Summary
Completion of DNA replication after replication stress depends on PCNA, which undergoes mono-ubiquitination to stimulate direct bypass of DNA lesions by specialized DNA polymerases or is poly-ubiquitinated to promote recombination dependent DNA synthesis across DNA lesions by template switching mechanisms. Here we report that the ZRANB3 translocase, a SNF2 family member related to the SIOD disorder SMARCAL1 protein, is recruited by poly-ubiquitinated PCNA to promote fork restart following replication arrest. ZRANB3 depletion in mammalian cells results in an increased frequency of sister chromatid exchange and DNA damage sensitivity after treatment with agents that cause replication stress. Using in vitro biochemical assays, we show that recombinant ZRANB3 remodels DNA structures mimicking stalled replication forks and disassembles recombination intermediates. We therefore propose that ZRANB3 maintains genomic stability at stalled or collapsed replication forks by facilitating fork restart and limiting inappropriate recombination that could occur during template switching events.
Introduction
Genomic integrity is constantly challenged by DNA damage either spontaneously generated or induced by environmental sources such as UV, ionizing radiation (IR) and chemical agents (Ciccia and Elledge, 2010). During DNA replication, DNA lesions can lead to the blockage or collapse of replication forks, thus resulting in the accumulation of extensive single-strand DNA (ssDNA) regions associated with the RPA complex (Byun et al., 2005). RPA can act as a loading platform for the recruitment of factors involved in activation of the DNA damage response, stabilization of stalled/collapsed replication forks and subsequent restart of DNA synthesis (Ciccia and Elledge, 2010). To restart stalled/collapsed forks, RPA directly recruits the SMARCAL1 translocase and interacts at forks with the RecQ helicases BLM, WRN and the Fanconi Anemia FANCM/FAAP24 complex (Bachrati and Hickson, 2008; Bansbach et al., 2009; Ciccia et al., 2009; Huang et al., 2010; Postow et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009).
The DNA polymerase sliding clamp PCNA can also function as a loading platform to recruit DDR factors that allow completion of DNA replication after DNA damage and promote post-replication repair (Moldovan et al., 2007). Following induction of replication stress, PCNA arrested at DNA lesions is mono-ubiquitinated by the RAD6/RAD18 ubiquitin ligase complex (Bergink and Jentsch, 2009). Mono-ubiquitinated PCNA can recruit translesion (TLS) polymerases, which are able to synthesize across DNA lesions in a potentially error prone manner (Sale et al., 2012). An error free pathway exists and requires Lys63 (K63)-linked poly-ubiquitination of PCNA, which is induced by UBC13/MMS2 in complex with Rad5 in yeast or HLTF and SHPRH in mammalian cells (Unk et al., 2010). This pathway, also known as template switching, employs recombination mechanisms to synthesize across the lesion using as a template the undamaged newly synthesized strand of the sister chromatid (Branzei, 2011).
Current models propose fork reversal or strand invasion as possible mechanisms employed by template switching to allow bypass of DNA lesions at replication forks. Fork reversal has been suggested to promote the annealing of the arrested DNA strand with the undamaged strand of the sister chromatid, thus allowing the blocked strand to restart DNA synthesis, whereas strand invasion could mediate the bypass of DNA lesions through the formation of sister chromatid junctions (Atkinson and McGlynn, 2009; Branzei, 2011). Sister chromatid junctions resemble double Holliday junctions, which during mitosis are primarily dissolved by the BLM/TOPOIIIα to prevent the formation of crossover events between sister chromatids, also known as sister chromatid exchanges (SCEs) (Wu and Hickson, 2003). Elevated crossover events are potentially deleterious since they can lead to chromosomal alterations and cause loss of heterozygosity (LOH), which can uncover recessive tumor suppressor mutations and predispose to cancer formation, as in Bloom syndrome patients carrying BLM mutations (Bachrati and Hickson, 2008).
The observation that deletions of genes involved in post-replication repair, such as RAD18 and the ATPase WRNIP1, increase SCE frequencies suggests that template switching events may limit inappropriate recombination events at replication forks (Hayashi et al., 2008; Szuts et al., 2006; Tateishi et al., 2003). In yeast Rad18, Rad5 and poly-ubiquitinated PCNA can regulate template switching in conjunction with SUMOylated PCNA, which recruits the anti-recombinase helicase Srs2 to suppress hyper-recombination at stalled replication forks (Branzei et al., 2008; Ulrich and Walden, 2010). In higher eukaryotes, the protein PARI has recently been shown to inhibit recombination following its association with SUMOylated PCNA in a manner similar to Srs2 (Moldovan et al., 2012). However, the role of PCNA poly-ubiquitination in regulating template switching events in higher eukaryotes to promote completion of DNA replication and prevent inappropriate recombination after replication stress is currently unknown. Moreover, no proteins have yet been shown to associate with poly-ubiquitinated PCNA in mammalian cells.
In this study we report that poly-ubiquitinated PCNA recruits the ZRANB3 translocase, also known as AH2, to sites of replication stress to promote genomic stability. ZRANB3 depletion leads to increased formation of SCEs and enhanced sensitivity to DNA damaging agents that cause replication stress. Moreover, we show that ZRANB3 promotes fork restart following replication arrest by associating with poly-ubiquitinated PCNA. In agreement with these observations, ZRANB3 catalyzes the regression of DNA substrates that mimic stalled replication forks and disassembles D-loop recombination intermediates in vitro. Altogether, these data allow us to propose that ZRANB3 cooperates with poly-ubiquitinated PCNA to maintain genomic stability and prevent inappropriate recombination that could take place during template switching events at sites of replication stress.
Results
ZRANB3 is recruited to sites of DNA damage in a PCNA-dependent manner
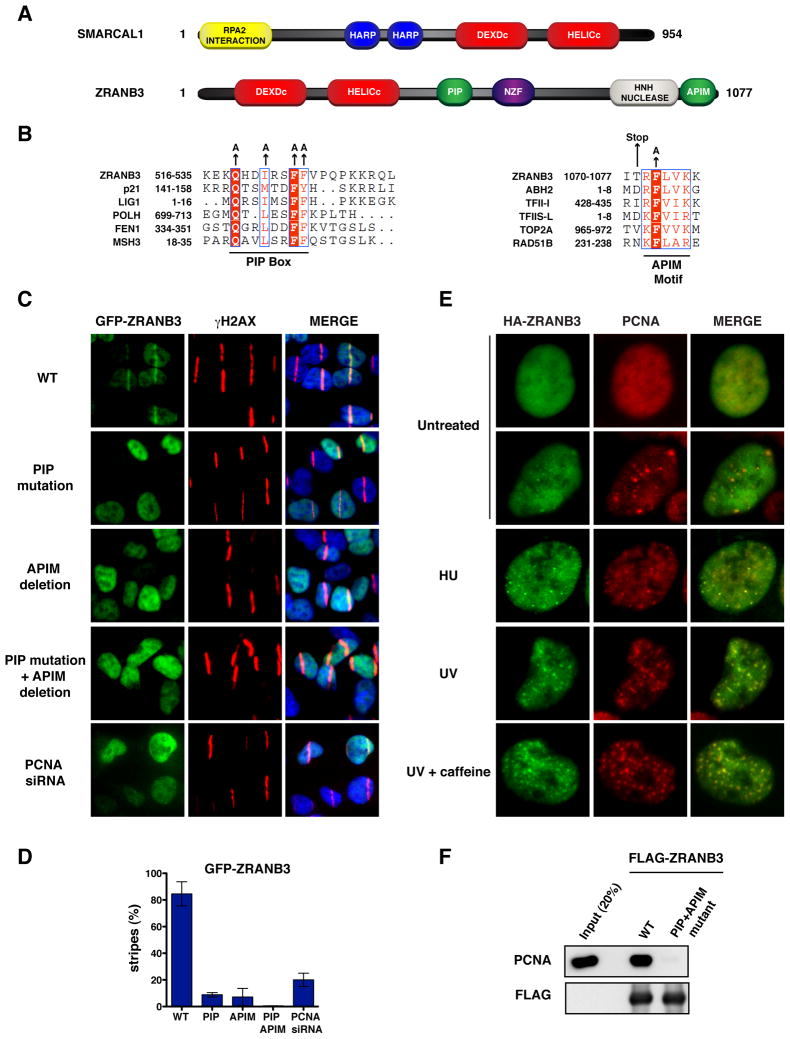
ZRANB3 is a member of the SNF2 family of proteins related to the SIOD disorder protein SMARCAL1 (Flaus et al., 2006; Yusufzai and Kadonaga, 2010). The similarity between ZRANB3 and SMARCAL1 is confined to their helicase domains, which share 44% identity (Figure 1A and Figure S1). Distinct from SMARCAL1, ZRANB3 possesses an NPL4 zinc finger (NZF) and an HNH nuclease motif (Figure 1A) (Flaus et al., 2006). The function of the HNH motif of ZRANB3 is currently unknown and no nuclease activity has been reported for ZRANB3 (Yusufzai and Kadonaga, 2010).
Figure 1. PCNA-dependent recruitment of ZRANB3 to DNA damage sites.
(A) Schematic representation of the protein domains of SMARCAL1 and ZRANB3. The helicase domains are indicated in red.
(B) Sequence alignments of the PIP box (left) and APIM (right) motifs of ZRANB3 with known PIP box and APIM motifs of other PCNA interacting proteins. The amino acids of the PIP box and APIM motifs that have been mutated are indicated by arrows.
(C) Localization of WT or mutant GFP-ZRANB3 to DNA damage sites generated by UV-laser microirradiation. U2OS cells expressing WT GFP-ZRANB3 are shown with or without PCNA siRNA treatment prior to UV microirradiation.
(D) Graphical representation of the percentage of U2OS cells that display co-localization of GFP-ZRANB3 with γH2AX at laser-generated stripes. The data represent the average and standard deviation of three independent experiments performed on cells expressing the GFP-constructs shown in (C).
(E) Co-localization of ZRANB3 with PCNA. U2OS cells expressing HA-ZRANB3 were left untreated or subjected to hydroxyurea (2 mM) or UV radiation (25 J/m2) treatment. Cells subjected to UV radiation were also treated with caffeine (2 mM). Cells were stained with anti-HA (green) and anti-PCNA (red) antibodies.
(F) Immunoprecipitation of FLAG-ZRANB3 with PCNA. FLAG-ZRANB3 either WT or mutated in the PIP and APIM motifs was purified from insect cells with anti-FLAG beads and then incubated with recombinant PCNA. Immunoprecipitated complexes were detected by western blotting.
To test whether ZRANB3 could be recruited to DNA damage sites like SMARCAL1, U2OS cells expressing GFP-tagged ZRANB3 were microirradiated with a UV-A laser following treatment with BrdU, which undergoes UV-mediated photolysis and facilitates the formation of DNA breaks at sites of laser irradiation. As shown in Figure S2A, GFP-ZRANB3 is recruited to laser-generated stripes, where it co-localizes with the DNA damage marker γH2AX. GFP-ZRANB3 localization to laser-generated stripes peaks between 5 and 15 minutes after microirradiation, diminishes by 30 min and disappears by 1 h post irradiation. In contrast, the association of GFP-SMARCAL1 to laser-generated stripes peaks 30 minutes following microirradiation, but is still prominent 1 h post irradiation (Figure S2B). These observations suggest that ZRANB3 and SMARCAL1 are recruited to DNA damage sites by distinct mechanisms.
SMARCAL1 recruitment to DNA damage is dependent on an RPA2 interaction motif located in the first 31 amino acids of SMARCAL1 (Bansbach et al., 2009; Ciccia et al., 2009; Postow et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009). ZRANB3 does not contain RPA interaction motifs and does not associate with RPA (Yusufzai and Kadonaga, 2010). However, based on similarity searches, we discovered that ZRANB3 possesses two putative PCNA interaction motifs, the PCNA-interacting protein (PIP) box (amino acids 519-526) and the AlkB homolog 2 PCNA-interaction motif (APIM) (amino acids 1070-1077), which have been shown to mediate the recruitment of repair factors to DNA damage sites in a PCNA dependent manner (Figure 1B) (Ciccia and Elledge, 2010). Whereas the amino acid sequence of the PIP box is well characterized, the APIM motif is determined by a shorter and more variable consensus sequence (Gilljam et al., 2009). To determine if the putative APIM motif of ZRANB3 could be recruited to sites of DNA damage, we generated U2OS cells expressing a C-terminal fragment of GFP-ZRANB3 containing the APIM motif (GFP-APIM, amino acids 1049-1077) fused to an NLS sequence. Following UV microirradiation, we observed that GFP-APIM localized to laser-generated stripes, while a GFP-APIM mutant harboring a single mutation in a highly conserved phenylalanine residue (F1073) of the APIM motif important for PCNA interaction was defective in localization to DNA damage sites, suggesting that GFP-APIM recruitment depends on an intact APIM motif (Figure S2C–D) (Gilljam et al., 2009). SiRNA depletion of PCNA greatly decreased the recruitment of GFP-APIM to laser stripes, further supporting the conclusion that PCNA mediates the localization of GFP-APIM to DNA damage sites (Figure S2C–D).
To determine if PCNA promotes the recruitment of full-length ZRANB3 to sites of DNA damage, we generated U2OS cells expressing GFP-ZRANB3 mutants containing PIP box mutations (PIP mutant) or a deletion of the APIM motif (APIM mutant). Following UV microirradiation, we observed that both individual GFP-ZRANB3 PIP or APIM mutants had impaired recruitment to laser-generated stripes (Figure 1C). PIP or APIM mutant GFP-ZRANB3 formed stripes in approximately 5% of UV irradiated cells, whereas GFP-ZRANB3 with both PIP and APIM mutations was completely defective in recruitment, indicating that the PIP and APIM motifs are both important for localization but can partially compensate for each other (Figure 1D). GFP-ZRANB3 displayed impaired recruitment after PCNA siRNA depletion, indicating that ZRANB3 localizes to sites of DNA damage in a PCNA dependent manner as predicted by the phenotype of the PIP and APIM mutants (Figures 1C–D).
During DNA replication and repair, PCNA accumulates in focal structures indicative of sites of DNA synthesis. To test whether ZRANB3 co-localizes with PCNA in these structures, U2OS cells expressing HA-tagged ZRANB3 were treated with a variety of DNA damaging agents and then co-stained with antibodies against HA-ZRANB3 and PCNA. In untreated cells HA-ZRANB3 displayed weak co-localization with PCNA in larger foci typical of late S-phase replication factories (Figures 1E and S3A). This co-localization was greatly enhanced by treatment with agents that induce replication stress, such as hydroxyurea, camptothecin, mitomycin C, cisplatin and UV radiation (Figures 1E and S3A).
The recruitment of many DDR factors to DNA damage sites is positively regulated by posttranslational modifications induced by the PI3K-related kinases (PIKKs) ATM and ATR (Ciccia and Elledge, 2010). To test if the recruitment of ZRANB3 to DNA damage sites was mediated by PIKKs, U2OS cells expressing HA-ZRANB3 were UV irradiated in the presence of caffeine, a known PIKK inhibitor. Rather than preventing ZRANB3 recruitment, caffeine treatment increased the formation of ZRANB3 foci following UV radiation (Figures 1E and S3). Similar results have been obtained by treating UV irradiated cells with ATM and ATR inhibitors, further suggesting that the recruitment of ZRANB3 to DNA damage sites can occur under conditions of reduced ATM and ATR activities (Figure S3).
Since ZRANB3 co-localizes with PCNA, we tested whether this association was direct. Purified PCNA was incubated with recombinant FLAG-ZRANB3 bound to anti-FLAG beads. As shown in Figure 1F, WT but not the double PIP and APIM mutant ZRANB3 associated with PCNA. Thus, ZRANB3 directly associates with PCNA through the PIP box and/or APIM motifs.
ZRANB3 preferentially associates with poly-ubiquitinated PCNA
Following treatment with DNA damaging agents that induce replication stress, PCNA is subjected to mono-ubiqutination and K63-linked poly-ubiquitination to promote TLS or template switching, respectively (Ulrich and Walden, 2010). These modifications serve as a signal for the recruitment of DDR factors containing ubiquitin-binding modules. Mono-ubiquitinated PCNA is recognized by TLS polymerases through ubiquitin zinc fingers (UBZ) or ubiquitin binding motifs (UBM) (Sale et al., 2012). In contrast, mammalian proteins able to bind poly-ubiquitinated PCNA and promote template switching have not yet been identified.
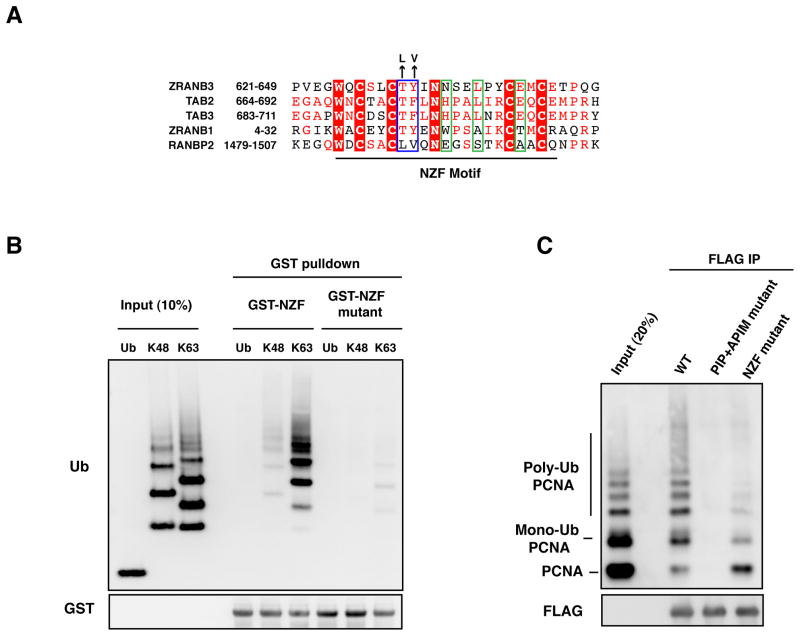
Bioinformatic analyses have revealed that the zinc finger of ZRANB3 is related to the NPL4 zinc finger (NZF) of the ubiquitin binding proteins TAB2 and TAB3, which associate with ubiquitinated components of NF-kB pathway (Figure 2A) (Kulathu et al., 2009). A NZF motif has also been identified in the APC deubiquitinase ZRANB1 and in the SUMO ligase RANBP2, among others (Figure 2A) (Alam et al., 2004; Kulathu et al., 2009). Whereas the NZF motif of TAB2, TAB3 and ZRANB1 binds ubiquitin though a conserved TY/F motif (T674, F675 in TAB2), the NZF motif of RANBP2 does not associate with ubiquitin, due to the divergence of the TY/F motif to LV (L1489, V1490) (Figure 2A, blue box) (Alam et al., 2004). The NZF motifs of ZRANB1, TAB2 and TAB3 have been shown to associate with K63-linked poly-ubiquitin chains (Komander et al., 2009; Kulathu et al., 2009; Tran et al., 2008). In particular, ZRANB1 possesses multiple NZFs, each of which able to bind a single ubiquitin moiety of the ubiquitin chain (Tran et al., 2008). In contrast, TAB2 and TAB3 contain single NZF motifs that can simultaneously bind two ubiquitin moieties (two-sided NZF motif), due to the presence of amino acid residues important for a second ubiquitin binding site (H678, L681, E685 in TAB2) in addition to the TY/F motif forming the first ubiquitin binding site (Figure 2A, blue and green boxes) (Kulathu et al., 2009). Like TAB2 and TAB3, ZRANB3 is likely to possess a two-sided NZF motif based on the conservation of residues required for the formation of two ubiquitin binding sites (T631, Y632, N635, L638, E642) (Figure 2A, blue and green boxes) (Kulathu et al., 2009). This, in principle, should provide poly-ubiquitin chain type specificity. To test this prediction, glutathione beads bound to a GST-tagged fragment of ZRANB3 containing the NZF motif (amino acids 500-800) were incubated with either free ubiquitin or K48- or K63-linked ubiquitin chains in vitro. The NZF motif of ZRANB3 preferentially associated with K63-linked ubiquitin chains (Figure 2B). This interaction was disrupted by mutating the ubiquitin binding site formed by the TY/F motif to LV, as in the non-ubiquitin binding protein RANBP2, thus indicating that the interaction with ubiquitin chains depends on a functional NZF motif (Figures 2A–B).
Figure 2. Interaction of ZRANB3 with ubiquitinated PCNA in vitro.
(A) Sequence alignment of the NZF motifs of ZRANB3, TAB2, TAB3, ZRANB1 and RANBP2. Residues involved in the binding of proximal and distal ubiquitin binding sites, as described in (Kulathu et al., 2009), are within green and blue boxes, respectively. NZF motif residues that have been mutated are indicated.
(B) Association of K63-linked ubiquitin chains with the ZRANB3 NZF motif. GST-ZRANB3 containing a WT or mutant NZF motif were affinity purified from bacteria and incubated with mono-ubiquitin or ubiquitin chains linked on lysine 48 (K48) or lysine 63 (K63). Complexes were detected with anti-ubiquitin and anti-GST antibodies after western blotting.
(C) Association of ZRANB3 with ubiquitinated PCNA. WT or mutant FLAG-ZRANB3 was immunoprecipitated with anti-FLAG beads from insect cells and incubated with a mixture of unmodified, mono- and poly-ubiquitinated PCNA. Immunoprecipitated PCNA and FLAG-ZRANB3 were detected by western blotting.
Given that ZRANB3 possesses both PCNA interaction motifs and K63-linked ubiquitin chain binding motifs, we wondered whether ZRANB3 might preferentially bind poly-ubiquitinated PCNA. To this end, recombinant PCNA was ubiquitinated in vitro and mixed with unmodified PCNA. This PCNA mixture was then incubated with anti-FLAG beads bound to recombinant FLAG-ZRANB3. As shown in Figure 2C, ZRANB3 displayed significantly higher affinity towards the poly-ubiquitinated forms of PCNA compared to mono-ubiquitinated or unmodified PCNA. Mutation of the NZF motif severely impaired the ability of ZRANB3 to bind ubiquitinated forms of PCNA, indicating that the NZF motif is important for this interaction (Figure 2C). In the context of the entire ZRANB3 protein, the presence of the NZF domain is not sufficient to allow association of ZRANB3 with ubiquitinated forms of PCNA because the double PIP and APIM mutant of ZRANB3 is completely defective for binding all forms of PCNA. Thus it appears that the PIP and APIM motifs provide an interaction surface required for the association of the NZF motif with the poly-ubiquitin chains on PCNA and it is likely that all three work cooperatively to bind poly-ubiquitinated PCNA (Figure 2C).
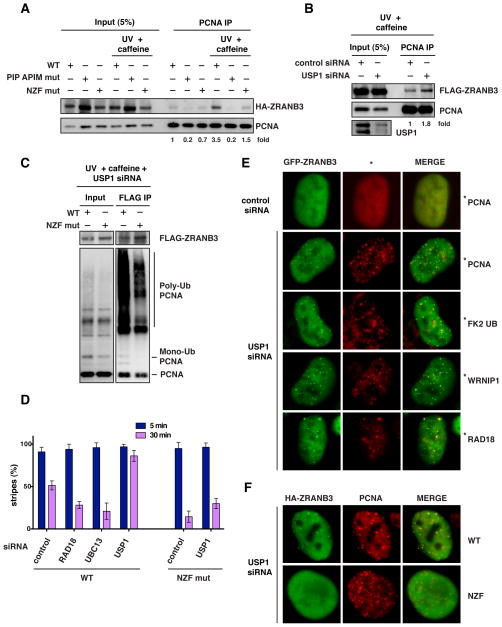
The ubiquitin binding motif of TLS polymerases, such as polη, has been suggested to facilitate their recruitment to DNA damage sites (Bienko et al., 2005; Plosky et al., 2006). To test if the NZF motif of ZRANB3 could promote the interaction with PCNA after DNA damage, U2OS cells expressing WT ZRANB3 or ZRANB3 mutated in the PIP and APIM motifs or in the NZF motif were untreated or treated with UV radiation in the presence of caffeine, which was used to facilitate the recruitment of ZRANB3 to damage sites, as discussed above. Protein complexes were then crosslinked with formaldehyde and immunoprecipitated with an anti-PCNA antibody, after which crosslinks were reversed. As shown in Figure 3A, treatment with UV and caffeine stimulated the association of WT ZRANB3 with PCNA, and this stimulation was reduced more than two-fold in the NZF mutant ZRANB3, suggesting that the NZF motif promotes the interaction between ZRANB3 and PCNA after DNA damage.
Figure 3. Association of ZRANB3 with ubiquitinated PCNA in mammalian cells.
(A) Association of ZRANB3 with PCNA after DNA damage. Protein complexes from U2OS cells expressing WT or mutant HA-ZRANB3 were crosslinked and immunoprecipitated with an anti-PCNA antibody +/− UV radiation (30 J/m2) and caffeine (2 mM). ZRANB3 and PCNA were detected with anti-HA and anti-PCNA antibodies. The fold change of immunoprecipitated HA-ZRANB3 is indicated.
(B) Depletion of USP1 increases the association of ZRANB3 with PCNA. 293T-Rex cells expressing FLAG-ZRANB3 were treated with control or USP1 siRNA prior to UV (30 J/m2) and caffeine (2 mM) treatment. Protein complexes were crosslinked, immunoprecipitated with an anti-PCNA antibody and immunoblotted with anti-FLAG and anti-PCNA antibodies. The fold change of immunoprecipitated FLAG-ZRANB3 is indicated.
(C) Interaction of ZRANB3 with ubiquitinated PCNA in mammalian cells. U2OS cells expressing FLAG-ZRANB3 were treated with USP1 siRNA prior to UV (30 J/m2) and caffeine (2 mM) treatment. Protein complexes were subjected to anti-FLAG immunoprecipitation after crosslinking and then detected by western blotting.
(D) Graphical representation of the percentage of cells that display co-localization of HA-ZRANB3, either WT or NZF mutant, with γH2AX at laser-generated stripes following treatment with siRNAs targeting RAD18, UBC13 or USP1. The time after microirradiation in which the samples were fixed is indicated. The data represent the average and standard deviation of three independent experiments.
(E) Formation of GFP-ZRANB3 foci after depletion of USP1. U2OS cells were treated with control or USP1 siRNA and stained with antibodies recognizing GFP (green) and PCNA, ubiquitinated proteins (FK2), WRNIP1 or RAD18 (all in red), as indicated.
(F) Foci formation after USP1 siRNA treatment of U2OS cells expressing either WT or NZF mutant HA-ZRANB3. Cells were stained with antibodies recognizing HA (green) and PCNA (red).
To test if the association between ZRANB3 and PCNA after UV radiation could be further stimulated by increasing the levels of the ubiquitinated forms of PCNA, 293T-Rex cells expressing FLAG-ZRANB3 were treated with siRNAs targeting the deubiquitinase USP1, which regulates the levels of mono- and poly-ubiquitinated forms of PCNA (Brun et al., 2010; Huang et al., 2006; Yang and Zou, 2009). Following crosslinking and immunoprecipitation of PCNA protein complexes, we observed that USP1 depletion led to an approximately two-fold increase in the interaction of FLAG-ZRANB3 with PCNA after UV radiation and caffeine treatment (Figure 3B). Moreover, USP1 siRNA depletion promoted to the association of multiple ubiquitinated forms of PCNA with FLAG-ZRANB3 following ZRANB3 immunoprecipitation from U2OS cells after UV and caffeine treatment (Figure 3C). However, mutation of the NZF motif of ZRANB3 led to defective interaction with poly-ubiquitinated PCNA, further confirming our in vitro interaction data.
ZRANB3 is transiently recruited to DNA damage sites showing reduced localization to laser-induced stripes 30 min after microirradiation (Figure 3D). Interestingly, USP1 siRNA depletion prevented the dissociation of ZRANB3 from DNA damage sites and led to the persistence of ZRANB3 stripes 30 min after microirradiation (Figures 3D and S4B). Conversely, siRNA depletion of RAD18 or UBC13 decreased the number of ZRANB3 stripes without affecting the initial recruitment of ZRANB3 to DNA damage sites (Figures 3D and S4A–B). In agreement with this, mutation of the NZF motif resulted in defective retention of ZRANB3 to laser-induced stripes with or without USP1 siRNA treatment (Figures 3D and S4C–D). The observation that the initial recruitment of ZRANB3 is not significantly affected by depletion of RAD18 or UBC13 and by mutation of the NZF motif suggests that ZRANB3 could be initially recruited to DNA damage sites independently of PCNA ubiquitination. The subsequent ubiquitination of PCNA by RAD18 and UBC13 at sites of DNA damage could then provide an additional interaction surface important for the retention of ZRANB3.
In agreement with the results described above, depletion of USP1 induced the formation of an extensive number of spontaneous ZRANB3 foci that co-localized with PCNA and ubiquitinated proteins (as detected by the FK2 antibody) (Figure 3E). Mutation of the NZF motif resulted in a 4-fold decrease in the number of cells with spontaneous ZRANB3 foci co-localizing with PCNA after USP1 depletion, further suggesting that the NZF motif is important for the association of ZRANB3 with ubiquitinated PCNA (Figures 3F and S4G). The spontaneous ZRANB3 foci induced by USP1 depletion co-localized also with WRNIP1 and RAD18 (Figure 3E). WRNIP1 is a RAD18- and WRN-interacting ATPase, which associates with sites of replication stress and binds ubiquitinated proteins through a UBZ motif (Bish and Myers, 2007; Crosetto et al., 2008; Kawabe et al., 2006; Yoshimura et al., 2009). As shown in Figure S4H, WRNIP1 was identified in ZRANB3 immunoprecipitates following USP1 depletion in combination with UV and caffeine treatment. Interestingly, Mgs1, the yeast ortholog of WRNIP1, has recently been reported to associate with ubiquitinated PCNA (Saugar et al., 2011). These observations suggest that both ZRANB3 and WRNIP1 may associate with poly-ubiquitinated PCNA.
ZRANB3 prevents sister chromatid exchange and promotes fork restart after replication stress
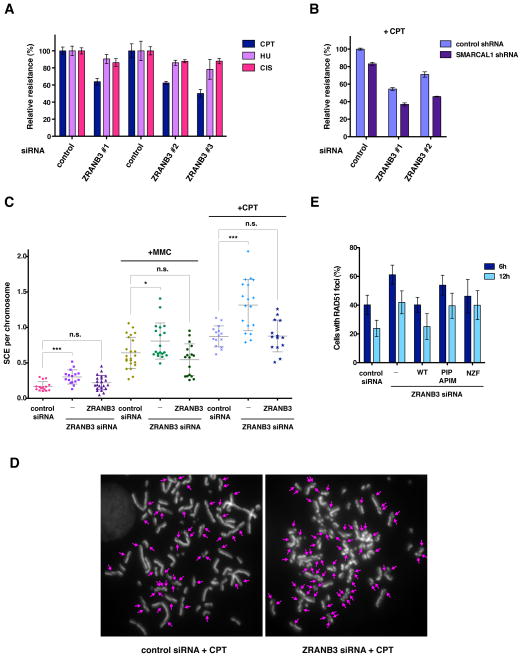
Since ZRANB3 associates with ubiquitinated forms of PCNA at sites of replication stress, we tested whether ZRANB3 depletion could sensitize cells to DNA damaging agents that arrest replication fork progression. To this end, U2OS cells transfected with control or three independent ZRANB3 siRNAs were mixed with control cells expressing GFP in a cell competition assay and then subjected to treatment with camptothecin, hydroxyurea or cisplatin (Figure 4A) (Smogorzewska et al., 2007). ZRANB3 depleted cells displayed marked sensitivity to camptothecin and milder sensitivity to hydroxyurea or cisplatin (Figure 4A). SMARCAL1 depletion also sensitizes cells to replication stress (Bansbach et al., 2009; Ciccia et al., 2009; Yuan et al., 2009). To test whether ZRANB3 and SMARCAL1 cooperate in protecting from replication stress, U2OS cells expressing either control or SMARCAL1 shRNAs were transfected with control or ZRANB3 siRNAs and then subjected to camptothecin treatment. Cells depleted of both ZRANB3 and SMARCAL1 displayed additive sensitivity to camptothecin (Figure 4B). Similar results were also obtained by using an additional ZRANB3 siRNA (data not shown), suggesting that ZRANB3 and SMARCAL1 function independently from each other.
Figure 4. Effects of ZRANB3 depletion in mammalian cells.
(A) Cell competition assay in U2OS cells treated with control siRNAs or three independent ZRANB3 siRNAs following treatment with camptothecin (CPT, 5 nM), hydroxyurea (HU, 2 mM) or cisplatin (CIS, 0.5 μM). The data represent the average and standard deviation of three independent experiments.
(B) Cell competition assay in U2OS cells expressing either control or SMARCAL1 shRNAs treated with control or ZRANB3 siRNAs following treatment with camptothecin (CPT, 5 nM). Error bars have been calculated as in (A).
(C) Graphical representation of the frequencies of sister chromatid exchanges (SCEs) of mitotic chromosomes isolated from U2OS cells transfected with control or ZRANB3 siRNA with or without mitomycin C (MMC, 20 nM) or camptothecin (CPT, 2.5 nM) treatment. The SCE frequencies of U2OS cells expressing a ZRANB3 cDNA clone resistant to ZRANB3 siRNA treatment are indicated. The average frequencies of SCEs and the standard deviation are indicated. Statistically significant p-values calculated using the Mann-Whitney test are indicated by asterisks (*p<0.05, ***p<0.001). n.s., not significant.
(D) Chromosome spreads from U2OS cells transfected with control or ZRANB3 siRNA after camptothecin (CPT, 2.5 nM) treatment. SCEs are indicated.
(E) Percentage of U2OS cells transfected with control or ZRANB3 siRNAs displaying more than 10 RAD51 foci. Cells were fixed 6 h or 12 h following 1 h camptothecin (CPT, 10 nM) treatment. The percentage of U2OS cells with more than 10 RAD51 foci that expressed siRNA resistant cDNA clones coding for either WT, PIP and APIM or NZF mutant ZRANB3 is also indicated. The data represent the average and standard deviation of three independent experiments in which 100 or more cells were counted.
Genomic stability after replication stress could be ensured by regulating homologous recombination between sister chromatids at stalled or collapsed replication forks. Recombination between sister chromatids can result in crossover events, which can be visualized on metaphase chromosomes as SCEs, or non-crossover events mediated by gene conversion. In order to determine if ZRANB3 could affect the outcome of such events, U2OS cells treated with control or ZRANB3 siRNA were incubated with BrdU for one round of DNA replication and subsequently subjected to MMC or camptothecin treatment during the second cell cycle. Following isolation of mitotic chromosomes, exchanges between sister chromatids were visualized with orange acridine staining, which differentially labels double-stranded DNA with one or two BrdU-incorporated strands. As indicated in Figure 4C–D, ZRANB3 depletion led to increased frequencies of SCEs following MMC or camptothecin treatment. The increase in SCE frequencies following treatment with ZRANB3 siRNA was reversed by the expression of a siRNA resistant ZRANB3 cDNA (Figure 4C). Similar results were obtained with two independent ZRANB3 shRNAs after MMC treatment (Figure S5E–G). Statistically significant increases in SCE frequencies were also observed in ZRANB3 siRNA depleted cells without treatment, suggesting that ZRANB3 could also prevent the formation of SCEs during unperturbed DNA replication (Figure 4C). In agreement with these data, ZRANB3 depletion resulted in an increased number of cells with more than 10 RAD51 foci after camptothecin treatment, further indicating that ZRANB3 prevents inappropriate recombination after replication stress (Figures 4E and S5D). This increase was significantly reversed by the expression of a siRNA resistant clone expressing wild-type, but not PIP and APIM or NZF mutant ZRANB3, suggesting a role for the interaction between ZRANB3 and ubiquitinated PCNA in limiting RAD51 accumulation after camptothecin treatment (Figure 4E). Increase in the number of cells with RAD51 foci was also observed after SMARCAL1 depletion (data not shown), as previously reported (Postow et al., 2009).
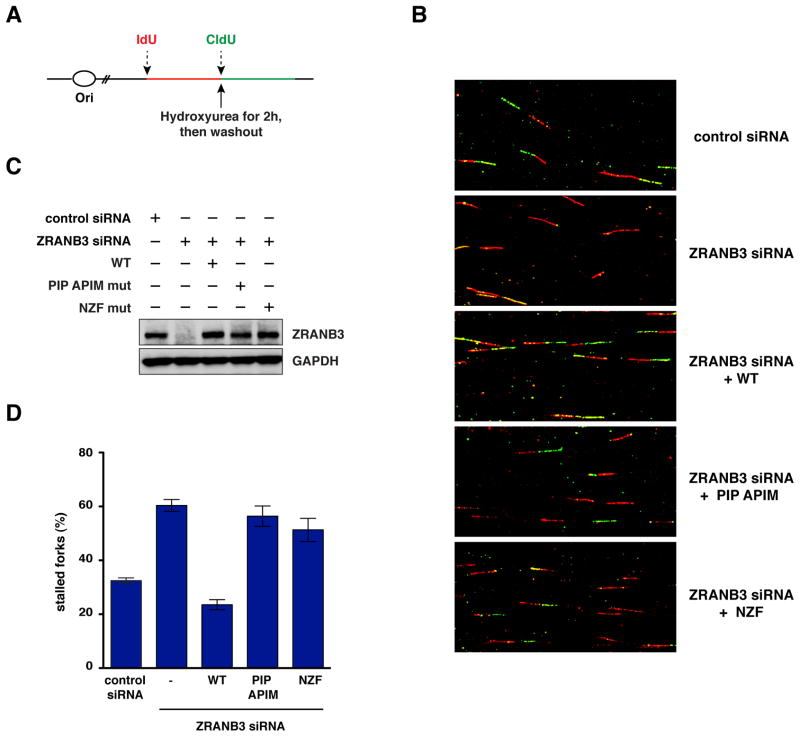
To test whether ZRANB3 depletion affects replication restart after replication stress, we performed single DNA fiber analysis. U2OS cells were pulse-labeled with the thymidine analog IdU for 25 min and subsequently incubated with hydroxyurea for 2h to arrest replication fork progression. Following hydroxyurea washout, cells were pulse-labeled with CldU, another thymidine analog, for additional 40 min and DNA fibers were then isolated and stained for IdU (red) and CldU (green) (Figure 5A). Cells treated with ZRANB3 siRNA displayed an approximately two-fold increase in the number of DNA fibers that had incorporated only Idu (red only tracts), indicating that ZRANB3 depletion leads to defective fork restart after replication arrest (Figure 5B–D). This defect could be reversed by the expression of a siRNA resistant clone coding for wild-type ZRANB3 but not mutants in the PIP and APIM motifs or the NZF motif (Figure 5B–D). Altogether, these results suggest that ZRANB3 maintains genomic stability at stalled or collapsed replication forks by promoting fork restart and limiting the formation of crossover events.
Figure 5. DNA fiber analysis after ZRANB3 depletion in mammalian cells.
(A) Schematics of the pulse-labeling experiment performed for DNA fiber analysis.
(B) Images of DNA fibers isolated from cells treated with control or ZRANB3 siRNAs following the expression of siRNA resistant WT, PIP and APIM or NZF mutant ZRANB3. DNA fibers were stained with antibodies that recognize IdU (red) and CldU (green).
(C) Detection by western blot of the ZRANB3 protein levels in the cells subjected to DNA fiber analysis shown in (B).
(D) Graphical representation of the percentage of stalled forks (red only tracts) from DNA fiber analyses of the samples shown in (B). The data represent the average and standard deviation of three independent experiments.
ZRANB3 remodels replication fork structures and disassembles D-loop intermediates
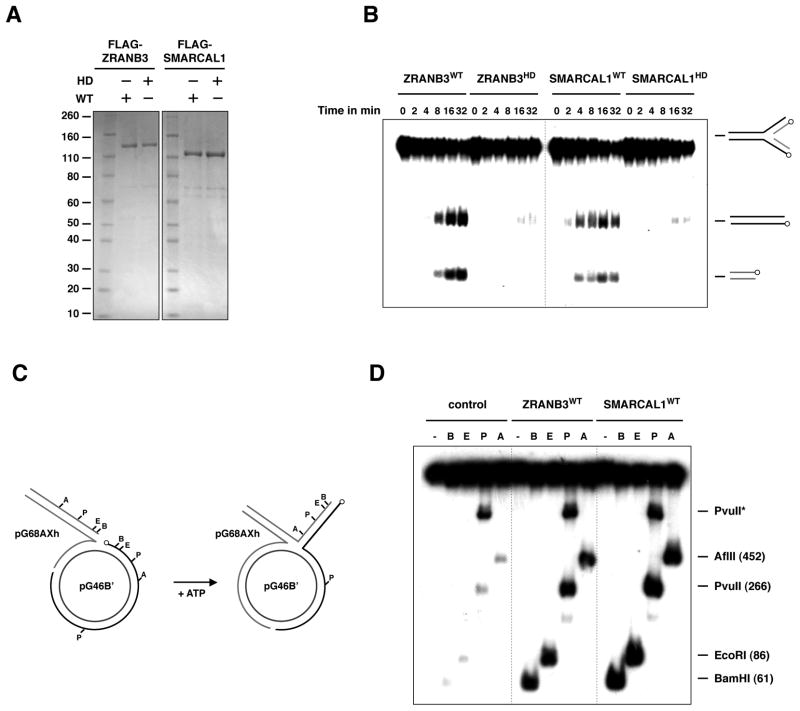
To determine the molecular mechanisms by which ZRANB3 could promote fork restart and limit crossovers, we purified ZRANB3 from insect cells and performed in vitro assays using DNA substrates mimicking replication and recombination DNA intermediates (Figure 6A). It has been suggested that upon replication fork arrest at DNA lesions, regression of the replication fork can lead to the annealing of the arrested DNA strand with the newly synthesized strand of the sister chromatid, thus allowing the blocked strand to restart DNA synthesis by template switching (Atkinson and McGlynn, 2009; Unk et al., 2010). To determine whether ZRANB3 promotes fork regression, we performed helicase assays on synthetic replication fork structures with homologous DNA strands. As shown in Figure 6B, ZRANB3 exhibited fork regression activity, which was abrogated by a mutation in the helicase domain.
Figure 6. Fork regression activities of ZRANB3 and SMARCAL1.
(A) Purification of FLAG-ZRANB3 and FLAG-SMARCAL1 from insect cells. WT and helicase-dead (HD) mutant proteins were subjected to CM sepharose chromatography, affinity purified with anti-FLAG beads and eluted with FLAG peptide. The eluates were analyzed by SDS-PAGE and visualized by Coomassie.
(B) Regression of synthetic fork substrates using ZRANB3 and SMARCAL1. 32P-labeled fork substrates were incubated with WT and mutant proteins in a time course reaction. DNA products were then analyzed by gel electrophoresis and visualized by autoradiography. The position of the 32P-labels on the DNA substrates is indicated by circles.
(C) Schematic representation of the plasmid based assay employed in (D). 32P-labels are indicated by circles.
(D) Regression of plasmid based replication forks by ZRANB3 and SMARCAL1. Following incubation with ZRANB3 or SMARCAL1, fork structures were digested with restriction enzymes that cleave the dsDNA generated by annealing of the regressed DNA strands. DNA products of the restriction digests were analyzed by gel electrophoresis and visualized by autoradiography.
Stalled replication forks generally have asymmetric DNA strands due to the uncoupling of leading- and lagging-strand synthesis. To determine if ZRANB3 can regress replication forks that more closely mimic stalled replication forks, we employed a strategy that utilizes a previously developed plasmid based DNA substrate (Figure 6C) (Blastyak et al., 2007). In this assay, a duplex DNA with a ssDNA tail is annealed to a gapped plasmid to form a joint molecule that resembles a stalled fork. Regression of the stalled fork can be monitored by restriction digestion of the 5′ labeled duplex DNA that is formed after annealing of the two regressed strands. As shown in Figure 6D, ZRANB3 promotes extensive regression of the replication fork, generating a regressed arm longer than 450 bp. Similar results on the synthetic fork structure and on the plasmid-based fork were also observed for recombinant FLAG-SMARCAL1, indicating that SMARCAL1 possesses fork remodeling activity, as recently reported (Figures 6B–D) (Betous et al., 2012). Together, these data suggest that ZRANB3 and SMARCAL1 could facilitate fork restart by remodeling stalled or blocked replication forks.
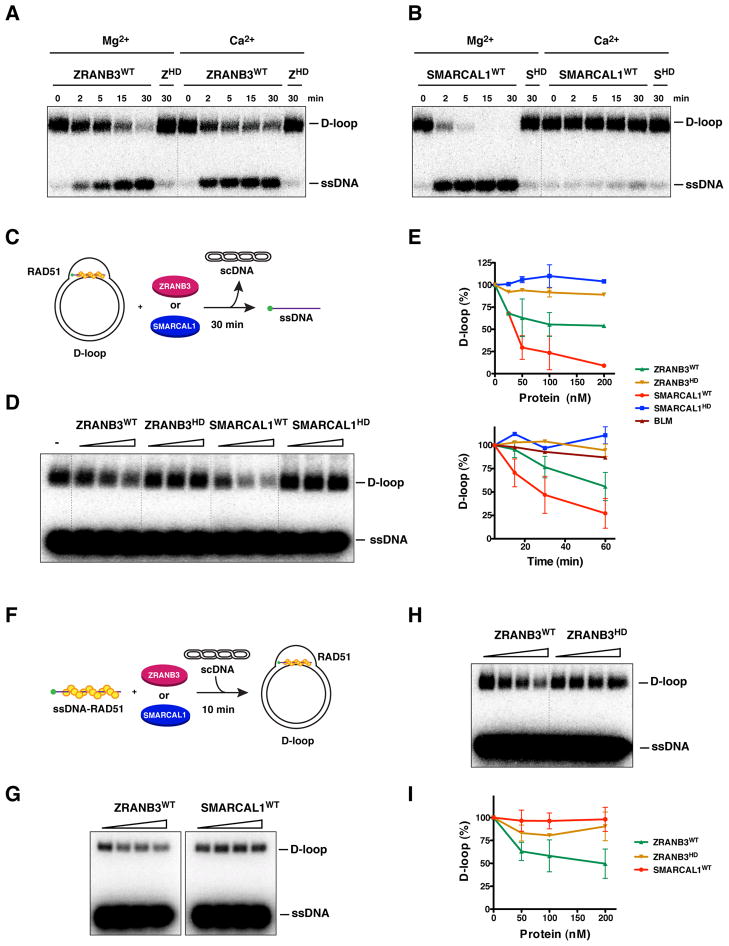
Suppression of SCEs by ZRANB3 after replication stress could be mediated by disassembling recombination intermediates, such as D-loop structures, that are generated during the repair of ssDNA gaps or DNA breaks formed at stalled replication forks. To test this hypothesis, we first generated a D-loop substrate using RecA protein to assimilate a complementary 100-mer ssDNA into supercoiled DNA (scDNA). The reaction was then deproteinized, purified and used to determine if ZRANB3 and SMARCAL1 can dismantle such structures. As shown in Figure 7A–B, both ZRANB3 and SMARCAL1 were able to disrupt D-loop structures in a manner dependent on functional helicase domains. In particular, SMARCAL1 exhibited stronger activity than ZRANB3 in magnesium containing buffer, whereas ZRANB3, but not SMARCAL1, was active in calcium containing buffer (Figures 7A–B and S6A). This suggests that the helicase domains of ZRANB3 and SMARCAL1 could interact with divalent ions in a distinct manner.
Figure 7. Disruption of D-loop structures by ZRANB3 and SMARCAL1.
(A) Dissociation of D-loops (400 nM) by ZRANB3, either WT or helicase-dead (HD) (100 nM) as a function of time. Experiments were performed in buffer containing either magnesium or calcium acetate (5 mM).
(B) D-loop structures generated as described in (A) were incubated in a time course reaction with either WT or helicase-dead (HD) SMARCAL1 (100 nM) in the presence of either magnesium or calcium acetate (5 mM).
(C) Schematics of the dissociation of preformed RAD51-containing D-loop structures by ZRANB3 and SMARCAL1.
(D) Increasing amounts of ZRANB3 and SMARCAL1 (25, 50 and 200 nM), either WT or mutant, were incubated with preformed RAD51-containing D-loops as depicted in (C).
(E) Quantification of the disruption of preformed RAD51-containing D-loop structures following addition of increasing (upper panel) or fixed amounts of ZRANB3 and SMARCAL1 proteins (100 nM) in a time course reaction (lower panel). The activity of BLM (100 nM) in the time course reaction is indicated. Points with error bars represent the average and standard deviation of three or more independent experiments.
(F) Schematics of the formation of RAD51-containing D-loop structures in the presence of ZRANB3 and SMARCAL1.
(G) Formation of RAD51-containing D-loop structures following addition of increasing amounts of ZRANB3 or SMARCAL1 proteins (50, 100 and 200 nM) as represented in the schematics in (F).
(H) Formation of RAD51-containing D-loop structures following addition of increasing amounts of either WT or mutant ZRANB3 (50, 100 and 200 nM).
(I) Quantification of the formation of RAD51-containing D-loops in the presence of ZRANB3 and SMARCAL1 proteins. Points with error bars represent the average and standard deviation of three or more independent experiments.
Having established that ZRANB3 and SMARCAL1 can disassemble purified D-loops, we wondered whether the two proteins possess a similar activity on RAD51 containing D-loops. We therefore generated D-loops using human RAD51 (hRAD51) and tested the effect of addition of ZRANB3 or SMARCAL1 to an ongoing hRAD51-mediated D-loop reaction (Figure 7C). As shown in Figure 7D–E, ZRANB3 or SMARCAL1 promoted disruption of D-loops in a concentration dependent manner. An extended time course at fixed concentrations of the translocases supported this observation (Figures 7E and S6B–C). SMARCAL1 exhibited more pronounced activity than ZRANB3 under these conditions (compare Figure 7A–B and Figures 7D–E and S6B–C). These reactions were performed in the presence of both magnesium and calcium, which is known to stabilize RAD51 filaments by inhibiting RAD51 ATPase activity (Bugreev and Mazin, 2004). Consistent with previous observations, BLM did not dissociate D-loop structures under these reaction conditions (Figure 7E) (Barber et al., 2008; Bugreev et al., 2007).
To investigate whether ZRANB3 and SMARCAL1 could prevent the formation of D-loop structures, ZRANB3 or SMARCAL1 were incubated with hRAD51 nucleoprotein filaments in the presence of magnesium and calcium before the addition of scDNA for 10 min prior to termination of the reaction (Figure 7F). Interestingly, ZRANB3, but not SMARCAL1, decreased the efficiency of D-loop formation (Figure 7G–I). Given that ZRANB3 does not appear to directly disrupt hRAD51 filaments on ssDNA (Figure S6D), we propose that this activity of ZRANB3 could be due to unwinding of the initial intermediates of strand invasion. Taken together, these data suggest that ZRANB3 and SMARCAL1 could suppress inappropriate recombination by either preventing or disrupting D-loop intermediates.
Discussion
The ability of cells to properly restart stalled or collapsed replication forks and limit inappropriate recombination after replication stress is critical to the survival of the organism. Limiting crossover events between homologous chromosomes prevents reduction to homozygosity, which can uncover recessive tumor suppressor mutations leading to cancer. In this study we discovered that the ZRANB3 translocase is recruited to sites of replication stress by poly-ubiquitinated PCNA to promote genomic stability by facilitating replication restart and limiting exchanges between sister chromatids. ZRANB3 contains two distinct PCNA interaction motifs, the PIP box and APIM motifs. Both are critical for the localization of ZRANB3 to sites of DNA damage, suggesting that they cooperate with each other in mediating the interaction with PCNA. ZRANB3 also contains a NZF zinc finger that preferentially binds K63-linked poly-ubiquitin chains and is required for efficient association with PCNA after DNA damage. Thus all three motifs of ZRANB3 contribute to localization to poly-ubiquitinated PCNA in response to damage. This is reminiscent of the ability of the PIP box and UBM/UBZ motifs of TLS polymerases to cooperatively interact with mono-ubiquitinated PCNA (Bienko et al., 2005; Plosky et al., 2006). In the case of ZRANB3, the PIP and APIM motif of ZRANB3 appear to play a primary role in the initial recruitment of ZRANB3 to DNA damage sites, whereas the NZF motif provides an interaction surface important for the temporal retention of ZRANB3 following PCNA ubiquitination by RAD18 and UBC13 at sites of DNA damage.
ZRANB3 remodels replication forks and promotes fork restart after replication arrest
In this study we show that ZRANB3, like SMARCAL1, facilitates fork restart and this activity depends on its interaction with poly-ubiquitinated PCNA. Poly-ubiquitination of PCNA is known to promote the template switching pathway, which could involve fork regression or strand invasion mechanisms to allow bypass of DNA lesions at or near stalled replication forks (Unk et al., 2010). Using biochemical assays, we found that ZRANB3 and SMARCAL1 are capable of regressing DNA structures mimicking stalled replication forks. This activity, which is distinct from the previously described annealing helicase activity of ZRANB3 and SMARCAL1, provides a possible explanation for their role in replication fork restart (Ciccia et al., 2009; Yusufzai and Kadonaga, 2008, 2010).
The activity of ZRANB3 and SMARCAL1 resembles the fork reversal activity of dsDNA translocases like FANCM and HLTF, and is mechanistically distinct from the fork regression activity of BLM, WRN and RECQ5, which, in contrast are also able to unwind forks with heterologous arms (Atkinson and McGlynn, 2009; Yusufzai and Kadonaga, 2008, 2010). The availability of several helicases/translocases displaying fork regression activities provides the cell with multiple options that could be employed in different contexts to maintain genomic stability. As in the case of ZRANB3 and SMARCAL1, the recruitment of the appropriate helicase/translocase needed to restart DNA synthesis can be mediated by different DDR factors, such as PCNA and RPA, which are able to recognize distinct DNA intermediates formed during replication fork stalling and therefore operate in response to a multiplicity of replication fork insults.
ZRANB3 can act as an anti-recombinase to dissociate D-loop intermediates and limit sister chromatid exchange after replication stress
ZRANB3 prevents SCEs after treatment with DNA damaging agents, suggesting that ZRANB3 can prevent inappropriate recombination at stalled/blocked replication forks. Suppression of SCEs after replication stress could be ensured by preserving the integrity of blocked replication forks, thereby limiting the generation of recombination intermediates leading to SCE formation. The fork remodeling activity of ZRANB3 could help stabilize the stalled fork by limiting the amount of ssDNA regions at the fork and by promoting the restart of the blocked strand, thus preventing replication fork collapse (Atkinson and McGlynn, 2009). It has recently been reported that fork reversal is induced to prevent fork breakage following treatment with low doses of camptothecin in U2OS cells (Ray Chaudhuri et al., 2012). The sensitivity of ZRANB3 and SMARCAL1 depleted cells to low doses of camptothecin could therefore be caused by defective fork reversal and increased fork collapse in the absence of ZRANB3 and SMARCAL1. ZRANB3 and SMARCAL1 could participate in alternative pathways involved in remodeling and protecting stalled forks following camptothecin treatment, as indicated by the additive sensitivity to camptothecin treatment of the double ZRANB3 and SMARCAL1 depletion compared to the respective individual depletions in U2OS cells.
The suppression of SCEs mediated by ZRANB3 could alternatively result from a direct inhibition of inappropriate recombination at stalled or collapsed forks. Indeed, we have shown that ZRANB3 is able to limit RAD51-mediated D-loop formation and can also dissociate preformed D-loop structures in vitro. Surprisingly, SMARCAL1 exhibited activity only towards preformed D-loops. This raises the possibility that ZRANB3 could play both an early and a late role in regulating RAD51-dependent strand invasion, whereas SMARCAL1 could function after strand invasion is completed. The mechanisms by which these activities are exerted are still unclear. We have not observed disruption of RAD51 nucleoprotein filaments by ZRANB3, suggesting that ZRANB3 does not act similarly to the recently described PARI anti-recombinase (Moldovan et al., 2012). Therefore ZRANB3 could limit the formation of D-loops possibly by dissociating early D-loop intermediates. This process could also be important to prevent recombination between partially homologous (homeologous) sequences.
With respect to helicases/translocases capable of disrupting D-loops, ZRANB3 and SMARCAL1 act on preformed D-loops similarly to RTEL1 and Mph1, which are able to dissociate preformed D-loops without disrupting RAD51 nucleoprotein filaments (Barber et al., 2008; Prakash et al., 2009). It should be noted that ZRANB3 and SMARCAL1 are not ssDNA or dsDNA dependent ATPases. The fact that their ATPase activity is stimulated by forked DNA structures indicates that ZRANB3 and SMARCAL1 operate by binding to the branch point of forked structures, where they can promote spooling and threading of the DNA arms of these substrates (Betous et al., 2012; Yusufzai and Kadonaga, 2008, 2010). This is similar to the action of E. coli RecG and phage T4 UvsW, which are able to catalyze both fork regression and D-loop disruption (Atkinson and McGlynn, 2009; Singleton et al., 2001). We therefore suggest that ZRANB3 and SMARCAL1 could be bona fide functional orthologs of RecG and UvsW.
ZRANB3 may participate in the template switching pathway of post-replication repair
Post-replication repair (PRR) promotes the bypass of DNA lesions during DNA replication. Several lines of evidence suggest that ZRANB3 could participate in the template switching pathway of PRR. First, ZRANB3 can associate with poly-ubiquitinated PCNA, which regulates template switching in yeast (Bergink and Jentsch, 2009; Ulrich and Walden, 2010). In addition, ZRANB3 associates with the WRNIP1 ATPase, which has been suggested to modulate the activity of polδ during template switching (Saugar et al., 2011; Tsurimoto et al., 2005). Moreover, ZRANB3 facilitates fork restart in a manner dependent on its association with poly-ubiquitinated PCNA and catalyzes fork regression, which has been proposed as a possible mechanism by which template switching could operate. Finally, ZRANB3 limits the frequency of SCEs, similar to RAD18 and WRNIP1, raising the possibility that these proteins may co-operate to limit formation of crossover events occurring during template switching events at stalled or collapsed replication forks (Hayashi et al., 2008; Szuts et al., 2006; Tateishi et al., 2003). Elucidating the molecular mechanisms by which this error free pathway of template switching functions to maintain genomic stability during DNA replication will have important implications in understanding how mutations and chromosomal rearrangements arise in genomic disorders and cancer.
Experimental Procedures
Construction of plasmid vectors, protein purification, in vitro pulldowns, immunoprecipitation of protein complexes, cell culture and biochemical assays were conducted as described in the Supplemental Experimental Procedures.
Antibodies
Rabbit polyclonal anti-GST (1:1000, Abcam, ab21070), anti-PCNA (1:1000, Abcam, ab18197), anti-ZRANB3 (1:1000, Bethyl, A303-033A), anti-USP1 (1:500, Proteintech, 14346-1-AP), anti-WRNIP1 (1:1000, Bethyl, A301-389A), anti-RAD18 (1:1000, Bethyl, A301-340A), anti-GAPDH (1:2000, Santa Cruz, sc-25778) and mouse monoclonal anti-FLAG (1:1000, Sigma, M2), anti-HA (1:1000, Covance, HA.11) anti-UBC13 (1:1000, Invitrogen, 37-1100) and anti-ubiquitin (1:1000, Millipore, Ubi-1) antibodies were used in western blot experiments.
RNAi treatment
PCNA siGenome siRNA pool (Dharmacon, MU-003289-02-0002), USP1 siGenome siRNA (Dharmacon, D-006061-03-0005), RAD18 siGenome siRNA pool (Dharmacon, MU-004591-00-0002), UBC13 siGenome siRNA pool (Dharmacon, MU-003920-01-0002), ZRANB3 siRNA #1 (Dharmacon, D-010025-03-0005), ZRANB3 siRNA #2 (Invitrogen, Silencer Select siRNA s105378), ZRANB3 siRNA #3 (Invitrogen, Silencer Select siRNA s105379) were used to transfect U2OS cells or 293T-Rex cells. U2OS cells with stable knockdown of ZRANB3 were obtained after infection with lentiviruses derived from the pGIPZ vector containing ZRANB3 shRNA #1 (Open Biosystems, V3LHS_357167, TGGTGTGTGTCAGCTCTGT) or ZRANB3 shRNA #2 (Open Biosystems, V2LHS_117955, CAAGAGATATCATCGATTA). The SMARCAL1 shRNA sequence has been previously described (Ciccia et al., 2009). Viruses carrying the firefly shRNA sequence (CCCGCCTGAAGTCTCTGATTAA) were used to generate control U2OS cells.
Sister chromatid exchange assay
U2OS cells were treated with control or ZRANB3 siRNA (50 nM) and 72 h after siRNA tranfection cells were incubated with BrdU (10 μM) and 24 h later, cells were treated with or without mitomycin C (20 nM) or camptothecin (2.5 nM) for another 24 h. U2OS cells expressing a ZRANB3 cDNA clone resistant to the ZRANB3 siRNA were subjected to similar treatment following ZRANB3 siRNA transfection. U2OS cells expressing FF shRNA and ZRANB3 shRNAs #1 and #2 were similarly treated with BrdU and mitomycin C. Isolation of metaphase chromosomes and staining of SCEs was performed as previously described (Hu et al., 2005). Statistical Mann-Whitney analyses were performed with the Prism software (GraphPad Software).
DNA fiber analysis
U2OS cells were transfected with control or ZRANB3 siRNA (50 nM) and 96 h after siRNA tranfection cells were incubated with 34 μM IdU for 25 min. Cells were then treated with 2 mM hydroxyurea for 2 h and incubated in 250 μM CldU for 40 min after washout of the drug. Spreading of DNA fibers on glass slides was done as reported (Jackson and Pombo, 1998). Glass slides were then washed in distilled water and in 2.5 M HCl for 45 min followed by three washes in PBS. The slides were incubated for 1 h in blocking buffer (PBS with 1% BSA and 0.1% NP40) and then for 2 h in rat anti-BrdU antibody (Abcam, ab6326) diluted 1:500 in blocking buffer. After washing with blocking buffer containing 500 mM NaCl, the slides were incubated for 2 h in goat anti-rat Alexa-488 antibody (1:250, Invitrogen). The slides were then washed with PBS/1% NP40 and then incubated for 2 h with mouse anti-BrdU antibody diluted in blocking buffer (1:100, BD Biosciences, 347580). Following an additional wash with PBS/1% NP40, the fibers were stained for 2 h with anti-mouse Alexa-594 (1:250, Invitrogen). Fibers were then analyzed using an Olympus confocal microscope. U2OS cells expressing WT or mutant ZRANB3 cDNA clones resistant to the ZRANB3 siRNA were subjected to similar treatment following ZRANB3 siRNA transfection.
Supplementary Material
Acknowledgments
We thank Andrea Bredemeyer Artyom Alekseyenko, Satya Prakash and Jung-Hoon Yoon for help and advice and Alan Lehmann for reagents. A.C. and I.H. are recipients of EMBO long-term fellowships. Y.H. was supported by a U.S. Department of Defense post-doctoral fellowship award (W81XWH-09-1-0632). This work was supported by grants from the National Institute of Health and the National Cancer Institute to S.J.E., D.M.L, J.C.Y., S.C.K. (grant GM62653) and by Howard Hughes Medical Institute Grant 55005612 and Hungarian Science Foundation Grants OTKA 77495 and TAMOP-4.2.2/08/1 to L.H. S.J.E. is an Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes & development. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes & development. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Bish RA, Myers MP. Werner helicase-interacting protein 1 binds polyubiquitin via its zinc finger domain. J Biol Chem. 2007;282:23184–23193. doi: 10.1074/jbc.M701042200. [DOI] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D. Ubiquitin family modifications and template switching. FEBS letters. 2011;585:2810–2817. doi: 10.1016/j.febslet.2011.04.053. [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Brun J, Chiu RK, Wouters BG, Gray DA. Regulation of PCNA polyubiquitination in human cells. BMC research notes. 2010;3:85. doi: 10.1186/1756-0500-3-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein RAD51 by modulating its ATPase activity. Proc Natl Acad Sci U S A. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes & development. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem. 2008;283:35173–35185. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilljam KM, Feyzi E, Aas PA, Sousa MM, Muller R, Vagbo CB, Catterall TC, Liabakk NB, Slupphaug G, Drablos F, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Seki M, Inoue E, Yoshimura A, Kusa Y, Tada S, Enomoto T. Vertebrate WRNIP1 and BLM are required for efficient maintenance of genome stability. Genes & genetic systems. 2008;83:95–100. doi: 10.1266/ggs.83.95. [DOI] [PubMed] [Google Scholar]
- Hu YD, Lu XC, Barnes E, Yan M, Lou H, Luo GB. RecQl5 and BLM RecQ DNA helicases have non-redundant roles in suppressing crossovers. Mol Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D’Andrea AD. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Seki M, Yoshimura A, Nishino K, Hayashi T, Takeuchi T, Iguchi S, Kusa Y, Ohtsuki M, Tsuyama T, et al. Analyses of the interaction of WRNIP1 with Werner syndrome protein (WRN) in vitro and in the cell. DNA Repair (Amst) 2006;5:816–828. doi: 10.1016/j.dnarep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16:1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Woo EM, Chait BT, Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J Biol Chem. 2009 doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes & development. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugar I, Parker JL, Zhao S, Ulrich HD. The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuts D, Simpson LJ, Kabani S, Yamazoe M, Sale JE. Role for RAD18 in homologous recombination in DT40 cells. Mol Cell Biol. 2006;26:8032–8041. doi: 10.1128/MCB.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S, Niwa H, Miyazaki J, Fujimoto S, Inoue H, Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol Cell Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Hamada F, Schwarz-Romond T, Bienz M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22:528–542. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Shinozaki A, Yano M, Seki M, Enomoto T. Human Werner helicase interacting protein 1 (WRNIP1) functions as a novel modulator for DNA polymerase delta. Genes to cells: devoted to molecular & cellular mechanisms. 2005;10:13–22. doi: 10.1111/j.1365-2443.2004.00812.x. [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Blastyak A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Yang XH, Zou L. Dual functions of DNA replication forks in checkpoint signaling and PCNA ubiquitination. Cell Cycle. 2009;8:191–194. doi: 10.4161/cc.8.2.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Seki M, Kanamori M, Tateishi S, Tsurimoto T, Tada S, Enomoto T. Physical and functional interaction between WRNIP1 and RAD18. Genes & genetic systems. 2009;84:171–178. doi: 10.1266/ggs.84.171. [DOI] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes & development. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20970–20973. doi: 10.1073/pnas.1011196107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes & development. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.