Summary
The mechanism by which cells decide to skip mitosis to become polyploid is largely undefined. Here we used a high-content image-based screen to identify small-molecule probes that induce polyploidization of megakaryocytic leukemia cells and serve as perturbagens to help understand this process. We found that dimethylfasudil (diMF, H-1152P) selectively increased polyploidization, mature cell-surface marker expression, and apoptosis of malignant megakaryocytes. A broadly applicable, highly integrated target identification approach employing proteomic and shRNA screening revealed that a major target of diMF is Aurora A kinase (AURKA), which has not been studied extensively in megakaryocytes. Moreover, we discovered that MLN8237 (Alisertib), a selective inhibitor of AURKA, induced polyploidization and expression of mature megakaryocyte markers in AMKL blasts and displayed potent anti-AMKL activity in vivo. This research provides the rationale to support clinical trials of MLN8237 and other inducers of polyploidization in AMKL. Finally, we have identified five networks of kinases that regulate the switch to polyploidy.
Introduction
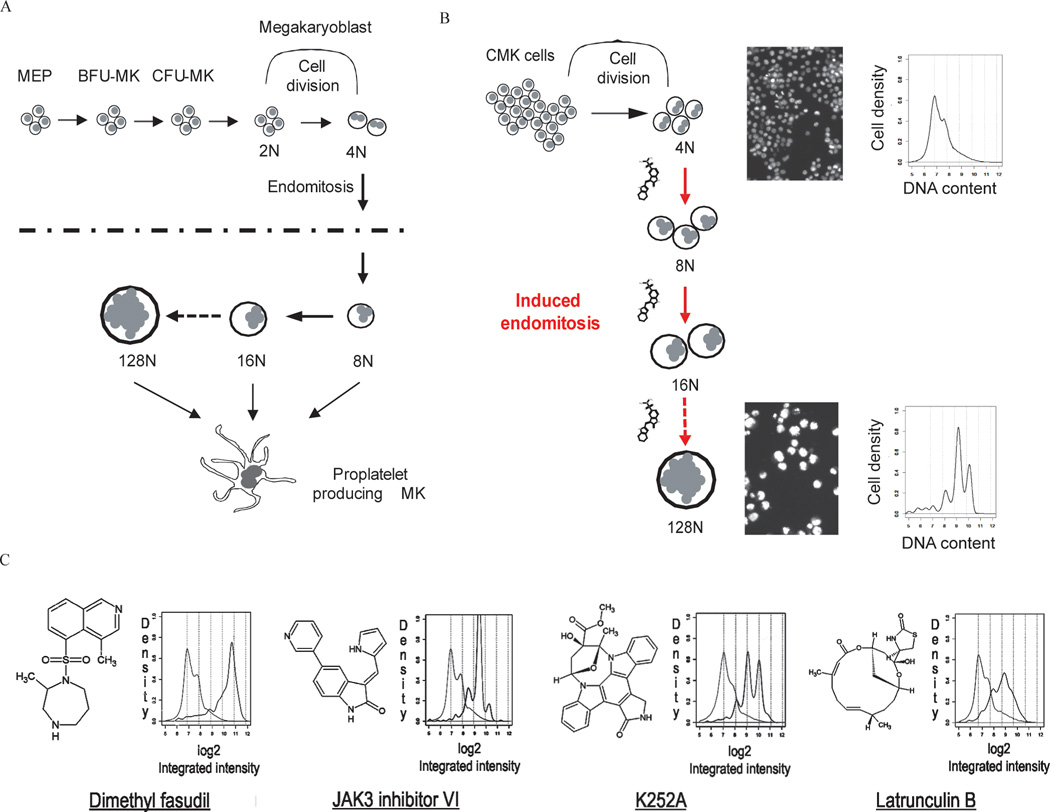
Megakaryocytes are one of the few cell types that undergo a modified form of the cell cycle termed endomitosis, in which cells skip the late stages of mitosis to become polyploid (Bluteau et al., 2009) (Fig 1A). Murine and human megakaryocytes commonly reach modal ploidy states of 32N and 16N, respectively, and can sometimes achieve DNA contents as high as 128N. Although the mechanism of polyploidization is still not well understood, altered expression of genes including D- and E-type cyclins, spindle checkpoint proteins, and chromosome passenger proteins have been implicated (Wen et al., 2011).
Figure 1. Cell-based, high-content imaging screen for compounds that induce megakaryocyte polyploidization.
(A) Schematic of megakaryocyte development. MEP, megakaryocyte-erythroid progenitor; BFU-MK, burst-forming unit-megakaryocyte; CFU-MK, colony-forming unit megakaryocyte. (B) Schematic of the image-based high-throughput screen to identify small molecules that induce polyploidization of leukemic megakaryocytes. (C) Structures of representative hit compounds and their effects on megakaryocyte polyploidization. Structures (left) and histograms of DNA content as measured by CellProfiler (right) are shown. Light gray lines depict DMSO control and black lines depict ploidy states of cells cultured with the respective compounds.
Acute megakaryoblastic leukemia (AMKL) is characterized by expansion of immature megakaryocytes and bone marrow myelofibrosis (Malinge et al., 2009). Pediatric AMKL is frequently associated with chromosomal abnormalities, including trisomy 21 in Down syndrome AMKL (DS-AMKL) and t(1;22), which leads to expression of the OTT-MAL fusion protein in non-DS-AMKL (Ma et al., 2001; Mercher et al., 2001). Mutations in GATA1, a transcription factor that is essential for proper growth and differentiation of megakaryocytes, are present in nearly all cases of DS-AMKL, while mutations in JAK3, MPL, KIT, and FLT3 are associated with a smaller subset of AMKL patients (Malinge et al., 2009; Wechsler et al., 2002). Although many DS-AMKL patients respond to current therapies, adults with non DS-AMKL have a very poor prognosis, with the vast majority relapsing within one year of the primary treatment (Tallman et al., 2000).
Given that leukemic blasts from AMKL patients are hyperproliferative and fail to undergo differentiation or polyploidization, we hypothesized that small-molecule inducers of polyploidization would drive these cells to exit the proliferative cell cycle and undergo terminal differentiation. These small-molecule probes could also serve to help understand the mechanism of polyploidization. Here we reveal that polyploidy inducers indeed show potent anti-leukemia activity in vitro and in vivo. Moreover, we report a successful and generally applicable multidisciplinary integrative approach to the target identification challenge. This approach allowed intelligent prioritization and testing of candidate targets, enabling us to show that AURKA kinase activity is an essential negative regulator of polyploidization in AMKL blasts and a therapeutic target for this subtype of leukemia. Moreover, the computational analysis led to the identification of five networks of kinases that control polyploidization. This new information will help reveal the mechanism by which cells decide to switch from a proliferative cell cycle to an endomitotic cycle and provide a mechanism-based rationale for therapy to treat AMKL.
Experimental Procedures
Compounds
JAK3 inhibitor VI, latrunculin B, K252a, PLK1 inhibitor, CDK1 inhibitor, CDK2 inhibitor and SU6656 were purchased from EMD Chemicals (Gibbstown, NJ). diMF, MLN8237, AZD1152, and AZD1152-HQPA (Mortlock et al., 2007) were prepared according to literature methods and characterization by 1H NMR (and optical rotation for diMF) was consistent with literature reports.
High-content chemical screening
4,000 CMK cells per well were seeded into black 384-well plates. Fifty nL of each compound was pin-transferred in duplicate into each well. After 72 hr, cells were fixed and stained with 1 µg/ml Hoechst 33342. The cells were imaged with a 20X objective at 9 sites within each well using ImageExpress Micro. CellProfiler was employed to identify isolated nuclei, and measure the integrated intensity of the DNA stain within each nucleus (Carpenter et al., 2006). The numerical data was analyzed using R Project to identify DNA content and to make histograms for each treatment. A compound was scored as a hit if the fraction of nuclear DNA content greater than the cutoff for the compound was significantly greater than that induced by DMSO. Confirmatory assays were conducted using 8 concentrations (0.16, 0.31, 0.63, 1.25, 2.5, 5, 10, and 20 µM) for each hit compound under the same imaging and data-analysis conditions.
Animal experiments
For drug treatment of non-transplanted mice, vehicle or test compound was given to mice by oral gavage twice a day for 7 or 14 days. Mice were sacrificed on day 14 after initiation of treatment. For the drug pre-treatment experiment, 6133/MPL cells were treated with vehicle or compound for 24 hr. Mice were sub-lethally irradiated at 600 cGy, and 106 live 6133/MPL cells were injected into the tail vein of each mouse. For drug treatment after transplantation, 1 million 6133/MPL cells were transplanted into sub-lethally irradiated mice. Forty-eight hr later, vehicle or compound was fed to mice by oral gavage twice a day. For drug treatment of mice transplanted with human AMKL blasts, bone marrow cells from a non DS-AMKL patient were injected into sub-lethally irradiated primary NSG (NOD/LtSz-scid IL2Rgc null) recipients at 300 cGy. Twelve weeks later, bone marrow cells were collected from primary NSG recipients and injected into sub-lethally irradiated secondary recipients. Five weeks after transplantation, the treatment was performed with vehicle, 30, or 60 mg/kg of diMF by oral gavage twice a day for 10 days and analyzed at the end of the treatment. Rock1-null mice have been previously described (Vemula et al., 2010). Aurora kinase A conditional knockout (Aurkaflox/flox) mice were generously provided by Dr. Terry Van Dyke of University of North Carolina at Chapel Hill (Cowley et al., 2009).
Affinity enrichment using SILAC (stable isotope-labeled amino acid in cell culture)-mediated quantitative proteomics
SILAC-labeled cells were used in affinity enrichment experiments using K252a-loaded affinity matrices and diMF as soluble competitor at 10- and 50-fold excess over K252a on bead. Proteins bound to the solid matrices were separated by SDS/PAGE and identified and quantified by high-performance mass spectrometry (MS). SILAC ratios from relative abundances of proteins enriched in the presence or absence of soluble competitor pull-down experiments were modeled using an empirical Bayes-based statistical framework to identify specific protein targets interacting with small molecules. Detailed methods for the experimental procedures are provided in the extended experimental procedures.
RNAi screen
For the RNAi screen, 1,000 CMK cells per well were seeded in 384-well plate format and treated with lentivirus for each individual shRNA. 24 hr post transduction, cells were selected with puromycin (3 µg/ml) and incubated for two days to allow knockdown. Vehicle control or 1 µM diMF was added to each well and incubated for 48 hr, following which cells were fixed and stained. The images were acquired and analyzed as in the small-molecule screen. Following Cell Profiler analysis of DNA content per cell, custom R scripts were designed to determine the relationship between Hoescht staining and DNA content. An shRNA was scored as a hit if the DNA was significantly greater than that induced by vector DNA in presence of DMSO or 1 µM diMF. A comparison was also made between these two conditions. To reduce the off-target effects of shRNA, a gene was called a hit only when two or more shRNAs of the gene scored as positive. The top 5% of genes in any one of three categories of comparison were considered to be hits.
Protein network analysis using Reactome
To assist with the interpretation of hits, we turned to network analysis using the protein-protein interaction database Reactome (Vastrik et al., 2007). The decision of which proteins to include was made for each component (SILAC, RNAi, KinomeScan) separately. Proteins detected by SILAC were analyzed using an empirical Bayesian method (Margolin et al., 2009) and we included those with false-discovery rates below 0.05. For RNAi, we included genes with p-values below 0.05 in any of the three modes (shRNA alone, shRNA with minimally effective dose of diMF, and difference between the two). For kinases profiled by Ambit KinomeScan we included those that had % activations reduced by diMF below 35%. We used random graphs with given expected degrees (Pradines et al., 2005) to assess the statistical significance and obtained a p-value of 7.1X10−64.
Statistics
For quantitative assays, treatment groups were reported as mean ± SD and compared using the unpaired Student’s t test. When multiple comparisons were necessary, one-way or two-way analysis of variance with post-test Bonferroni correction was used. Statistical significance was established at p less than or equal to 0.05, labeled as *, p < 0.05 and **, p < 0.01. Mouse survival data were evaluated by log-rank analysis, adjusted by multiple comparison test when necessary. The analysis was performed using GraphPad Prism Version 4.01 for Windows (GraphPad Software).
Results
A high-content screen identifies small molecules inducing polyploidization in human megakaryocytes
To identify compounds that induce megakaryocyte polyploidization, we developed an imaging assay capable of measuring DNA content in cultured AMKL cell lines (Fig 1). Small-molecule screening was performed using a human cell line derived from a patient with DS-AMKL (CMK) (Sato et al., 1989) and a chemically diverse small-molecule library. Following a three-day incubation, cells were fixed and stained with Hoechst dye, individual wells were imaged by automated epifluorescence microscopy (ImageXpress Micro; Molecular Devices), and then analyzed for nuclear morphology and fluorescence intensity with CellProfiler (Carpenter et al., 2006). SU6656, a Src kinase inhibitor known to induce polyploidization of a wide spectrum of cells was used as a positive control (Lannutti et al., 2005). This assay had an average Z’ factor (Zhang et al., 1999) >0.7 over the study. Among approximately 9,000 compounds screened, we identified 206 positives that significantly increased the fraction of cells with DNA content beyond a cutoff (between 4N and 8N) as compared to DMSO (Fig 1C and Table S1). Among these compounds were the expected microtubule-disrupting and -stabilizing agents, and actin-disrupting agents, including cytochalasin B and latrunculin B, which were predicted to cause alterations in spindle formation or cytokinesis and result in polyploidization. In addition, a number of compounds annotated as kinase inhibitors were identified, including dimethylfasudil (diMF, H1152P, BRD4911), reversine, K252a, and JAK3 inhibitor VI. Dose-ranging secondary studies of assay positives found 149 of the 206 compounds to exhibit an EC50 ≤ 100µM (Table S1).
A subset of assay positives induces both polyploidization and features of megakaryocyte differentiation
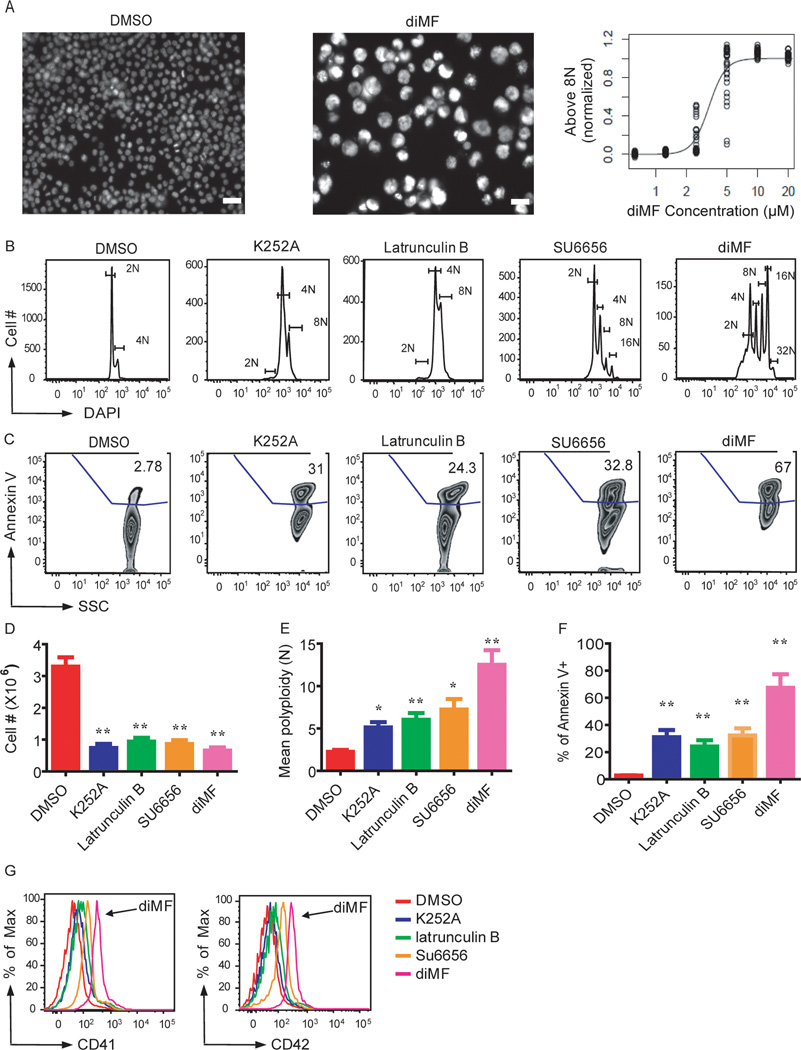
Despite the temporal association between polyploidization and up-regulation of megakaryocyte-specific genes, recent findings suggest that these two processes can be uncoupled (Huang et al., 2007). In order to assess whether small-molecule inducers of polyploidization also induced megakaryocyte maturation, we selected several compounds for more detailed analysis. This list included diMF and K252a; latrunculin B, an actin polymerization inhibitor; JAK3 inhibitor VI; reversine, a molecule that induces de-differentiation and polyploidization (D'Alise et al., 2008); NP003964, a natural product; and SU6656. Compared to SU6656, diMF induced much higher polyploidization in CMK cells over a much wider dose range (Fig 2B and data not shown). Although each of these compounds inhibited proliferation while inducing polyploidization and apoptosis, only diMF and NP003964 also induced expression of the megakaryocyte specific markers CD41 and CD42 (Fig 2 and data not shown).
Figure 2. Lead compounds induce polyploidization, expression of differentiation markers, and apoptosis of a human megakaryocytic cell line.
(A) Left, images of Hoechst-stained CMK cells treated with DMSO or diMF. Right, EC50 determination for diMF induction of polyploidization > 8N. Scale bar: 50 µm. (B–G) K252a (5 µM), latrunculin B (5 µM), SU6656 (4 µM) and diMF (5 µM) induced polyploidization (B,E), apoptosis (C,F), proliferative arrest (D) and expression of CD41 and CD42 (G) in CMK cells 72 hr after treatment. Representative flow cytometry plots are shown. Bar graphs depict mean ± SD of two independent experiments conducted in triplicate; * p<0.05, ** p<0.01
diMF was tested against a panel of megakaryocytic cell lines and found to induce robust polyploidization and expression of differentiation markers in every line tested, including CMS, CHRF, Meg01, K562, Y10 and G1ME (Stachura et al., 2006) cells (Fig S1A). diMF also induced polyploidization and CD41 expression in an AMKL cell line derived from ERG transgenic mice (tg-ERG) (L.G., Y.B, and S.I.; unpublished data), which express the ERG transcription factor under the control of the vav promoter (Fig S1B).
Next, we examined the effect of diMF on primary murine hematopoietic progenitor cells cultured in the presence of thrombopoietin (THPO). Treatment with diMF led to a dose-dependent increase in polyploidization and up-regulation of CD41 and CD42 expression in murine bone marrow-derived megakaryocytes (Fig S2A–C). At 5 µM, diMF upregulated CD41 and CD42 expression by 9- and 18-fold respectively, while increasing the mean polyploidy of CD41+ cells from 5.8N to 10.7N. diMF also induced polyploidization and up-regulation of CD41 and CD42 in GATA1s-KI fetal liver megakaryocytes, which mimic DS-AMKL in that they express the shortened leukemic isoform of GATA-1 called GATA-1s (Li et al., 2005; Wechsler et al., 2002) (Fig S2D and data not shown). Moreover, when human primary bone marrow mononuclear cells were cultured in the presence of THPO, diMF induced polyploidization of CD41+ cells, but not the CD41- fraction (Fig S2E). Thus, diMF selectively induces polyploidization of the megakaryocyte lineage and can override the proliferative defect caused by mutations in GATA1.
diMF has anti-leukemia activity
A subset of pediatric non-DS-AMKL patients harbors the (1;22)(p13;q13) chromosomal translocation, which results in expression of the OTT-MAL fusion protein. Transgenic mice that express OTT-MAL develop AMKL with a low penetrance and long latency, while recipients of OTT-MAL bone marrow expressing MPLW515L rapidly develop AMKL with high penetrance (Mercher et al., 2009). 6133/MPL cells, which were derived from OTT-MAL transgenic mice and engineered to express MPLW515L, are quite sensitive to diMF: treatment of 6133/MPL cells with 3 µM diMF led to strong up-regulation of CD41 and CD42, polyploidy, and apoptosis (Fig 3A–B and data not shown).
Figure 3. diMF displays anti-leukemic activity both in vitro and in vivo.

(A,B) diMF-induced polyploidization (A) and expression of CD41 and CD42 (B) in 6133/MPL cells 48 hr after treatment. Data are representative of two independent experiments. (C) Transplantation of 6133/MPL cells causes AMKL in sub-lethally irradiated recipient mice. H&E-stained spleen sections revealed massive infiltration of tumor cells in transplanted mice, but not control mice. Scale bar: 50 µm. (D) Survival curve of mice transplanted with 6133/MPL cells pretreated with vehicle or 10 µM diMF for 24 hr. N=7 mice per group; p=0.0002. (E) Measurement of drug concentration in plasma after a single dose of diMF. C57Bl/6 mice were dosed orally with 66 mg/kg of diMF, and plasma concentrations of the drug were assessed at multiple time points post-treatment; n=3 animals per time point. The insert depicts decay over 2 hr. (F) Survival curve of mice transplanted with 1 million 6133/MPL cells and treated with vehicle or diMF at 33 or 66 mg/kg for 10 days, beginning two days after transplantation; n=7 mice per group. Results are representative of two independent experiments. (G) diMF induction of polyploidization of 6133/MPL cells in vivo. Forty-eight hr after transplantation, mice were fed vehicle or 66 mg/kg diMF by oral gavage twice a day for 3 days and the DNA content of the transplanted cells in bone marrow was evaluated by flow cytometry; n=3 animals per group. (H) diMF induced polyploidization of human non-DS-AMKL blasts. Human CD41+ non-DS-AMKL blasts from primary NSG recipients were treated with vehicle or 5 µM diMF for 6 days. (I, J, K) diMF reduced tumor burden and induced polyploidization of NSG mice transplanted with human non-DS-AMKL blasts from primary NSG recipients. Secondary NSG recipients were treated with vehicle or diMF at 30 or 60 mg/kg for 10 days. diMF reduced human CD41+ cells in spleen (I), peripheral blood (J), and induced polyploidization (K) of human CD41+ cells in the spleen. (L) diMF inhibition of DS-AMKL blast colony formation. Bone marrow specimens from pediatric patients with DS-AMKL were cultured in Megacult-C media with THPO and either DMSO or 5 µM diMF for 10–12 days. Representative images of anti-CD41 antibody stained colonies are shown; scale bar: 100 µm. Bar graph depicts the mean ± SD of relative colony numbers following treatment; * p<0.05.
To assess whether diMF induces an irreversible proliferative arrest that would prevent development of leukemia in transplant recipients, we treated 6133/MPL cells with vehicle or 10µM diMF for 24 hr and then transplanted 1 million viable cells into recipient mice. Transplantation of vehicle-treated 6133/MPL cells into sub-lethally irradiated C57Bl/6 mice led to a fulminant AMKL with a short latency, with all of the animals developing leukemia and dying within 3 weeks (Fig 3C–D). The disease was characterized by splenomegaly, disruption of splenic architecture and a high proportion of GFP-labeled 6133/MPL cells in the peripheral blood, bone marrow, spleen, and liver (data not shown). Strikingly, none of the mice transplanted with diMF-pre-treated 6133/MPL cells developed leukemia (Fig 3D). Thus, brief exposure of 6133/MPL cells to diMF is sufficient to block their leukemia-inducing activity.
Next, we assessed the oral pharmacokinetics of diMF in vivo. C57Bl/6 mice were fed a single dose of 66 mg/kg diMF and plasma concentrations were measured. The mean maximum concentration achieved was 5.8 µM (Cmax) at 5 min (Tmax) (Fig 3E), which is higher than the dose of diMF necessary to induce maximum polyploidization of 6133/MPL cells. Having shown biologically active concentrations of diMF were achieved in vivo after oral administration, we fed healthy C57Bl/6 mice with vehicle or diMF by oral gavage twice a day for 7 days and evaluated their body weight and hematopoietic indices. diMF was well tolerated: we did not observe significant changes in body weight or peripheral blood indices, including platelet counts (Fig S3A–E).
Having established that diMF is well tolerated in vivo, we transplanted sub-lethally irradiated recipient C57Bl/6 mice with 1 million 6133/MPL cells. Forty-eight hr after transplantation we detected GFP positive cells in the spleen and bone marrow of recipient mice, confirming engraftment of the 6133/MPL cells. Mice were then fed vehicle or diMF twice a day for 10 days. Whereas all the vehicle-treated mice died within 20 days, 43% of the mice treated with 66 mg/kg diMF and 29% of mice fed 33 mg/kg diMF survived at least 70 days post transplantation (Fig 3F). diMF also significantly increased the survival of mice when treatment was initiated 7 days after transplantation, when the percentage of GFP positive cells in the peripheral blood was near 20% (data not shown). Importantly, we observed a significant increase in DNA content, expression of CD41, and apoptosis of 6133/MPL cells in the bone marrows of recipient mice 72 hr after treatment with 66 mg/kg diMF (Fig 3G and Fig S3F–G). We also noted that diMF reduced the number of GFP positive 6133/MPL cells in recipient animals (Fig S3H–K). These findings reveal that diMF promotes the survival of transplanted mice by inducing polyploidization of 6133/MPL cells in vivo.
We next treated human non-DS-AMKL blasts isolated from primary immunodeficient (NOD/LtSz-scid IL2Rgc null; NSG) recipient mice with vehicle or diMF. In vitro, 5 µM diMF significantly increased polyploidization and inhibited proliferation of the AMKL blasts (Fig 3H). Secondary recipient NSG mice transplanted with non-DS-AMKL blasts showed decreased tumor burden of human CD41+ cells (Fig 3I–J) when treated with diMF at 30 mg/kg or 60 mg/kg. diMF also significantly increased the polyploidization of human CD41+ cells in the spleens of recipient mice (Fig 3K). Finally, we cultured primary human DS-AMKL bone marrow specimens with DMSO or diMF and monitored the growth of the leukemic cells in colony assays. In all 5 patient samples studied, diMF significantly reduced the colony formation of AMKL blasts (Fig 3L).
An integrated, multidisciplinary target identification method identifies candidate physiologic targets of diMF in AMKL
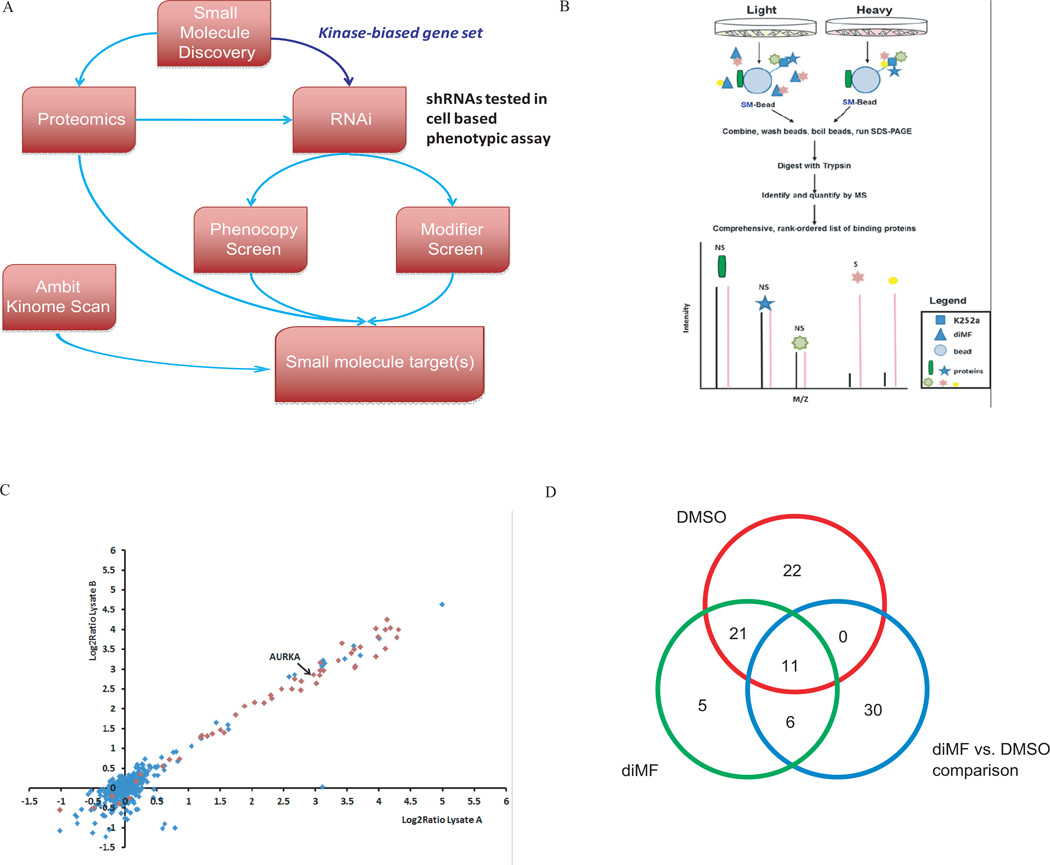
To address the general challenge of determining the mode of action of small molecules identified from phenotypic screens (Terstappen et al., 2007), we implemented a broadly applicable, multidisciplinary, integrative approach for identifying the targets of diMF (Fig 4A). diMF is an ATP competitive inhibitor of several kinases that include the Rho kinase family (Ikenoya et al., 2002), but other Rho kinase inhibitors such as fasudil did not fully recapitulate the strong phenotype induced by diMF in AMKL-derived cells (data not shown). Thus we began with the hypothesis that diMF produced this phenotype by acting as a kinase inhibitor, but perhaps not exclusively through Rho kinase family members.
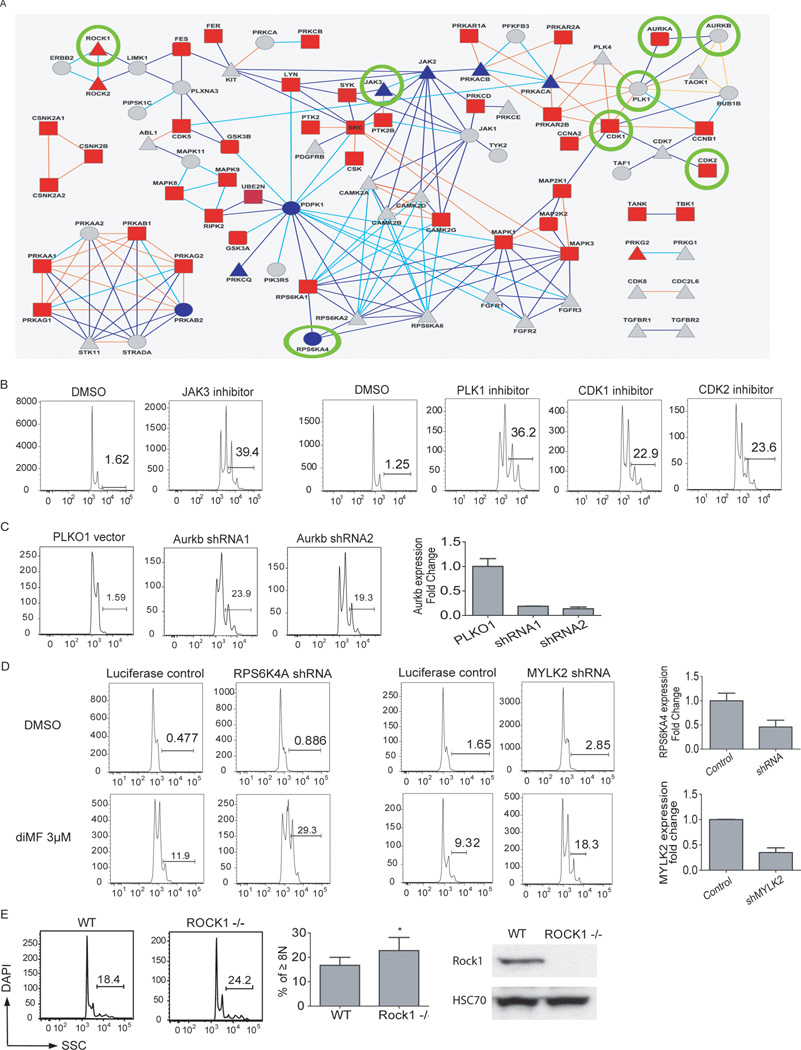
Figure 4. An integrated target ID approach identifies Aurora kinase A as a target of diMF in AMKL.
(A) Schematic representation of the integrated target identification workflow. CMK cells were transduced with shRNAs targeting the human kinome and the effects of knockdown were studied in the presence of DMSO (phenocopy screen) or a minimally effective dose of diMF at 1 µM (modifier screen). (B) Quantitative proteomic strategy for identification of specific diMF-protein interactions. Proteins in cell populations were fully metabolically labeled with light (yellow) and heavy amino acids lysine and arginine (red) using SILAC methodology. Cell lysates were incubated either with K252a-loaded beads (K252a-Beads) and excess soluble diMF competitor or K252a-Beads alone. Proteins interacting directly with diMF or via secondary and/or higher-order interactions (marked “S” for specific) were enriched in the heavy state over the light and identified with differential ratios in the mass spectrometer. Nonspecific (via binding to the bead) or K252a (NS) interactions of proteins were enriched equally in both states and have ratios close to unity. (C) Identification of significant targets of diMF using affinity proteomics with SILAC. Scatter plot of two replicate experiments of diMF at 50-fold excess over K252a on beads. Each data point is a single protein with kinases (Manning et al., 2002), represented as red diamonds and blue diamonds denoting non-kinases. Six hundred ninety-eight proteins were identified and quantified in at least three experiments, resulting in 68 proteins with a combined q-value < 0.05. (D) Venn diagram of genes scored as hits in each type of comparisons.
First, we performed an Ambit KinomeScan analysis and compared the kinase binding profiles of diMF to those of the closely related compound, fasudil (Fabian et al., 2005; Karaman et al., 2008). From 402 purified kinases tested, we identified 117 kinases whose binding to the immobilized ligand was inhibited by more than 65% in the presence of 5 µM diMF relative to the control that contained no competing ligand (Table S2). In contrast, only 27 kinases were inhibited to a similar extent when fasudil was used as the competing ligand. Among the differentially affected kinases, the Aurora kinase family (A, B, and C) was notable for being strongly inhibited by diMF, but not at all by fasudil.
To identify protein binders of diMF in CMK cells, we used a modified version of our previously published method (Ong et al., 2009) (Fig 4 B). Using the broad specificity kinase ligand, K252a (shown to induce polyploidization; Fig 2B), immobilized on beads as bait (Ong et al., 2009), and pre-incubating CMK cell lysates with excess soluble diMF, we identified 68 proteins that were significantly and specifically competed away from K252a by diMF (Table S3, Fig 4B–C). The majority of these proteins are kinases or known to associate with kinases.
To obtain an orthogonal dataset to the proteomic and biochemical methods, we performed an RNAi screen to identify kinases whose knockdowns induced polyploidization in CMK cells, either on their own (phenocopy screen) or in conjunction with a dose of diMF that produces 5–10% of its maximum polyploidization induction effect (1 µM; modifier screen) (Fig 4A, Fig S4). The modifier screen complements the phenocopy screen in that combining a low dose of diMF with RNAi-based gene knockdown may provide selectivity for genes directly involved in the diMF mechanism of action. Large increases in high-ploidy cells were produced by hit shRNAs; in untreated control wells, 3–5% of cells were high-ploidy whereas the top 2% of shRNAs produced wells with 30–80% high-ploidy cells in both the DMSO and diMF screens. Genes were ranked for the effect on ploidy of their top two scoring shRNAs (see Methods for details). Knockdown of 54 kinases increased the fraction of high-ploidy cells in DMSO treatment. In cells treated with 1 µM diMF, knockdown of 43 kinases increased the fraction of high-ploidy cells versus diMF treatment alone. We also ranked shRNAs by their differential effect in the two screens; 47 genes showed significant increase in induction of polyploidy upon knockdown under diMF treatment versus vehicle alone (Table S4 and Fig 4D). Using these three criteria, a total of 95 distinct genes were selected for integrated analyses.
Aurora A kinase is a target of diMF and a mediator of polyploidization of malignant megakaryocytes
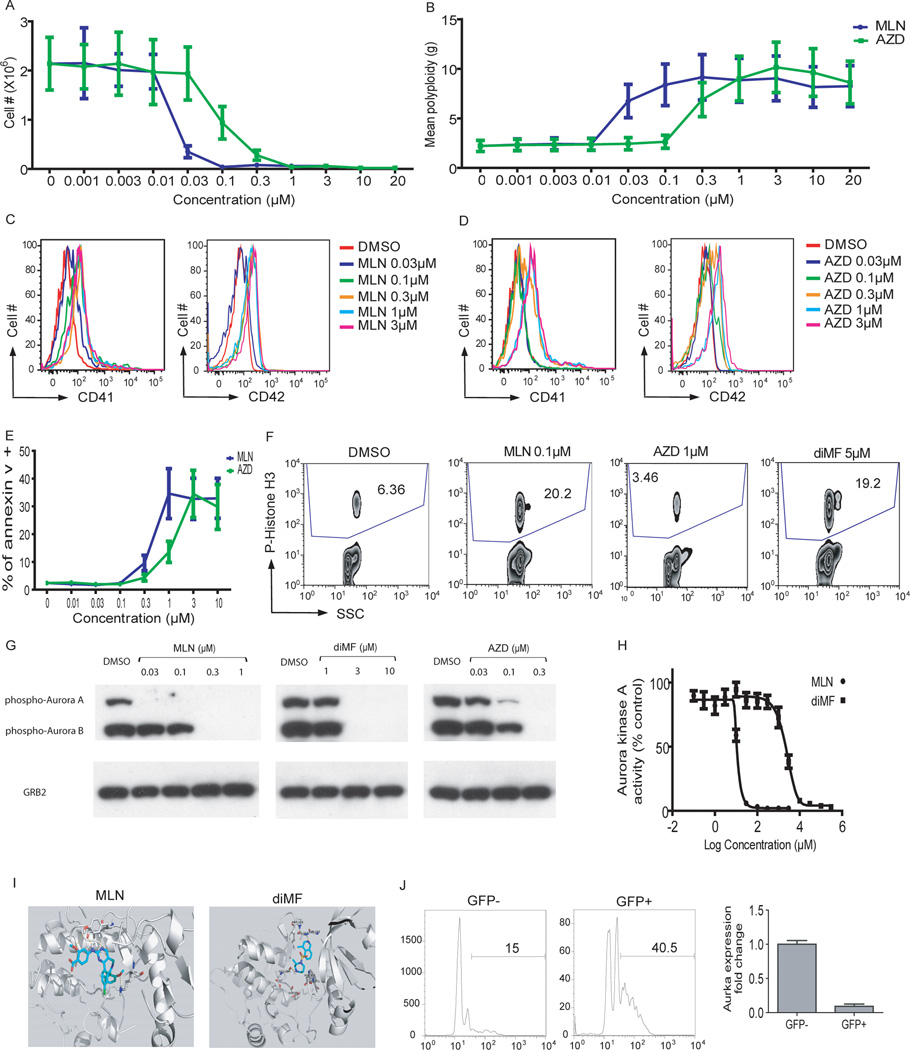
We performed an integrated analysis of the results of the KinomeScan, the SILAC-based protein binding assay, and the RNAi screen for polyploidization. We assigned combined p-value scores based on the p-values of each individual approach and evidence counts, and identified 15 kinases with scores less than 0.05 (Table S5). We prioritized the candidates for follow-up studies based upon the availability of biologically annotated, highly selective small-molecule kinase inhibitors. For example, although little is known about AURKA with respect to polyploidization, it is known to regulate microtubule-organizing center localization, chromosome dynamics, and histone H3 phosphorylation in oocytes (Ding et al., 2011), and is required for bipolar spindle formation and early development (Cowley et al., 2009). Several small-molecule inhibitors of AURKA have been developed, including the highly selective compound MLN8237, which displays 200-fold selectivity for AURKA relative to AURKB in cells (Gorgun et al., 2010; Manfredi et al., 2011). Expression of MLN8237-resistant AURKA mutants has previously validated AURKA as the target of this molecule in cells (Sloane et al., 2010). Of note, AURKB ranked highly in the biochemical and RNAi screens, but was not detected in the quantitative proteomics experiment.
To determine if inhibition of Aurora kinases could phenocopy diMF, we treated CMK cells with their inhibitors and assayed proliferation, survival, and megakaryocyte cell-surface marker expression. Both MLN8237, a specific inhibitor of Aurora A kinase, and AZD1152-HQPA, a specific inhibitor of Aurora B kinase, restricted proliferation, induced robust polyploidization, differentiation as assessed by CD41 and CD42 expression, and apoptosis of CMK cells (Fig 5A–E). In the 6133/MPL murine cell line, they increased polyploidization, CD41 and CD42 expression, and apoptosis (Fig S5A–D). Moreover, both compounds induced proliferation arrest and megakaryocyte lineage-specific surface marker expression of tg-ERG cells (data not shown). MLN8237 induced polyploidization and expression of CD41 and CD42 in primary mouse bone marrow cells cultured ex vivo (Fig S5E–G), and both MLN8237 and AZD1152-HQPA induced robust and selective polyploidization of primary human megakaryocytes expanded from human CD34+ cells (Fig S5H and data not shown).
Figure 5. Inhibition of Aurora kinase A phenocopies diMF.
(A–E) MLN8237 and AZD1152-HQPA induced proliferation arrest (A), polyploidization (B), expression of CD41 and CD42 (C,D), and apoptosis (E) in CMK cells 72 hr after treatment. Data are representative of two experiments conducted in duplicate. Line graphs depict mean ± SD. (F) MLN8237 and diMF increased the phosphorylation of histone H3, while AZD1152-HQPA decreased its levels. (G) MLN8237, AZD1152-HQPA, and diMF differentially inhibited phosphorylation of Aurora kinases. CMK cells were incubated with 0.1 µM paclitaxel for 18 hr, then DMSO, MLN8237, AZD1152-HQPA, or diMF was added and incubated for 2 hr. The degree of phosphorylation of the Aurora kinases in each sample was determined by Western blot. Treatment of cells with 1 µM AZD1152-HQPA also led to complete loss of phospho-AURKA and AURKB (data not shown). (H) MLN8237 and diMF inhibit AURKA. Purified Aurora kinase A was incubated with MLN8237 or diMF and the change of AURKA phosphorylation was determined by spectrophotometry. Data are representative of two experiments conducted in triplicate. (I) Docking studies were performed to evaluate the binding of MLN8237 and diMF to Aurora kinase A using Schrodinger software. Both MLN8237 and diMF showed a strong hydrogen-bond network with the hinge residues of AURKA. (J) Excision of Aurka leads to enhanced polyploidization of megakaryocytes. Bone marrow cells from Aurkaflox/flox mice were transduced with MIGR1-Cre-IRES-GFP and the cells were cultured in the presence of THPO for 72 hours. (Left) The DNA contents of CD41+ cells from GFP+ (Cre-expressing) or GFP- (without Cre expression) populations of the same culture are shown. (Right) The levels of Aurka mRNA in sorted GFP-positive or GFP-negative cells were assayed by qRT-PCR. Data are representative of two experiments.
We next compared the effects of diMF, AZD1152-HQPA, and MLN8237 on cellular biomarkers. diMF and MLN8237 induced a similar degree of accumulation of phospho-histone H3, and both compounds reduced AURKA autophosphorylation (Fig 5F–G), two hallmarks of selective AURKA inhibition (Carmena and Earnshaw, 2003; Lok et al., 2010). Importantly, 0.1 µM MLN8237, which induced polyploidization, proliferation arrest, up-regulation of megakaryocyte lineage specific markers, and apoptosis of CMK cells, inhibited autophosphorylation of AURKA, but not AURKB. This result indicates that MLN8237 inhibits AMKL cell growth and induces polyploidization by selective inhibition of AURKA. In contrast, AZD1152-HQPA led to a reduction in phospho-histone H3 and reduced phosphorylation of both Aurora A and B kinases at the concentrations that inhibit AMKL cell growth (Fig 5F–G and data not shown). Of note, diMF inhibited phosphorylation of both Aurora A and B kinases, but led to a dramatic increase in the levels of phospho-histone H3 (Fig 5F), consistent with a dominant inhibition of AURKA. Inhibition of AURKB phosphorylation by diMF at 3 and 5µM parallels the finding that diMF binds to Aurora kinase B in the Ambit KinomeScan assay (Table S2). A purified kinase assay further confirmed that diMF inhibits AURKA, albeit more weakly than MLN8237 (Fig 5H). Although RNAi-targeted knockdown of AURKB or pharmacologic inhibition by AZD1152-HQPA of its encoded kinase induces polyploidization in megakaryocyte precursors, this phenotype is not exclusive to the megakaryocyte lineage (Wilkinson et al., 2007). The lineage-selective induction of polyploidization appears, however, to result from inhibition of AURKA, a major target of diMF mode of action in megakaryocyte precursors.
We also performed in silico docking studies to compare the binding of diMF and MLN8237 to Rho kinase I (ROCK1), a known target of diMF, and to Aurora A kinase. Using the LigPrep, Macromodel, and Glide-XP (Extra Precision) modules incorporated in Schrodinger software package, we found both MLN8237 and diMF are predicted to form a strong hydrogen-bond network with the hinge residues of the ATP-binding site of Aurora kinase A (Fig. 5I). In contrast, diMF, but not MLN8237, appears to bind to the active site of ROCK1. This failure of MLN8237 to dock with ROCK1 further supports that Aurora A kinase is a common target for both diMF and MLN8237.
To further confirm that loss of function of AURKA promotes polyploidization of megakaryocytes, we assayed the effect of Aurka deletion on megakaryocytes. Bone marrow cells from Aurka conditional knockout mice (Aurkaflox/flox) were infected with a retrovirus harboring Cre recombinase (MIGR1-Cre-IRES-GFP) and then cultured for 72h in the presence of THPO to foster megakaryocyte development. As predicted, depletion of AURKA by expression of Cre (GFP+ fraction) led to a marked increase in the degree of polyploidization of CD41+ megakaryocytes (Fig 5J), reminiscent of the phenotypes induced by MLN8237 and diMF in primary mouse bone marrow cells (Fig S2A and Fig 5G). No appreciable depletion of Aurkb mRNA was detected in GFP+ cells (data not shown). Furthermore, expression of Cre in wild-type bone marrow cells does not alter polyploidization of megakaryocytes (Wen et al., 2011; Wen et al., 2009).
MLN8237 shows potent anti-leukemia activity in vitro and in vivo
Given that MLN8237 and AZD1152-HQPA, acting by distinct mechanisms, affected the in vitro growth of CMK and 6133/MPL cells in a manner similar to diMF, we investigated the extent to which these AURK-specific compounds could be used as anti-megakaryocytic leukemia agents. As seen with diMF (Fig 3C–D), 24-hr pre-treatment of 6133/MPL cells with MLN8237 significantly reduced their ability to induce leukemia in recipient mice, with 80% of the animals surviving up to 120 days (Fig 6A). In contrast, 24-hr pre-treatment with AZD1152-HQPA failed to significantly interfere with leukemia development in vivo (Fig 6A). Pharmacokinetic studies, performed in C57Bl/6 mice following a single oral administration of 15 mg/kg MLN8237 revealed excellent bioavailability (Fig 6B). Rapid absorption was observed reaching the peak concentration at 0.5 hr (Tmax) with a maximum concentration of 34.8 µM (Cmax). Mean plasma concentrations remained above 0.5 µM for at least 12 hr with a moderate elimination (terminal half-life of 3.1 hr). These results indicate that MLN8237 is easily absorbed orally, has very high exposure in circulation, and demonstrates moderate metabolism in vivo. Moreover, MLN8237 is well tolerated in mice (Maris et al., 2010). Healthy animals fed 15 mg/kg MLN8237 twice a day for 2 weeks using a dosing regimen of five days on, two days off, showed no changes in body weight or peripheral blood counts (Fig S6).
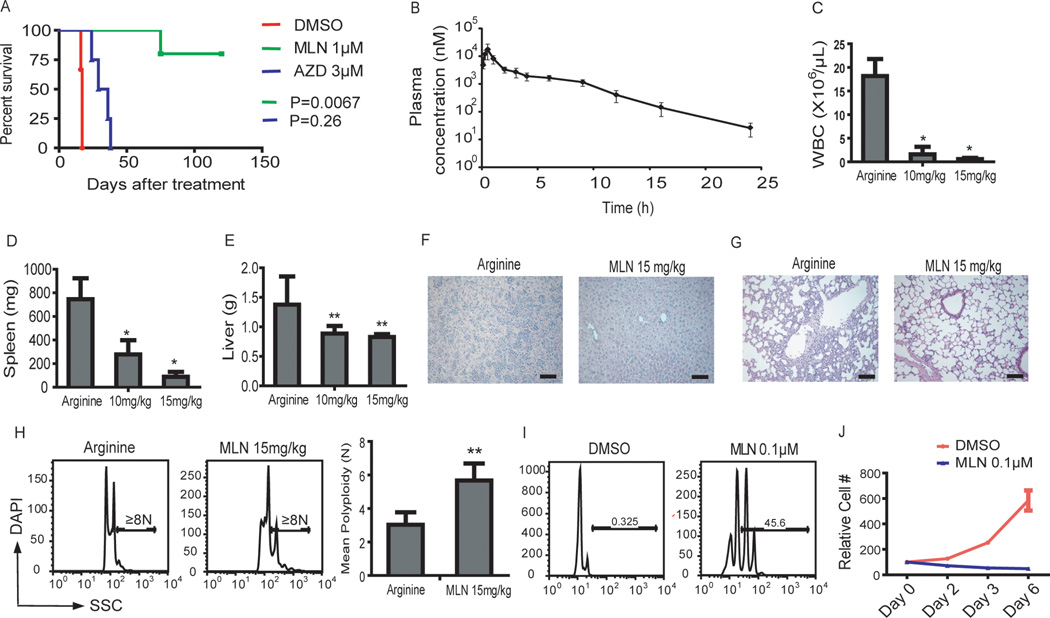
Figure 6. MLN8237 shows anti-leukemic activity in vitro and in vivo.
(A) Pretreatment with MLN8237, but not AZD1152-HQPA, impaired the ability of 6133/MPL cells to induce leukemia. 6133/MPL cells were incubated with 1 µM MLN8237 or 3 µM AZD1152-HQPA. One million live cells were transplanted to mice and survival of the mice was monitored. n=6 (DMSO), n=5 (MLN8237) and n= 4 (AZD1152-HQPA). MLN vs DMSO, p=0.067; AZD vs DMSO, p=0.26. (B) Measurement of drug concentration in plasma after a single dose of MLN8237. C57Bl/6 mice were dosed orally with 15 mg/kg of MLN8237, and plasma concentrations of the drug were assessed at different time points post-treatment; n=3 animals per time point. (C–G) MLN8237 reduced tumor load of 6133/MPL cell transplanted mice. Forty-eight hr after transplantation of 6133/MPL cells, mice were fed arginine (solvent for MLN8237) or MLN8237 at 10 and 15 mg/kg by oral gavage twice a day for 10 days. MLN8237 reduced white cell count in the peripheral blood (C) and decreased spleen and liver weight of transplanted mice (D,E). MLN8237 reduced infiltration of megakaryoblasts in the liver (F) and lung (G) in transplanted mice detected by H&E staining of tissue sections. (H) MLN8237 induced polyploidization of 6133/MPL cells in vivo. Forty-eight hr after transplantation of 6133/MPL cells, mice were given arginine or 15 mg/kg MLN8237 by oral gavage twice a day for 3 days and the DNA content of the transplanted cells in bone marrow was evaluated by flow cytometry. Left, representative flow plots. Right, Bar graph of mean ± SD; ** p<0.01. n=4 animals per group. (I, J) MLN8237 induced polyploidization and inhibited proliferation of human non DS-AMKL blasts. Human CD41+ non-DS-AMKL blasts isolated from primary NSG recipients were treated with DMSO or 0.1 µM MLN8237 for 6 days. Results are representative of two independent experiments in duplicate. Error bars represent mean ± SD; * p<0.05, ** p<0.01.
We treated 6133/MPL-transplanted mice with 15 mg/kg MLN8237 for 2 weeks and compared leukemia burden with animals treated with vehicle alone. MLN8237 significantly reduced peripheral white blood cell count and spleen and liver weights of transplanted animals (Fig 6C–E). There was also a striking reduction of infiltration of leukemic cells (Fig 6F–G). Similar to diMF, MLN8237 induced polyploidization of the malignant cells in vivo (Fig 6H). MLN8237 also induced both polyploidization and proliferative arrest of human non-DS AMKL cells cultured ex vivo (Fig 6I–J). Taken together, MLN8237, like diMF, displayed potent anti-megakaryocytic leukemia activity both in vitro and in vivo.
Network analysis reveals five networks that control polyploidization
Next, we sought to use our experimental data from the proteomic, biochemical and functional screens to infer the broader network of interacting proteins that lead to megakaryocyte polyploidy and differentiation. We performed a network analysis using the protein-protein interaction database Reactome (Vastrik et al., 2007). Reactome analysis integrating the data from the three approaches yielded 117 proteins that were mapped to 5 networks with 116 nodes and 194 connections in the Reactome database (Fig 7A). As expected, genes that control cytokinesis, including ROCK1, ROCK2, Aurora B, and polo-like kinases, were present. Known negative regulators of megakaryopoiesis, the SRC kinase LYN and PTK2 (focal adhesion kinase, FAK) (Hitchcock et al., 2008; Lannutti et al., 2006) were also evident, along with known mediators of thrombopoietin signaling, the JAK kinases. Unexpected factors included several members of the MAP kinase pathway, the CAM kinase family, and Aurora kinase A.
Fig 7. Pathways that regulate polyploidization of megakaryocytes.
(A). Reactome analysis integrating the data from the KinomeScan, SILAC, and RNAi screen yielded 117 proteins that were mapped to 116 nodes and 194 connections. In the protein network, shapes of nodes correspond to the source; squares for SILAC, circles for RNAi, rounded squares for both SILAC and RNAi, triangles for Kinome scan only. Colors of the nodes correspond to false-discovery rate of SILAC ratio in the range 0.05 (red) – 1.0 (blue) or gray for proteins not detected by SILAC. Colors of connections correspond to the type of interaction: direct complex (orange), indirect complex (yellow), reaction (blue), neighboring reaction (cyan). Thick connections mark pairs of proteins not distinguished from each other by SILAC. Kinases that were validated in separate experiments are circled. (B–E). Validation of kinases in the network. (B) Induction of CMK polyploidization after 72 hours of treatment with JAK3 inhibitor VI (1 µM), PLK1 inhibitor (1 µM), CDK1 inhibitor (3 µM) and CDK2 inhibitor (3 µM) is shown. (C) Knockdown of Aurkb induced polyploidization of megakaryocytes. 6133/MPL cells were transduced with PLKO1 control vector or shRNA against Aurkb. (D) Knockdown of RPS6KA4 or MYLK2 sensitized CMK cells to diMF treatment. CMK cells transduced with luciferase control viruses or shRNAs against RPS6KA4 or MYLK2 were cultured with DMSO or diMF (3 µM) for 72 hr. The extent of gene knockdown as assessed by qRT-PCR is shown. Results are representative of two independent experiments performed in duplicate. (E) (Left) Megakaryocytes (CD41+) derived from the bone marrow of Rock1-null mice showed increased degree of polyploidization relative to megakaryocytes from their wild-type littermates. (Middle) Bar graphs depict the percentages of cells with DNA contents ≥8N. Error bars represent mean ± SD; * p<0.05, n=5 mice per group. (Right) Expression of ROCK1 was assessed by western blot in extracts from murine bone marrow mononuclear cells.
We confirmed the polyploidy-inducing effects of inhibition or knock-down of a subset of kinases. First, small-molecule inhibitors of JAK3, PLK1, CDK1 and CDK2 induced polyploidization of CMK cells (Fig 7B). Second, knockdown of Aurora kinase B was observed to cause a robust increase in the extent of polyploidization of megakaryocytes (Fig 7C). Third, although knockdown of RPS6KA4 or MYLK2 did not induce polyploidy on their own, reduced expression of these kinases sensitized CMK cells to diMF treatment (Fig 7D). Finally, analysis of the ploidy state of megakaryocytes derived from Rock1-null mice and their wild-type littermates demonstrated that loss of Rock1 leads to increased polyploidization in vivo (Fig 7E). Together, these data reveal that the protein network, which is based on the experimental data from our proteomic and genomic studies, can serve as road map for further studies of function of genes in megakaryocyte polyploidization and development and their potential application to AMKL.
Discussion
In general, cell-based, phenotypic approaches for initial discovery of novel probes provides a rich dataset, but efforts to determine how promising leads might be exploited for therapy is often complicated by uncertainty regarding the cellular target. This problem is clearly evident in the case of diMF, which is a broad kinase inhibitor. Each of the individual approaches to target identification used in this study identified a large number of possible targets that might not have been tractable or warranted follow-up given only one line of evidence. We show that the ability to integrate approaches and organize the disparate data types in a disciplined and rigorous manner led to testable hypotheses and not only identified the physiologically relevant target of diMF, but also a potential therapeutic target for AMKL. Moreover, network analysis of these data constitutes the first comprehensive analysis of kinases that control the endomitotic process.
Small molecules that induce megakaryocyte polyploidization, such as diMF, appear to be promising therapeutic agents for AMKL for multiple reasons. First, since diMF targets polyploidization, a normal element of megakaryocyte differentiation and maturation, we predict that it would be active against all subtypes of AMKL regardless of their genetic alterations. Indeed, we show that diMF inhibits proliferation and induces polyploidization and up-regulation of megakaryocyte markers of cells with GATA-1 and MPL mutations as well as cells harboring +21 or the (1;22) translocation. However, diMF did not induce platelet production of GATA-1 mutant cells. This finding suggests that diMF cannot overcome all of the requirements for key developmental regulators in terminal differentiation. A second advantage to the use of these compounds in AMKL is illustrated by the ability of diMF and MLN8237 to block the growth of cells that express the MPLW515L-activating allele associated with human myeloproliferative disorders. Thus, we predict that polyploidization therapy may also be useful for disorders that involve hyperproliferation of megakaryocytes, such as essential thrombocytosis (ET) and primary myelofibrosis (PMF). The third benefit lies in the propensity of megakaryocytes to become polyploid. diMF and MLN8237 induced robust polyploidization of the CD41+, but not CD41-negative cells, reflecting the inherent susceptibility of megakaryocytes to polyploidization-inducing agents.
A recent study has demonstrated that ROCK1 is required for the survival and proliferation of leukemia blasts that harbor activated oncogenic forms of KIT, FLT3, and BCR-ABL (Mali et al., 2011). Knock-down of ROCK1, or inhibition with diMF or fasudil, restricted the growth of these leukemia cells both in vivo and in vitro. It is interesting to note that diMF thus shows activity against multiple forms of AML through distinct targets; ROCK1 in non-megakaryocytic AML blasts that bear activated KIT, FLT3, or BCR-ABL, and AURKA in megakaryocytic AML. The lack of activity of fasudil in AMKL provides further evidence that diMF inhibits different kinase pathways in the two subtypes. Of note, the small-molecule inhibitor MLN8237 is under clinical investigation for a variety of tumors, including acute myeloid leukemia. Despite the notion that Aurora kinase inhibitors should broadly be considered for treatment of AML, however, our studies are the first to suggest that MLN8237 (Alisertib) would be especially effective against the megakaryocytic leukemia sub-type.
Supplementary Material
Highlights.
-
○
Novel screening strategy identified over 200 compounds and five kinase networks that regulate polyploidization.
-
○
Implemented a broadly applicable novel multidisciplinary integrated target identification approach.
-
○
Aurora kinase A activity mediates megakaryocyte polyploidization and induces markers of differentiation.
-
○
Induction of polyploidization and differentiation is a promising new strategy for treating AMKL.
Acknowledgements
The authors thank Sandeep Gurbuxani, Alex Minella, and Lou Doré for critical reading of the manuscript, and Bang Wong for valuable advice on figures of the manuscript. This research was funded by grants from the Samuel Waxman Cancer Research Foundation (JDC and SI), the US Israel Binational Science Foundation (to SI and JC), the Leukemia and Lymphoma Society Translational Research Program (JDC), the Children with Leukaemia UK (SI), the Leukemia Research Foundation (YB), and by NIH grants CA101774 (JDC), HL077177 (RK), HL075816 (RK) and HL081111 (RK). Other support included an NIH grant to AEC supporting CellProfiler (GM089652), an NIH grant supporting screening informatics (U54 HG005032), NIH Genomics Based Drug Discovery U54 grants Discovery Pipeline RL1-CA133834 and Driving Medical Projects RL1-GM084437, administratively linked to NIH grants RL1-HG004671 and UL1-DE019585 (AEC, PAC, VD, CM, AS, CAS, and MS). YB is a European Hematology Association Fellow. In vivo treatment of human AMKL samples was supported by Foundation Gustave Roussy and José Carreras Leukemia Foundation- European Hematology Association (TM), CEA-EA, Ligue Nationale Contre le Cancer (FP), Association Laurette Fugain (FP), Société Française d’Hématologie (BG, FP), Foundation pour la Recherche Médicale (CT). The authors would also like to thank Jason Berman, Soheil Meshinchi, Todd Alonzo, and Sommer Castro and the Children’s Oncology Group (COG) for their assistance with DS-AMKL specimens. Research with DS-AMKL samples was supported by the Chair’s Grant U10 CA98543 (to COG) from the National Cancer Institute (NCI). The project has also been funded in part with Federal funds from the NCI’s Initiative for Chemical Genetics under Contract N01-CO-12400. The content of this publication is solely the responsibility of the authors and does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. A part of this work was performed by the Northwestern University ChemCore at the Center for Molecular Innovation and Drug Discovery (CMIDD), which is funded by the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bluteau D, Lordier L, Di Stefano A, Chang Y, Raslova H, Debili N, Vainchenker W. Regulation of megakaryocyte maturation and platelet formation. J Thromb Haemost. 2009;7(Suppl 1):227–234. doi: 10.1111/j.1538-7836.2009.03398.x. [DOI] [PubMed] [Google Scholar]
- Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley DO, Rivera-Perez JA, Schliekelman M, He YJ, Oliver TG, Lu L, O'Quinn R, Salmon ED, Magnuson T, Van Dyke T. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol Cell Biol. 2009;29:1059–1071. doi: 10.1128/MCB.01062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alise AM, Amabile G, Iovino M, Di Giorgio FP, Bartiromo M, Sessa F, Villa F, Musacchio A, Cortese R. Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Mol Cancer Ther. 2008;7:1140–1149. doi: 10.1158/1535-7163.MCT-07-2051. [DOI] [PubMed] [Google Scholar]
- Ding J, Swain JE, Smith GD. Aurora kinase-A regulates microtubule organizing center (MTOC) localization, chromosome dynamics, and histone-H3 phosphorylation in mouse oocytes. Mol Reprod Dev. 2011;78:80–90. doi: 10.1002/mrd.21272. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Gorgun G, Calabrese E, Hideshima T, Ecsedy J, Perrone G, Mani M, Ikeda H, Bianchi G, Hu Y, Cirstea D, et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood. 2010;115:5202–5213. doi: 10.1182/blood-2009-12-259523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock IS, Fox NE, Prevost N, Sear K, Shattil SJ, Kaushansky K. Roles of focal adhesion kinase (FAK) in megakaryopoiesis and platelet function: studies using a megakaryocyte lineage specific FAK knockout. Blood. 2008;111:596–604. doi: 10.1182/blood-2007-05-089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Richmond TD, Muntean AG, Barber DL, Weiss MJ, Crispino JD. STAT1 promotes megakaryopoiesis downstream of GATA-1 in mice. J Clin Invest. 2007;117:3890–3899. doi: 10.1172/JCI33010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoya M, Hidaka H, Hosoya T, Suzuki M, Yamamoto N, Sasaki Y. Inhibition of rho-kinase-induced myristoylated alanine-rich C kinase substrate (MARCKS) phosphorylation in human neuronal cells by H-1152, a novel and specific Rho-kinase inhibitor. J Neurochem. 2002;81:9–16. doi: 10.1046/j.1471-4159.2002.00801.x. [DOI] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Lannutti BJ, Blake N, Gandhi MJ, Reems JA, Drachman JG. Induction of polyploidization in leukemic cell lines and primary bone marrow by Src kinase inhibitor SU6656. Blood. 2005;105:3875–3878. doi: 10.1182/blood-2004-10-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannutti BJ, Minear J, Blake N, Drachman JG. Increased megakaryocytopoiesis in Lyn-deficient mice. Oncogene. 2006;25:3316–3324. doi: 10.1038/sj.onc.1209351. [DOI] [PubMed] [Google Scholar]
- Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- Lok W, Klein RQ, Saif MW. Aurora kinase inhibitors as anti-cancer therapy. Anticancer Drugs. 2010;21:339–350. doi: 10.1097/CAD.0b013e3283350dd1. [DOI] [PubMed] [Google Scholar]
- Lordier L, Chang Y, Jalil A, Aurade F, Garcon L, Lecluse Y, Larbret F, Kawashima T, Kitamura T, Larghero J, et al. Aurora B is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood. 2010;116:2345–2355. doi: 10.1182/blood-2010-01-265785. [DOI] [PubMed] [Google Scholar]
- Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- Mali RS, Ramdas B, Ma P, Shi J, Munugalavadla V, Sims E, Wei L, Vemula S, Nabinger SC, Goodwin CB, et al. Rho kinase regulates the survival and transformation of cells bearing oncogenic forms of KIT FLT3, and BCR-ABL. Cancer Cell. 2011;20:357–369. doi: 10.1016/j.ccr.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinge S, Izraeli S, Crispino JD. Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood. 2009;113:2619–2628. doi: 10.1182/blood-2008-11-163501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, Stroud SG, Chen W, Shindi V, Huck JJ, et al. Characterization of Alisertib (MLN8237), An Investigational Small Molecule Inhibitor of Aurora A Kinase Using Novel In Vivo Pharmacodynamic Assays. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Margolin AA, Ong SE, Schenone M, Gould R, Schreiber SL, Carr SA, Golub TR. Empirical Bayes analysis of quantitative proteomics experiments. PLoS ONE. 2009;4:e7454. doi: 10.1371/journal.pone.0007454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM, Morton CL, Gorlick R, Kolb EA, Lock R, Carol H, Keir ST, Reynolds CP, Kang MH, Wu J, et al. Initial testing of the aurora kinase A inhibitor MLN8237 by the Pediatric Preclinical Testing Program (PPTP) Pediatr Blood Cancer. 2010;55:26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T, Coniat MB, Monni R, Mauchauffe M, Nguyen Khac F, Gressin L, Mugneret F, Leblanc T, Dastugue N, Berger R, et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U S A. 2001;98:5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T, Raffel GD, Moore SA, Cornejo MG, Baudry-Bluteau D, Cagnard N, Jesneck JL, Pikman Y, Cullen D, Williams IR, et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J Clin Invest. 2009;119:852–864. doi: 10.1172/JCI35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock AA, Foote KM, Heron NM, Jung FH, Pasquet G, Lohmann JJ, Warin N, Renaud F, De Savi C, Roberts NJ, et al. Discovery, synthesis, and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of aurora B kinase. J Med Chem. 2007;50:2213–2224. doi: 10.1021/jm061335f. [DOI] [PubMed] [Google Scholar]
- Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradines JR, Farutin V, Rowley S, Dancik V. Analyzing protein lists with large networks: edge-count probabilities in random graphs with given expected degrees. J Comput Biol. 2005;12:113–128. doi: 10.1089/cmb.2005.12.113. [DOI] [PubMed] [Google Scholar]
- Sato T, Fuse A, Eguchi M, Hayashi Y, Ryo R, Adachi M, Kishimoto Y, Teramura M, Mizoguchi H, Shima Y, et al. Establishment of a human leukaemic cell line (CMK) with megakaryocytic characteristics from a Down's syndrome patient with acute megakaryoblastic leukaemia. Br J Haematol. 1989;72:184–190. doi: 10.1111/j.1365-2141.1989.tb07681.x. [DOI] [PubMed] [Google Scholar]
- Sloane DA, Trikic MZ, Chu ML, Lamers MB, Mason CS, Mueller I, Savory WJ, Williams DH, Eyers PA. Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem Biol. 2010;5:563–576. doi: 10.1021/cb100053q. [DOI] [PubMed] [Google Scholar]
- Stachura DL, Chou ST, Weiss MJ. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood. 2006;107:87–97. doi: 10.1182/blood-2005-07-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallman MS, Neuberg D, Bennett JM, Francois CJ, Paietta E, Wiernik PH, Dewald G, Cassileth PA, Oken MM, Rowe JM. Acute megakaryocytic leukemia: the Eastern Cooperative Oncology Group experience. Blood. 2000;96:2405–2411. [PubMed] [Google Scholar]
- Terstappen GC, Schlupen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula S, Shi J, Hanneman P, Wei L, Kapur R. ROCK1 functions as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability. Blood. 2010;115:1785–1796. doi: 10.1182/blood-2009-08-237222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- Wen Q, Goldenson B, Crispino JD. Normal and malignant megakaryopoiesis. Expert Rev Mol Med. 2011:e32. doi: 10.1017/S1462399411002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q, Leung C, Huang Z, Small S, Reddi AL, Licht JD, Crispino JD. Survivin is not required for the endomitotic cell cycle of megakaryocytes. Blood. 2009;114:153–156. doi: 10.1182/blood-2008-11-190801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RW, Odedra R, Heaton SP, Wedge SR, Keen NJ, Crafter C, Foster JR, Brady MC, Bigley A, Brown E, et al. AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clin Cancer Res. 2007;13:3682–3688. doi: 10.1158/1078-0432.CCR-06-2979. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.