces major alterations of key pathways for feto-placental lipid fluxes with differential activation in GDM and T1DM.
Subjects, Material and Methods
Study subjects
The protocol was approved by the institutional review boards of MHMC and Hospital Cochin and volunteers gave informed written consent in accordance with each institution guidelines. Women with no pregnancy complication (n=5), GDM (n=9) or type 1 diabetes (n=6) were recruited at the time of delivery in term pregnancy. GDM was defined as an abnormal glucose tolerance during the third trimester according to the criteria defined by Carpenter and Coustan (18). All GDM subjects required insulin therapy for glucose control. Glycemic control of the diabetic women was considered satisfactory based on an average %HbA1C <6.3 (Table 1). Placenta, maternal and cord blood samples were obtained at time of term elective C-section. Maternal anthropometrics were recorded at the first antenatal visit. At term women with T1DM were hyperglycemic and hyperinsulinemic. No significant hyperlipidemia was noted in mothers or fetuses although there was a tendency to slightly higher plasma NEFA in women with GDM. Neonatal anthropometric measurements were performed at delivery. Fetuses of both GDM and type 1 diabetic women were hyperinsulinemic and macrosomic with increased adiposity at birth based on higher ponderal index (Table 1).
Table 1.
anthropometrics and metabolic data of the study population Results are means ±SD.
| Control (5) | GDM (9) | T1DM (6) | |
|---|---|---|---|
| maternal age at delivery | 28.0±2.4 | 29.7±2.2 | 31.7±4.3 |
| gestational age (weeks) | 38.7±0.7 | 38.5±1.0 | 37.3±0.5 |
| maternal pre-gravid BMI | 20.7±1.2 | 42.7±6.0** | 21.8±1.6 |
| maternal glucose (mg/dl) | 80.8±10.4 | 90.7±23.7 | 126±19.4** |
| maternal insulin (mU/ml) | 22.9±1.1 | 53.7±36.7* | 14.0±3.5 |
| maternal HbA1C (%) | 5.3±0.1 | 6.1±0.2* | 6.2±0.28* |
| maternal triglycerides (mg/dl) | 184±9 | 198±21 | 189±18 |
| maternal NEFA (mM) | 0.67±0.03 | 0.81±0.08 | 0.68±0.12 |
| placenta weight (g) | 568±90 | 703±107* | 658±72 |
| birth weight (g) | 3440±340 | 3679±499* | 3840±620* |
| neonatal ponderal index (g/cm3) | 2.62 ± 0.07 | 3.02±0.4* | 3.01±0.4* |
| umbilical cord insulin (mU/ml) | 12.6±2.7 | 23.2±2.1** | 49.6±19.9** |
| umbilical cord glucose (mg/dl) | 77±6.6 | 87±6.2* | 89±10.9* |
| umbilical cord triglycerides (mg/dl) | 34.3±8.5 | 27.9±1.9 | 33.5±5.9 |
| umbilical cord NEFA (mM) | 0.18±0.01 | 0.16±0.01 | 0.17±0.05 |
p<0.01 and
p<0.001 vs controls.
GDM: gestational diabetes, T1DM: Type 1 diabetes, NEFA: Non esterified fatty acids.
Analysis of glucose, lipids and insulin
Plasma glucose was measured by the glucose oxidase method with a glucose analyzer (Yellow Springs Instrument, Yellow Springs, OH). Plasma FFA concentrations were determined with an enzymatic method (WAKO chemicals, Neuss, Germany). Plasma triglyceride concentrations were determined with an enzymatic method (Sigma). Plasma insulin was assayed by radioimmunoassay (Linco, St louis, MO).
Tissue and microarray processing
For each placenta, six biopsies (approximately 1 cm3) were sampled within 5 distinct cotyledons (30 fragments total) after dissecting out the basal and the chorial plates and snap-frozen in liquid nitrogen. Total RNA was prepared from 12 out of 30 randomly selected tissue samples using CsCl gradient (19) and electrophoresed to verify integrity. Aliquots of the twelve RNA samples prepared from each placenta were pooled and reversed transcribed into cDNA using Superscript first strand synthesis (Invitrogen, CA). The pooling strategy was used to minimize inter and intra-placenta structural variations inherent to human placental heterogeneity. Biotinylated cRNA where generated by in vitro transcription (ENZO kit, Affymetrix) and evaluated with test-microarrays (Test-3array, Affymetrix). The quality of samples was considered satisfactory with the presence of bioC, bioD and cre and a 3’/5’ ratio of the polyA controls < 3. Each cRNA were run on duplicate microarrays (human U133 Affymetrix) according to manufacturer’s instructions. Signals scanning and analysis were performed with Affymetrix equipments (Fluidics station, HPgene array scanner, MAS 5.0 microarray suite). Results were scaled to an average signal intensity of 1500 to correct for hybridization efficiency.
Data analysis
Selection of the significantly modified transcripts was performed by applying the following filter strategy. All probe sets with intensity signal below the probe pair threshold were excluded. Among the genes showing an absolute call of present (P- calls) according to MAS 5 algorithm, we selected the genes with a difference in signal detection at least 4.5 times the average background minus the scaled noise. The genes having satisfied these criteria were then selected based on a fold change ≥ 1.52 or ≤ −1.52 consistent in at least two comparisons. The genes who did not meet the fold change criteria were referred to as not significantly changed (nsc). Genes related to glucose and lipid metabolism were identified according to the function of their putative encoded proteins from public databases. Fold changes were validated by real-time PCR measurements on ten selected genes (Table 2). The hybridization intensities of significantly modified genes were examined by hierarchical cluster analysis using genespring and treeview.
Table 2.
relative abundance of metabolic genes in pregnancy with type 1 and gestational diabetes. The signal intensities were obtained after normalization with MAS5.0. Numbers in parentheses show fold changes between diabetic and control groups estimated by realtime RT-PCR. Fold changes between diabetic and control groups were significant at p<0.005. nsc; not statistically significant. T1DM type 1 diabetes, GDM: gestational diabetes.
| NCBI Accession N° |
Gene | GDM signal intensity |
GDM fold change |
T1DM signal intensity |
T1DM fold change |
description | ||
|---|---|---|---|---|---|---|---|---|
|
lipid transport and activation (9) | ||||||||
| NM_001444 | FABP5 | 5412±2099 | 2.2 | (1.9) | 5328±623 | 1.7 | (1.8) | Fatty acid binding protein 5 |
| NM_001442 | FABP4 | 6091±2391 | 2.3 | (1.8) | 6218±1641 | 2.4 | (1.5) | Fatty acid binding protein 4 |
| NM_006033 | LIPG | 5560±448 | 1.3 | 6864±1248 | 1.9 | Endothelial lipase | ||
| NM_000237 | LPL | 4704±1273 | −2.7 | (−1.8) | 3247±129 | −3.4 | (−2.8) | Lipoprotein lipase |
| NM_004457 | FACL3 | 1097±333 | 2.3 | (4.6) | 1743±138 | 1.7 | Fatty-acid-Coenzyme A ligase, long-chain 3 | |
| NM_021122 | FACL2 | 1840±806 | 2.3 | 853±293 | nsc | Fatty-acid-Coenzyme A ligase, long-chain 2 | ||
| NM_022977 | FACL4 | 3178±1706 | 2.4 | 1783±631 | nsc | Fatty-acid-Coenzyme A ligase, long-chain 4 | ||
| NM_000041.1 | APOE | 1419±374 | 1.52 | 898±187 | 2.3 | Apolipoprotein E | ||
| M64497.1 | ARP-1 | 4432±1389 | 1.52 | 4270±835 | 2.8 | Apolipoprotein AI regulatory protein | ||
| lipid metabolism (23) | ||||||||
| NM_021105 | PLSCR1 | 1190±551 | 1.74 | 1580±144 | nsc | Phospholipide srcamblase | ||
| NM_000929 | PLA2G5 | 2713±217 | 2.6 | (2.2) | 1111±331 | nsc | (nsc) | Phospholipase A2, group V |
| NM_006745 | SC4MOL | 1202±793 | 2.1 | 881±226 | nsc | Sterol-C4-methyl oxidase-like | ||
| NM_002979.1 | SCP2 | 2840±325 | 1.52 | 2061±245 | nsc | Sterol carrier protein | ||
| NM_006579 | EBP | 170±99 | nsc | 1488±27 | 13.8 | Emopamil sterol isomerase | ||
| AF_148464.1 | PCYT1B | 470±176 | nsc | 58±33 | nsc | Phosphatidylcholine cytidyltransferase | ||
| NM_021213 | PCTP | 2433±459 | 2.3 | 1225±305 | nsc | Phosphatidylcholine transfer protein | ||
| NM_006227 | PLTP | 10983±4081 | 1.74 | 7565±2502 | 1.52 | Phospholipid transfer protein | ||
| NM_002543.1 | OLR1 | 3635±1460 | 1.74 | 3817±304 | 2 | Oxidised LDL receptor | ||
| NM_002332.1 | LRP1 | 1135±85 | nsc | 3159±485 | 2.3 | LDL-related protein 1 | ||
| NM_000859 | HMGCR | 1331±812 | 2.5 | (2.6) | 826±317 | nsc | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | |
| NM_002130 | HMGCS1 | 874±37 | 2 | (2.0) | 1686±141 | nsc | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase | |
| NM_005063.1 | SCD | 7420±951 | 8 | (3.6) | 1443±440 | nsc | (nsc) | Stearoyl-CoA desaturase |
| NM_004733 | ACATN | 531±205 | 1.62 | 417±82 | nsc | AcetylcoA transporter | ||
| NM_000182 | HADHA | 3936+1542 | −2 | 4778+781 | nsc | Hydroxyacyl-Coenzyme A dehydrogenase | ||
| NM_003129 | SQLE | 4792±1516 | 2 | (4.6) | 4864±835 | nsc | (nsc) | Squalene epoxidase |
| NM_006412 | AGPAT2 | 288±135 | −2 | 1454±226 | nsc | Acylglycerol-3-phosphate O-acyltransferase 2 | ||
| BC_005127 | ADRP | 37780±4883 | 1.74 | 15067±7516 | nsc | (nsc) | Adipose differentiation-related protein, | |
| M 22921 | HUMGSTE | 1426±324 | −2 | 2348±80 | nsc | Galactosyl-transferase | ||
| NM_000255 | MUT | 1639±336 | 1.62 | 1082±269 | nsc | MethylmalonyCoA mutase | ||
| AK022765 | AMACR | 132±27 | nsc | 395±64 | 4.2 | MethylacylcoA racemase | ||
| NM_005327 | HADHSC | 663±15 | 1.9 | 783±131 | 4.5 | HydroxyacylCoA dehydrogenase | ||
| NM_001805.1 | CEBP | 283±124 | 1.9 | 1297±268 | 1.8 | CCAAT enhancer binding protein | ||
| glucose metabolism (6) | ||||||||
| NM_030807 | GLUT10 | 1047±201 | nsc | 2354±310 | 1.6 | Facilitative hexose transporter | ||
| NM_002300 | LDHB | 12603±4442 | 2 | 11704±683 | nsc | Lactate dehydrogenase B | ||
| NM_000158 | GBE1 | 4098±1512 | 1.74 | 3417±681 | nsc | Glycogen branching | ||
| NM_000645 | AGL | 2466±734 | 1.87 | 1484±290 | nsc | Amylo1-6-glucosidase | ||
| NM-003918 | GYG | 1405±657 | 2.3 | (1.9) | 604±107 | nsc | (nsc) | Glycogenin2. DVS27related protein |
| NM_002438.1 | MRC1 | 3492±142 | 1.62 | 2558±659 | nsc | Mannose receptor | ||
| hexose monophosphate/glycosylation (11) | ||||||||
| NM_005907.1 | MAN1A1 | 1084±290 | nsc | 1308±585 | 2.2 | Mannosidase alpha1 | ||
| NM_003605.2 | OGT | 1067±352 | nsc | 2692±705 | 2.3 | UDP-N-acetylglucosamine transferase | ||
| NM_005476.2 | GNE | 5249±1768 | −1.5 | 4094±1060 | −2.1 | UDP-N-acetylglucosamine-2-epimerase | ||
| NM_002406.2 | MGAT1 | 9798±1643 | 1.6 | 6320±1311 | 8.0 | Mannosyl-acetylglucosaminyltransferase | ||
| NM_012269 | HYAl4 | 143±48 | nsc | 579±173 | 5.4 | Hyaluronoglucosaminidase | ||
| NM_002408 | MGAT2 | 91±29 | nsc | 221±153 | 4.3 | Mannosylacetylglucosamine transferase | ||
| NM_002034 | FUT5 | 59±34 | nsc | 564±148 | 6.3 | Fucosyltransferase | ||
| NM_000229 | SIAT4A | 626±145 | nsc | 2343±99 | 3.2 | Beta galactosidase syalyltransferase | ||
| BC00355 | TGM2 | 7972±1561 | nsc | 7240+498 | 1.7 | Transglutaminase 2 | ||
| U10473 | B4GALT1 | 585±271 | nsc | 2483±188 | 3.2 | Sialyltransferase 4A | ||
| NM-003033 | B4GALT7 | 626±145 | nsc | 174±49 | 1.7 | Galactosyltransferase | ||
Isolation of primary placental cells, culture and immunofluorescence
Placental cells were isolated through sequential DNase digestion and percoll gradient centrifugation as previously described (20). Approximately 30 to 45 g of villous tissue was dissected within 10 min of delivery, blotted on sterile gauze and washed with saline to remove excess blood. The tissue was then digested three times at 37°C in a shaking water bath for 30 min each with 0.25 % trypsin and 300 U/ml DNAse I in Hanks’ balanced salt solution, pH 7.4. Cytotrophoblasts were separated from mononuclear cells by Percoll (Amersham Pharmacia Biotech, NJ, USA) gradient centrifugation. Cells were counted and seeded in Iscove medium supplemented with 10% FCS in 12 well plates at a density of 2.5 × 106 cells per well.
Cell culture and placental lipid analysis
After overnight incubation non adherent cells were washed twice with serum free standard DMEM. Cells were cultured with serum free DMEM medium containing 5.5 mM glucose and 0.15 mM BSA (control) or supplemented with 10 mM glucose or 400 nmol/L oleic acid complexed to BSA (Sigma, MO) for 48 hours. For RNA extraction, cells were lysed with 0.5 ml Trizol per well. For lipid staining, cells were washed with PBS then fixed in 4% paraformaldehyde for 30 min at room temperature. Cells were exposed for 30 min to BODIPY 493/503 (Molecular Probes, CA). Nuclei were counterstained with DAPI (Millipore, MA). Slides were mounted in permanent aqueous mounting medium (DAKO, CA) and viewed with a confocal laser scanning microscope (Zeiss, model LSM Meta 510) and nuclei were counted manually from 6 random fields /slide with 15 nuclei per field. Lipid droplets were counted and quantified from digital images using MetaMorph imaging software (Molecular Devices, PA). Each experiment was performed in duplicate.
Real time PCR
Expression of stearoyl CoA desaturase (SCD), 3-hydroxy-methylglutaryl-CoA reductase (HMGCR) and 3-hydroxy-methylglutaryl-CoA synthase 1 (HMGCS) was quantified by real time RT-PCR (Lightcycler, Roche Molecular Diagnosis, IN) in primary cultured trophoblast cells incubated with glucose and oleate. Results were normalized for beta-actin. SCD primers: forward: TGTGTCCCAGATGCTGTCAT, reverse; CCCACCCAATCACAGAAAAG (Genebank accession number NM_005063). HMGCR primers: forward: TGTTTCAGATTGCAAGGTAATATGT, reverse: CCTGGCACCCAAAAGTTAAA (Genebank accession number NM_000859). HMGCS primers: forward: TTAGCTTTCTCAGGGGGTCA, reverse: CCATGGTTTCCTGGAATGTT (Genebank accession number NM_002130).
Statistical analysis
Data are presented as mean ± SEM. Comparisons between groups were performed using one-way analysis of variance (ANOVA). Significance for statistical differences was calculated using a two-tailed Student’s t–test for unpaired data. Differences were considered significant when p <0.05.
Results
Clusters for metabolic genes in placenta of gestational and type 1 diabetes
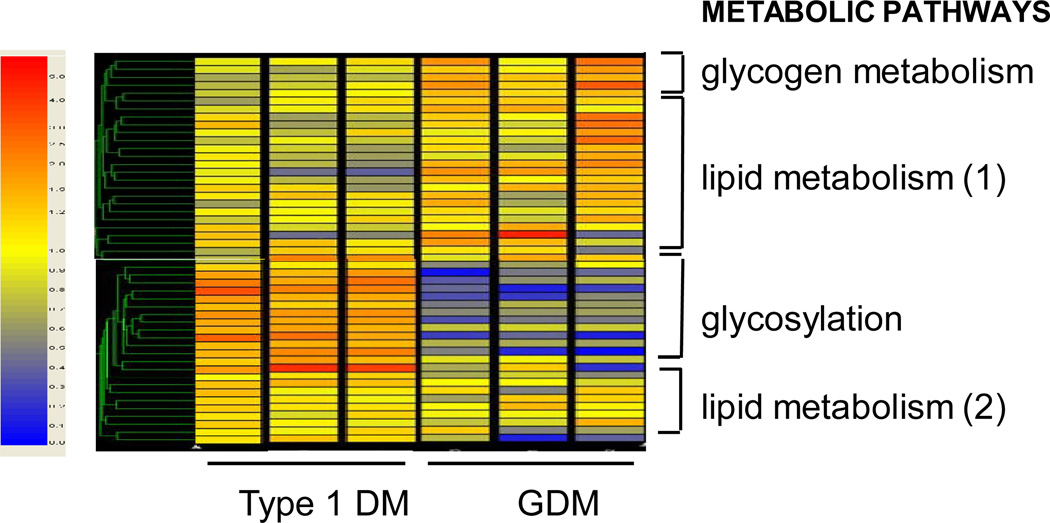
Hierarchical cluster analysis showed enhancement of lipid pathways in both GDM and T1DM (Figure 1). Among the 49 genes related to lipid and glucose energy pathways which were significantly modified, 45 were up-regulated and 4 down- regulated. Four distinct categories of genes were identified based on the functional property of their putative encoded proteins 1- lipid transport and activation, 2- lipid metabolism, 3- glycogen metabolism and 4- hexosamine pathways (Table 2). The vast majority of the activated genes (67 %) were implicated in lipid pathways and only 9 % were related to glucose energy metabolism with a clear dichotomy between T1DM and GDM. Genes at key steps of fatty acid uptake, transport and activation pathways were similarly up-regulated in pregnancy with GDM and T1DM (Table 2). However, pathways for triglyceride and cholesterol biosynthesis were selectively enhanced in GDM and obesity. There were marked differences in clusters of glucose-related genes with pathways for glycogen turnover primarily enhanced in GDM and hexosamine pathway and glycosylation reactions essentially recruited in T1DM (Figure 1, Table 2).
Figure 1. Placental metabolic genes differentially regulated in diabetic pregnancy.
Unsupervised hierarchical clustering shows expression data for the metabolic genes modified in pregnancy with GDM and type 1 diabetes. The prominent clusters shown on the right are based on the putative functional characteristics of the genes. The color tag and intensity represent the expression level from lowest (blue) to highest (red). Type 1 DM: Type 1 diabetes mellitus, GDM: gestational diabetes. Shown are data from 6 representative arrays run in duplicate.
Genes for lipid transport and metabolism
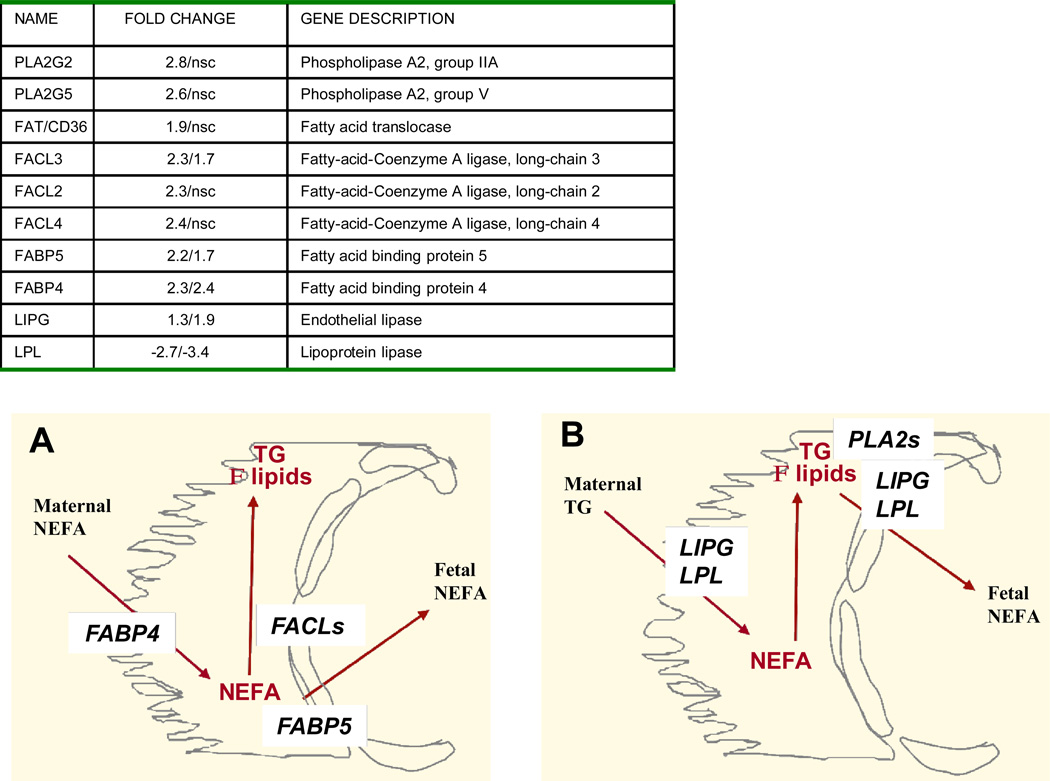
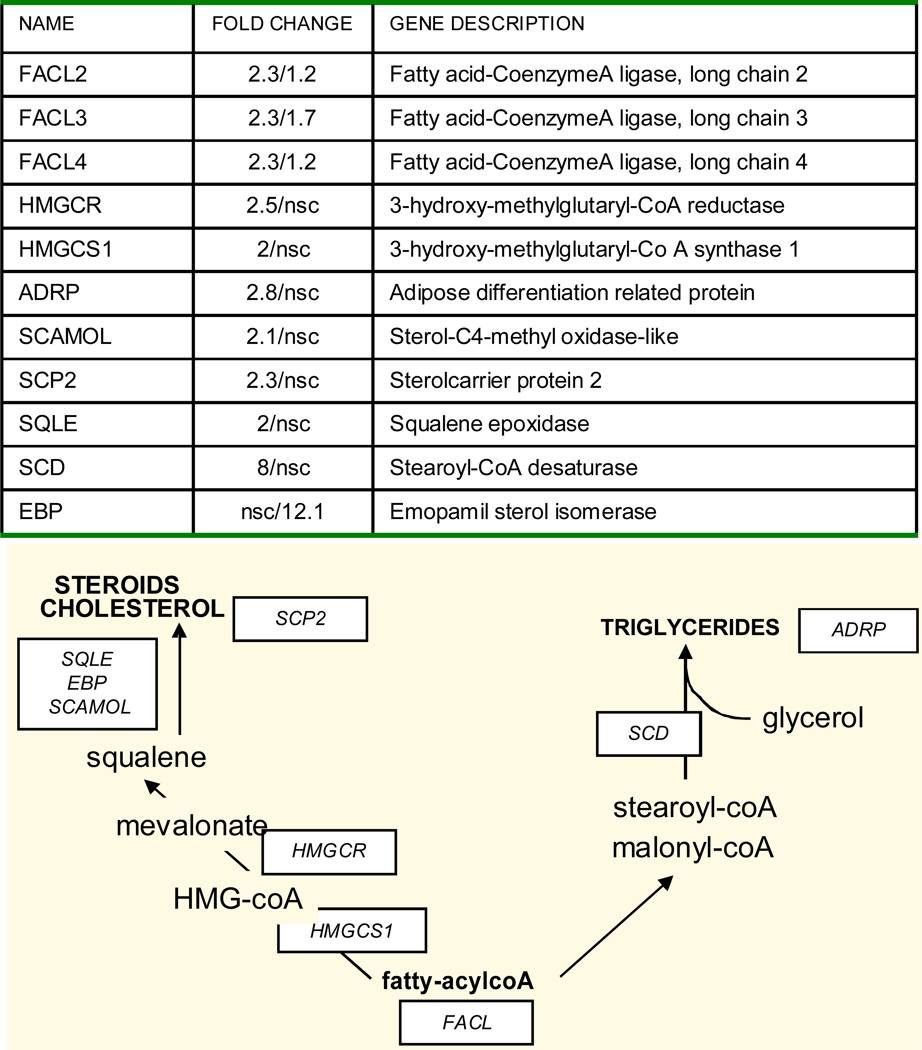
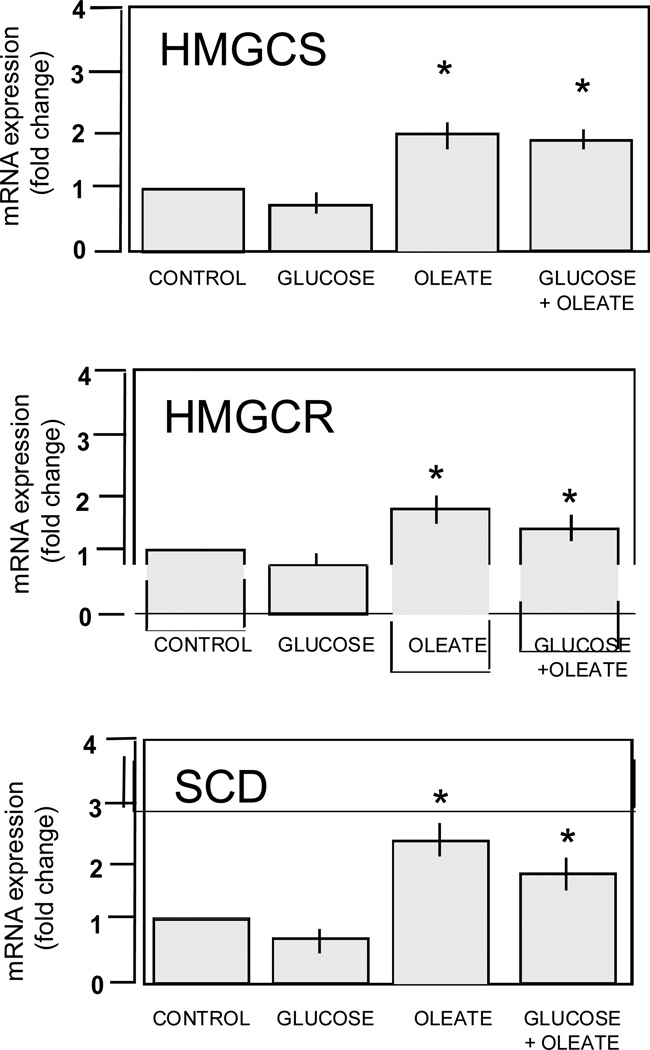
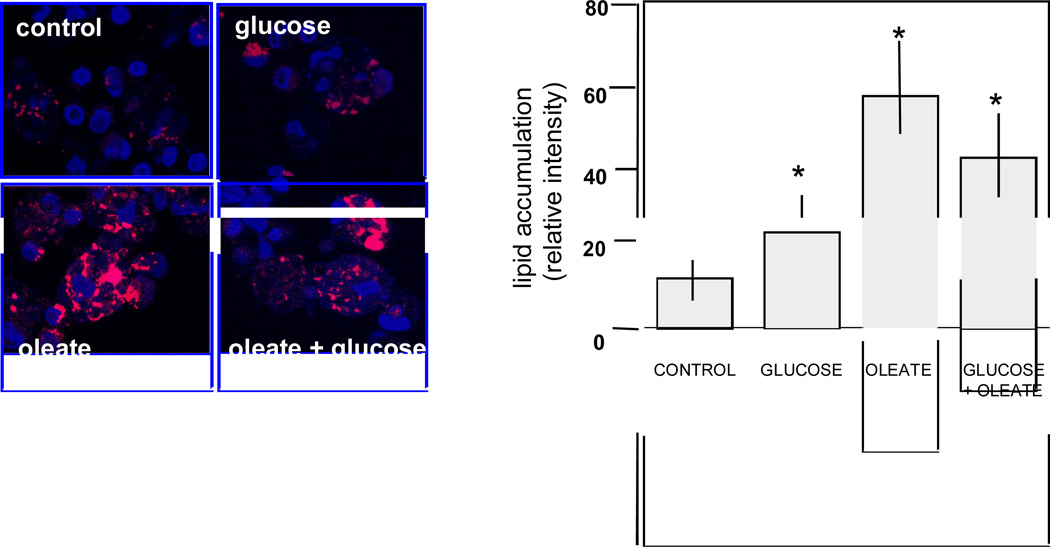
The pattern of genes for fatty transport and activation showed similarities in GDM and T1DM. FABP4 and FABP5, two isoforms of the fatty acid binding proteins family for facilitated transport of FFA were up-regulated 2-fold compared to control (Figure 2A). The three isoforms of the fatty acylcoA ligases (FACL2,3,4) which catalyze the conversion of fatty acids into fatty acyl-CoA esters to serve as precursors for the synthesis of triglycerides, cholesterol and membrane phospholipids were also activated. The endothelial lipase LIPG which hydrolyzes complex lipids into FFA before they can be taken up by the placental cells was also higher in diabetes (Figure 2B). A robust activation of lipid metabolism essentially observed in GDM was supported by the up-regulation of several genes in lipid biosynthetic pathways (Figure 3). The 8-fold increase in stearoylcoA desaturase (SCD) which introduce double bonds into growing fatty acid chains indicated a recruitment of cellular pathways leading to triglyceride synthesis. Activation of cholesterol synthesis and steroidogenesis was suggested by the coordinated up- regulation of 3-hydroxy-methylglutaryl-CoA reductase (HMGCR), 3-hydroxy-methylglutaryl-CoA synthase 1 (HMGCS), emopamil sterol isomerase (EBP), squalene epoxidase (SQLE) and sterol 4 methyl oxidase (SCAMOL). The link between the up-regulation of lipid pathways and lipid storage capacity of placental cells was assessed in vitro under conditions of high glucose and lipid availability. The expression of SCD, HMGCS and HMGCR was activated 1.5 to 2.7-fold (p<0.001) by culture of freshly isolated trophoblast cells in the presence of 400 nMol/L oleate but not with 10 mMol/L glucose (Figure 4). The size of the lipid droplets was increased 4-fold (p<0.002) upon addition of oleate and 1.9-fold (p<0.05) with 10 mMol/L glucose in the culture medium (Figure 5). The stimulatory effect of oleate was not further enhanced by glucose.
Figure 2. Changes in pathways for placental lipid transport in diabetes.
Upper panel: fold changes of regulated genes related to lipid transport in GDM (first number) and type 1 diabetes (second number). Lower panel: schematic diagram of the molecules related to placental lipid transport. The placental barrier is formed by the syncytiotrophoblast cells (yellow) facing the maternal side and the endothelial cells (red) lining the fetal vascular system. Panel A: the fatty acid binding proteins FABPs located on both sides of the syncytial cells bind fatty acids for import from the maternal circulation and export from the placenta to the fetal circulation. The fatty acylcoA ligases FACL process the first step of fatty acid elongation towards esterification of FFA into triglycerides. B: The lipases LIPG and LPL breakdown triglycerides and complex lipids before uptake by trophoblast cells or export to the fetal circulation. The phospholipases PLA2s contribute to mobilization of phospholipids. TG: triglycerides, FFA: free (non esterified) fatty acids.
Figure 3. Changes in placental lipid biosynthetic pathways in diabetes.
Fold changes and schematic representation of selected genes in pathways regulating the synthesis of triglycerides, cholesterol and phospholipids in GDM (first number) and type 1 diabetes (second number).
Figure 4. In vitro regulation of genes for placental lipid synthesis in primary placental cells.
mRNA expression of stearoylcoA desaturase (SCD), 3-hydroxy-methylglutaryl-CoA reductase (HMGCoAR) and 3-hydroxy-methylglutaryl-CoA synthase (HMGCoAR) limiting steps for triglyceride and cholesterol synthesis. Freshly isolated trophoblast cells were cultured for 48 hrs with no addition (control) or addition of glucose (10 mMol/L), oleate (400 nMol/L) or the combination of both. Results are means ± SE of 4 independent experiments with duplicate culture wells. * p< 0.005.
Figure 5. In vitro regulation of lipid accumulation in primary placental cells.
Left panel: visualization of lipid droplets in primary term placental cells. Freshly isolated trophoblast cells were cultured for 48 hrs with no addition (control) or addition of glucose (10 mMol/L), oleate (400 nMol/L) or the combination of both. Right panel: quantification of lipid accumulation by scanning densitometry. Results are means ± SE of 4–6 independent experiments with duplicate culture wells. * p< 0.0001.
Genes for Glucose metabolism
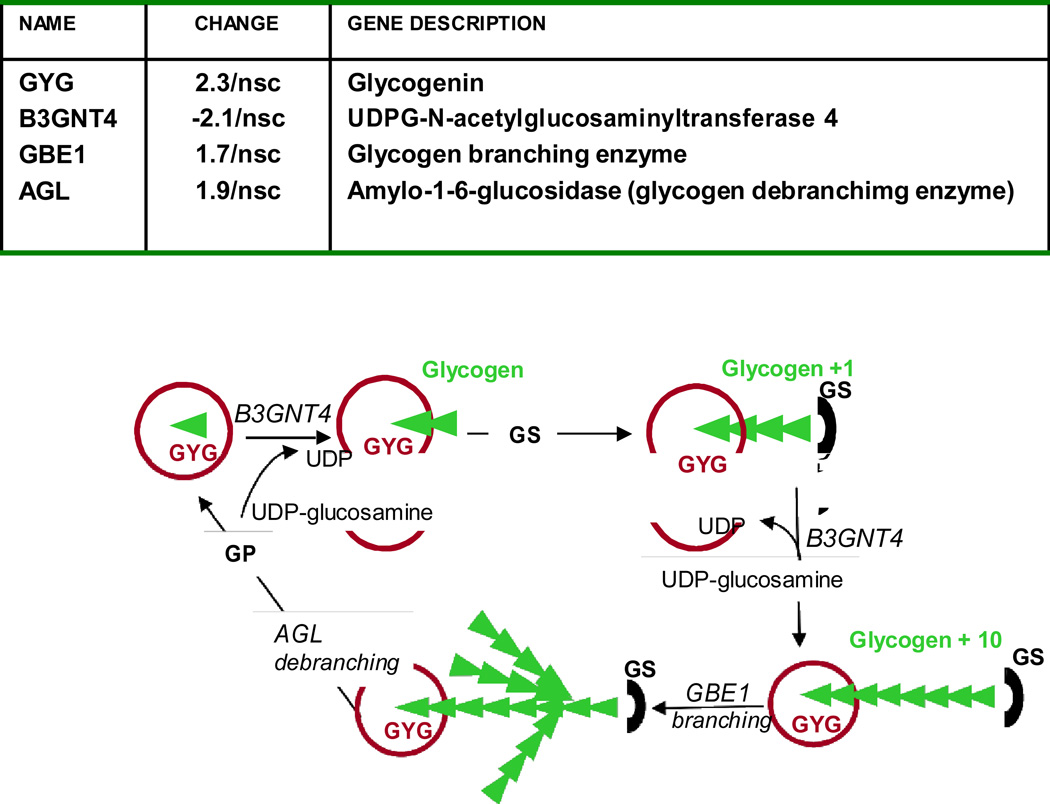
In contrast to the prominent activation of lipid pathways, genes related to glucose metabolism represented a smaller proportion (9 vs 67 %) of the recruited genes. The activation of glycogenin, the protein priming glycogen synthesis from UDPG-glucose, the glycogen branching enzyme GBE1 and amylo1-6 glucosidase AGL indicated active glycogen turnover in GDM only (Table 2, Figure 6). Ten genes (22% of diabetes modified genes) involved in glycosylation, acylation processes and the hexosamine synthetic pathway were exclusively recruited in placenta of T1DM (Table 2). The 5-fold activation of the fucosyltransferase FUT5 gene and the hyaluronoglucosaminidase HYAI4 gene were indicative of modifications in the carbohydrate content of glycoproteins.
Figure 6. Changes in placental pathways for glycogen turnover in pregnancy with gestational diabetes.
Glycogenin initiates glycogen synthesis by attaching C-1 of a UDP-glucose to a tyrosine residue on the enzyme. The attached glucose serves as the primer required by glycogen synthase to attach additional glucose molecules. Expression of glycogen synthase (GS) or glycogen phosphorylase (GP) was not modified in diabetes.
Comment
The most significant findings of this study were that diabetes induced an over- representation of genes implicated in placental lipid pathways compared to genes related to glucose metabolism. Furthermore there was a clear dichotomy between GDM and T1DM as regards to lipid and glucose metabolism. Genes for transport and activation of fatty acids and LDL were up-regulated to similar extent in GDM and T1DM reflecting the increased availability of lipid substrates at the maternal- placental interface. Pathways for intracellular lipid biosynthesis were selectively up- regulated in pregnancy with GDM and obesity. The robust activation of stearoyl-CoA desaturase (SCD) indicates an enhancement in the anabolic pathways leading to accumulation of triglycerides. High desaturase activities have also been associated with insulin resistance and dyslipidemia in obese rodents (22). Fatty acid desaturases are rate-limiting enzymes for triglyceride synthesis which are highly sensitive to dietary changes (23, 24). The ability of oleate to stimulate the expression of SCD and lipid droplet formation in vitro delineates fatty acids as lipogenic substrates as previously suggested (25). The enhanced expression of ADRP and SCP-2, which facilitate storage of FFA and cholesterol into membrane and neutral lipids further highlights that coordinated mechanisms are set to favor lipid accumulation in a diabetic environment (26). Our data support the original hypotheses by Szabo (27) and Shafrir (15, 21) that accumulation of placental triglycerides represent a regulatory step towards excess fetal adiposity. Additionally, our results emphasize that glucose plays only a minor role in triglyceride accumulation through de novo lipid synthesis in agreement with the absence of acetylcoA carboxylase and fatty acid synthase in placental cells (28).
The second lipid pathway that we found up-regulated in GDM leads to cholesterol synthesis through activation of HMG-coA reductase and HMGcoA synthetase. This is further supported by the 2 and 12–fold up-regulation of squalene epoxidase and emopamil binding protein, which catalyze distal steps of cholesterol biosynthesis (29). As primary constituents of cell membranes, phospholipids and cholesterol regulate membrane lipid composition and consequently changes in membrane fluidity. Phosphatidylcholine cytidyltransferase PCYT1B and sterol carrier protein SCP-2 which enhance the ratio of phospholipids/cholesterol are classically associated with fluidization and altered functions of membrane proteins (30). The phospholipid composition of the monolayer profoundly affects droplet morphology and lipid utilization (31). Hence, the molecular changes observed may result in higher membrane fluidity as well as ubiquitous lipid storage, two potential interim steps towards increased transplacental transfer of lipids.
Our overall concept is that maternal environmental factors contribute to the regulation of genes for lipid transport and metabolism in placenta of GDM women. Because maternal FFA and triglyceride plasma concentrations were not modified in lean type 1 diabetic or obese GDM women this raises the question of whether changes in maternal lipid fluxes contribute to the observed placental changes. Recent findings in a population of well-controlled GDM pregnancies in overweight women showing that circulating maternal lipids, but not glucose, correlate with fetal growth provides strong support to this view (32). Preliminary studies from our group indicate that obesity in pregnancy, without maternal hyperlipidemia, enhances expression of similar placental lipid gene clusters (shm unpublished). This raises the possibility that the environment of obesity may be critical for the regulation of placental lipid pathways. Whether an excess placental lipids are mobilized and exported into the fetal circulation is critical information for understanding the regulation of fetal adiposity and is not yet known with certainty. The positive materno-fetal FFA concentration gradient in humans reflects FFA taken up by all fetal tissues and not only by adipose tissue. Hence, fetal FFA concentrations cannot be used as markers of fat deposition in utero. The up-regulation of the lipid carriers FABP4 and FABP5 suggests an enhancement of placental fatty acid fluxes. The location of FABPs on both sides of the bipolar syncytiotrophoblast cells may favor transport of free fatty acids in both directions i.e. from mother to fetus and vice versa (33). Increased expression of the endothelial lipase LIPG2 provides additional evidence for mobilization of lipids for export to the fetal circulation (34, 35). Taken together our findings point to the recruitment of three coordinated pathways which control the amount of fatty acids available for the fetus in response to gestational diabetes i.e their rate of uptake from maternal circulation, the esterification/storage capacity and the mobilizing activity for export.
In addition to lipid genes, our cluster analysis has highlighted that changes in pathways of glycogen turnover were enhanced in GDM, whereas pathways for glycosylation and acylation were exclusively recruited in T1DM. The up-regulation of glycogenin, the glycogen branching enzyme GBE1 and of amylo1-6 glucosidase AGL are molecular markers for the acceleration of glucose fluxes into glycogen, a well documented feature of diabetic pregnancy in humans and rodents (36–40). The lack of glucose-6-phosphatase and the co-localization of glycogen and glycogenin in endothelial cells lining the fetal circulation were key observations for the concept that it is the excess glucose from the fetus rather than from the mother which may being stored as glycogen (41, 42). The increased expression of 10 genes for glycosylation and acylation reactions in T1DM suggested that hexosamine pathways may be recruited when tissue glycogen storage capacities are exceeded (43). GLUT10, a facilitative glucose transporter candidate gene for Type 2 diabetes (44) was the only gene related to glucose transfer to be up-regulated in our data set.
Taken together, these data point to a pronounced modification of fetal-placental lipid pathways and selective activation of transplacental lipid fluxes in pregnancy with gestational diabetes and obesity. Our findings have potentially important clinical implication in highlighting the importance of alterations of lipid homeostasis besides glucose changes as potential contributors to fetal macrosomia.
Acknowledgments
The authors wish to thank Dr P. Leahy, genetic core at Case Western Reserve University for valuable help and input with microarray analysis. The excellent technical assistance of Judi Minium is acknowledged.
Grant support: NIH RO-1 HD22965, DAGC grant-in-aid 484-04 and ADA1-04-tlg-01.
References
- 1.Catalano PM, Thomas A, Huston-Presley L, Amini SB. Increased fetal adiposity: a very sensitive marker of abnormal in utero development. Am J Obstet Gynecol. 2003;189:1698–1704. doi: 10.1016/s0002-9378(03)00828-7. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen J. Glucose content of the amniotic fluid in diabetic pregnancies; correlations with the maternal blood sugar. Acta Endocrinol (Copenh) 1954;15:342–354. doi: 10.1530/acta.0.0150342. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz R, Gruppuso PA, Petzold K, Brambilla D, Hiilesmaa V, Teramo KA. Hyperinsulinemia and macrosomia in the fetus of the diabetic mother. Diabetes Care. 1994;17:640–648. doi: 10.2337/diacare.17.7.640. [DOI] [PubMed] [Google Scholar]
- 4.Lepercq J, Taupin P, Dubois-Laforgue D, Duranteau L, Lahlou N, Boitard C, Landais P, Hauguel-De Mouzon S, Timsit J. Heterogeneity of fetal growth in type 1 diabetic pregnancy. Diabetes Metab. 2001;27:339–344. [PubMed] [Google Scholar]
- 5.Evers IM, de Valk HW, Mol BW, ter Braak EW, Visser GH. Macrosomia despite good glycaemic control in Type I diabetic pregnancy; results of a nationwide study in The Netherlands. Diabetologia. 2002;45:1484–1489. doi: 10.1007/s00125-002-0958-7. [DOI] [PubMed] [Google Scholar]
- 6.Sewell MF, Huston-Presley L, Super DM, Catalano P. Increased neonatal fat mass, not lean body mass, is associated with maternal obesity. Am J Obstet Gynecol. 2006;195:1100–1103. doi: 10.1016/j.ajog.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Tallarigo L, Giampietro O, Penno G, Miccoli R, Gregori G, Navalesi R. Relation of glucose tolerance to complications of pregnancy in nondiabetic women. N Engl J Med. 1986;315:989–992. doi: 10.1056/NEJM198610163151603. [DOI] [PubMed] [Google Scholar]
- 8.HAPO Study Cooperative Research Group. Metzer BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, Coustan DR, Hadden DR, McCance DR, Hod M, McIntyre HD, Oats JJ, Persson B, Rogers MS, Sacks DA. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2008;358:1991–2002. doi: 10.1056/NEJMoa0707943. [DOI] [PubMed] [Google Scholar]
- 9.Metzger BE, Phelps RL, Freinkel N, Navickas IA. Effects of gestational diabetes on diurnal profiles of plasma glucose, lipids, and individual amino acids. Diabetes Care. 1980;3:402–409. doi: 10.2337/diacare.3.3.402. [DOI] [PubMed] [Google Scholar]
- 10.Montelongo A, Lasunción MA, Pallardo LF, Herrera E. Longitudinal study of plasma lipoproteins and hormones during pregnancy in normal and diabetic women. Diabetes. 1992;41:1651–1659. doi: 10.2337/diab.41.12.1651. [DOI] [PubMed] [Google Scholar]
- 11.Freinkel N. Banting Lecture. Of pregnancy and progeny. Diabetes. 1980;29:1023–1035. doi: 10.2337/diab.29.12.1023. [DOI] [PubMed] [Google Scholar]
- 12.Hauguel S, Challier JC, Cedard L, Olive G. Metabolism of the human placenta perfused in vitro: glucose transfer and utilization, O2 consumption, lactate and ammonia production. Pediatr Res. 1983;17:729–732. doi: 10.1203/00006450-198309000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Haggarty P, Page K, Abramovich DR, Ashton J, Brown D. Long-chain polyunsaturated fatty acid transport across the perfused human placenta. Placenta. 1997;18:635–642. doi: 10.1016/s0143-4004(97)90004-7. [DOI] [PubMed] [Google Scholar]
- 14.Lindegaard ML, Olivecrona G, Christoffersen C, Kratky D, Hannibal J, Petersen BL, Zechner R, Damm P, Nielsen LB. Endothelial and lipoprotein lipases in human and mouse placenta. J Lipid Res. 2005;46:2339–2346. doi: 10.1194/jlr.M500277-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Shafrir E, Khassis S. Maternal-fetal transport vs new fat syntesis in the pregnant diabetic rat. Diabetologia. 1982;22:11–117. doi: 10.1007/BF00254839. [DOI] [PubMed] [Google Scholar]
- 16.Bitsanis D, Ghebremeskel K, Moodley T, Crawford MA. Gestational diabetes mellitus enhances arachidonic and docosahexaenoic acids in placental phospholipids. Lipids. 2006;41:341–346. doi: 10.1007/s11745-006-5104-8. [DOI] [PubMed] [Google Scholar]
- 17.Buchanan TA, Xiang A, Kjos SL, Watanabe R. What is gestational diabetes? Diabetes Care. 2007;30(Suppl 2):S105–S111. doi: 10.2337/dc07-s201. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter M, Coustan D. Criteria for screening tests for gestational diabetes. Am J. Obstet Gynecol. 1982;144:768–773. doi: 10.1016/0002-9378(82)90349-0. [DOI] [PubMed] [Google Scholar]
- 19.Chirgwin JM, Przybyla AE, McDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 20.Varastehpour A, Radaelli T, Minium J, Ortega H, Herrera E, Catalano P, Hauguel-de Mouzon S. Activation of phospholipase A2 is associated with generation of placental lipid signals and fetal obesity. J Clin Endocrinol Metab. 2006;91:248–255. doi: 10.1210/jc.2005-0873. [DOI] [PubMed] [Google Scholar]
- 21.Diamant YZ, Metzger BE, Freinkel N, Shafrir E. Placental lipid and glycogen content in human and experimental diabetes mellitus. Am J Obstet Gynecol. 1982;144:5–11. doi: 10.1016/0002-9378(82)90385-4. [DOI] [PubMed] [Google Scholar]
- 22.Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, Song Y, Cohen P, Friedman JM, Attie AD. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci USA. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampath H, Miyazaki M, Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase-1 mediates the pro-lipogenic effects of dietary saturated fat. J Biol Chem. 2007;282:2483–2493. doi: 10.1074/jbc.M610158200. [DOI] [PubMed] [Google Scholar]
- 24.Sjögren P, Sierra-Johnson J, Gertow K, Rosell M, Vessby B, de Faire U, Hamsten A, Hellenius ML, Fisher RM. Parallel activation of de novo lipogenesis and stearoyl-CoA desaturase activity after 3 days of high-carbohydrate feeding. Am J Clin Nutr. 2008;87:817–823. doi: 10.1093/ajcn/87.4.817. [DOI] [PubMed] [Google Scholar]
- 25.Elchalal U, Schaiff WT, Smith SD, Rimon E, Bildirici I, Nelson DM, Sadovsky Y. Insulin and fatty acids regulate the expression of the fat droplet-associated protein adipophilin in primary human trophoblasts. Am J Obstet Gynecol. 2005;193:1716–1723. doi: 10.1016/j.ajog.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Tobin KA, Harsem NK, Dalen KT, Staff AC, Nebb HI, Duttaroy AK. Regulation of ADRP expression by long-chain polyunsaturated fatty acids in BeWo cells, a human placental choriocarcinoma cell line. J Lipid Res. 2006;47:815–823. doi: 10.1194/jlr.M500527-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Szabo AJ, Szabo O. Placental free-fatty-acid transfer and fetal adipose-tissue development: an explanation of fetal adiposity in infants of diabetic mothers. Lancet. 1974;2:498–499. doi: 10.1016/s0140-6736(74)92020-0. [DOI] [PubMed] [Google Scholar]
- 28.Coltart TM, Bateman C. Carbohydrate-induced lipogenesis in the human placenta of normal and diabetic pregnancies. Br J Obstet Gynaecol. 1975;82:471–475. doi: 10.1111/j.1471-0528.1975.tb00672.x. [DOI] [PubMed] [Google Scholar]
- 29.Marseille-Tremblay C, Ethier-Chiasson M, Forest JC, Giguère Y, Masse A, Mounier C, Lafond J. Impact of maternal circulating cholesterol and gestational diabetes mellitus on lipid metabolism in human term placenta. Mol Reprod Dev. 2008;75:1054–1062. doi: 10.1002/mrd.20842. [DOI] [PubMed] [Google Scholar]
- 30.Atshaves BP, Storey SM, McIntosh AL, Petrescu AD, Lyuksyutova OI, Greenberg AS, Schroeder F. Sterol carrier protein-2 expression modulates protein and lipid composition of lipid droplets. J Biol Chem. 2001;276:25324–25335. doi: 10.1074/jbc.M100560200. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer-Graf UM, Graf K, Kulbacka I, Kjos SL, Dudenhausen J, Vetter K, Herrera E. Maternal lipids as strong determinants of fetal environment and growth in pregnancies with gestational diabetes mellitus. Diabetes Care. 2008;31:1858–1863. doi: 10.2337/dc08-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell FM, Bush PG, Veerkamp JH, Dutta-Roy AK. Detection and cellular localization of plasma membrane-associated and cytoplasmic fatty acid-binding proteins in human placenta. Placenta. 1998;19:409–415. doi: 10.1016/s0143-4004(98)90081-9. [DOI] [PubMed] [Google Scholar]
- 34.Masouyé I, Hagens G, Van Kuppevelt TH, Madsen P, Saurat JH, Veerkamp JH, Siegenthaler G. Endothelial cells of the human microvasculature express epidermal fatty acid-binding protein. Circ Res. 1997;81:297–303. doi: 10.1161/01.res.81.3.297. [DOI] [PubMed] [Google Scholar]
- 35.Lindegaard ML, Damm P, Mathiesen ER, Nielsen LB. Placental triglyceride accumulation in maternal type 1 diabetes is associated with increased lipase gene expression. J Lipid Res. 2006;47:2581–2588. doi: 10.1194/jlr.M600236-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Villee CA. The metabolism of human placenta in vitro. J Biol Chem. 1953;205:113–123. [PubMed] [Google Scholar]
- 37.Gabbe SG, Demers LM, Greep RO, Villee CA. Placental glycogen metabolism in diabetes mellitus. Diabetes. 1972;21:1185–1191. doi: 10.2337/diab.21.12.1185. [DOI] [PubMed] [Google Scholar]
- 38.Desoye G, Hofmann HH, Weiss PA. Insulin binding to trophoblast plasma membranes and placental glycogen content in well-controlled gestational diabetic women treated with diet or insulin, in well-controlled overt diabetic patients and in healthy control subjects. Diabetologia. 1992;35:45–55. doi: 10.1007/BF00400851. [DOI] [PubMed] [Google Scholar]
- 39.Jones CJ, Desoye G. Glycogen distribution in the capillaries of the placental villus in normal, overt and gestational diabetic pregnancy. Placenta. 1993;14:505–517. doi: 10.1016/s0143-4004(05)80204-8. [DOI] [PubMed] [Google Scholar]
- 40.Barash V, Riskin A, Shafrir E, Waddell ID, Burchell A. Kinetic and immunologic evidence for the absence of glucose-6-phosphatase in early human chorionic villi and term placenta. Biochim Biophys Acta. 1991;23(1073(1)):161–197. doi: 10.1016/0304-4165(91)90197-o. [DOI] [PubMed] [Google Scholar]
- 41.Barash V, Shafrir E. Mobilization of placental glycogen in diabetic rats. Placenta. 1990;11:515–521. doi: 10.1016/s0143-4004(05)80197-3. [DOI] [PubMed] [Google Scholar]
- 42.Desoye G, Shafrir E. Placental metabolism and its regulation in health and diabetes. Mol Aspects Med. 1994;15:505–682. doi: 10.1016/0098-2997(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 43.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 44.Dawson PA, Mychaleckyj JC, Fossey SC, Mihic SJ, Craddock AL, Bowden DW. Sequence and functional analysis of GLUT10: a glucose transporter in the Type 2 diabetes-linked region of chromosome 20q12-13.1. Mol Genet Metab. 2001;74:186–199. doi: 10.1006/mgme.2001.3212. [DOI] [PubMed] [Google Scholar]