Abstract
BM mesenchymal stromal cells (BM-MSCs) support multiple myeloma (MM) cell growth, but little is known about the putative mechanisms by which the BM microenvironment plays an oncogenic role in this disease. Cell-cell communication is mediated by exosomes. In this study, we showed that MM BM-MSCs release exosomes that are transferred to MM cells, thereby resulting in modulation of tumor growth in vivo. Exosomal microRNA (miR) content differed between MM and normal BM-MSCs, with a lower content of the tumor suppressor miR-15a. In addition, MM BM-MSC–derived exosomes had higher levels of oncogenic proteins, cytokines, and adhesion molecules compared with exosomes from the cells of origin. Importantly, whereas MM BM-MSC–derived exosomes promoted MM tumor growth, normal BM-MSC exosomes inhibited the growth of MM cells. In summary, these in vitro and in vivo studies demonstrated that exosome transfer from BM-MSCs to clonal plasma cells represents a previously undescribed and unique mechanism that highlights the contribution of BM-MSCs to MM disease progression.
Introduction
The BM microenvironment plays a crucial role in multiple myeloma (MM) pathogenesis by supporting plasma cell growth, survival, and drug resistance, which has been partially attributed to the ability of MM BM mesenchymal stromal cells (BM-MSCs) to secrete growth factors and cytokines such as IL-6, IGF-1, VEGF, and many others (1–3). These observations are indicative of paracrine growth circuits between BM-MSCs and clonal plasma cells and vice versa, which suggests that the BM niche provides an optimal substrate for MM cell localization and growth. Nevertheless, little is known about the putative mechanisms by which the BM microenvironment can lead to initiation or progression of oncogenesis in this disease.
It was recently reported that cell-cell communication is mediated by exosomes. Exosomes are small nanometer-sized (50–100 nm) vesicles of endocytic origin that are released in the extracellular milieu by several cell types (4–11) under physiological and pathological conditions, including antigen presentation, transmission of infectious agents, and tumors (12, 13). The role of exosomes in tumor progression is due to the ability of tumor cell–derived exosomes to modulate and mold the host microenvironment, thereby promoting tumor cell growth and disease progression (14–17).
The question of whether BM-MSCs transfer information to the tumor clone remains unanswered. We therefore chose MM as a model disease to examine the mechanism by which the BM microenvironment may induce genetic changes in the tumor clone that lead to tumor progression and dissemination. We first characterized BM-MSC–derived exosomes as key regulators of direct interaction with the clonal MM plasma cells. Moreover, although microRNAs (miRs) may be transferred via exosomes, as has been shown in mast cells, the same phenomenon has not yet been described in tumors. Notably, miRs characteristic of certain solid tumors were detected in serum-derived exosomes from patients with glioblastomas, ovarian cancer, and lung cancer (18–21). Therefore, we hypothesized that miRs present in the exosomes may mediate epigenetic transfer from the BM-MSCs to the MM tumor clone.
In the present study, we showed that BM-MSCs release exosomes and that exosomal miR content may be transferred to MM cells. We found that exosomes isolated from BM-MSCs of patients with MM induced MM tumor growth in vivo and promoted dissemination of tumor cells to the BM in an in vivo translational model of MM. Moreover, the profile of miR exosomal content varied between normal and MM BM-MSC–derived exosomes: for example, levels of exosomal miR-15a were significantly increased in normal versus MM BM-MSC–derived exosomes, suggestive of a tumor-suppressive role of MSC-derived exosomal miR-15a in MM. In addition, depletion of miR-15a in primary normal BM-MSCs and in the related exosomal content enhanced MM cell growth. However, the lack of tumor suppressor miRs in exosomes was not sufficient to explain the induction of tumor proliferation that we observed with MM BM-MSC–derived exosomes. Therefore, we investigated the proteomic content of exosomes and found that normal and MM BM-MSC–derived exosomes also differed in their protein content, with higher expression of oncogenic proteins, cytokines, and protein kinases in MM BM-MSC–derived exosomes. These findings demonstrated the existence of exosome-driven interactions between the BM microenvironment and MM cells and that exosomes constitute a novel mechanism for intercellular transfer of genetic and protein information in hematological malignancies such as MM.
Results
BM-MSC–derived exosomes are transferred to MM cells.
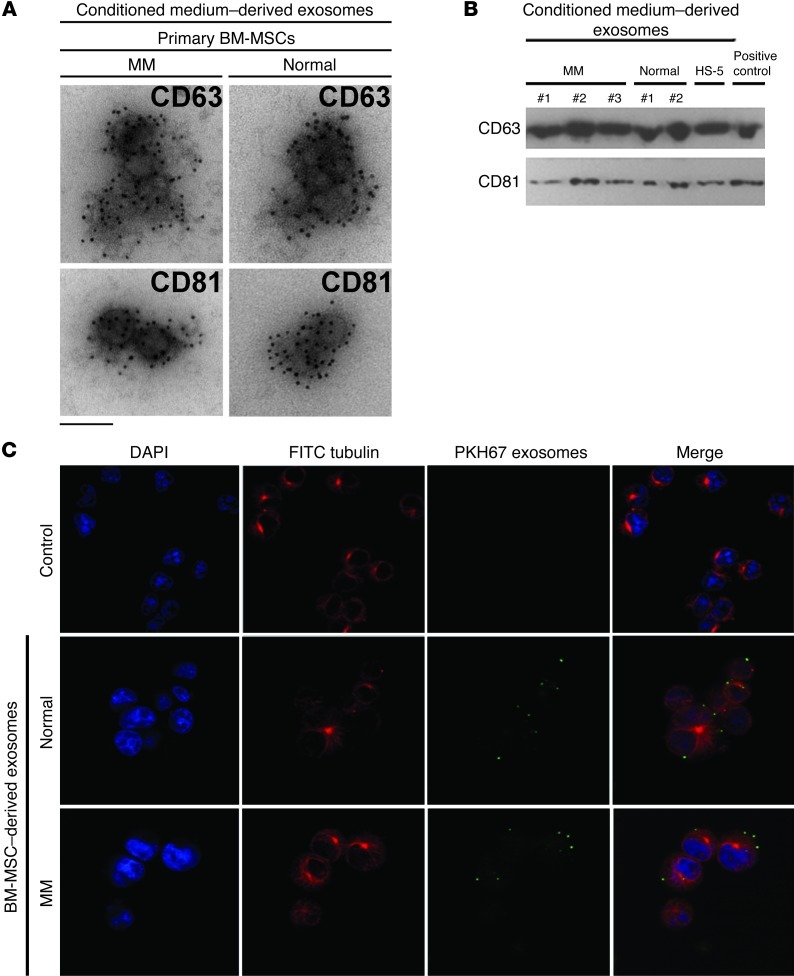
Exosomes were isolated from primary BM-MSC conditioned medium obtained from either patients with MM or healthy individuals, as described previously (18, 22, 23). The purity of BM-MSCs, evaluated as previously described (24, 25), was greater than 95% (Supplemental Table 1 and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI66517DS1), indicative of multipotent MSC origin. Isolated exosomes were studied by electron microscopy, which demonstrated rounded particles with approximately 100 nm in size and a membrane-like bilayer, as usually observed in exosomes (Supplemental Figure 2A). The exosomal expression of CD63 and CD81, surface antigens commonly used as exosomal markers (18, 23, 26), was subsequently evaluated and confirmed using immunogold labeling and Western blot (Figure 1, A and B). Similar results were obtained when we compared the morphology and immunophenotype of exosomes isolated using the 2 procedures adopted (i.e., with and without ExoQuik solution; Supplemental Figure 2B).
Figure 1. Characterization of BM-MSC–derived exosomes and their ability to be transferred to MM cells.
(A) Primary BM-MSCs are able to release exosomes. Exosomes were immunogold labeled with anti-CD63 and anti-CD81. Scale bar: 100 nm. (B) Western blot on MM (n = 3) and normal (n = 2) BM-MSC–derived exosome proteins using anti-CD63 and anti-CD81 antibodies. The normal stromal cell line HS-5 is also shown. Lysates obtained from human CD63– or human CD81–transfected 293T cells served as positive controls. (C) MM.1S cells were cultured in the absence (control) or presence of normal or MM BM-MSC–derived PKH67-labeled exosomes for 30 minutes. Exosomes were uptaken from MM cells, as shown using a confocal microscope (original magnification, ×100). MM cells were stained using DAPI (nuclei) and FITC-conjugated anti-tubulin antibody.
We next examined whether BM-MSC–derived exosomes could be transferred to MM cells. PKH67 fluorescently labeled exosomes isolated from both normal and MM BM-MSC conditioned medium were cultured with MM cells. We confirmed the ability of MM cells to uptake exosomes using confocal microscopy (Figure 1C). These findings were validated by reading the fluorescence signal of MM cells exposed to fluorescently labeled exosomes (Supplemental Figure 2C). These results indicate that normal and MM BM-MSC–derived exosomes can be transferred into MM cells, suggestive of a potential role in regulating MM biology.
Functional sequelae of exosomes on MM cell proliferation and homing to the BM.
The interaction between the BM microenvironment and MM plasma cells plays a pivotal role in supporting MM pathogenesis and progression (27). Our findings of exosomal transfer between BM-MSCs and MM cells support the hypothesis that exosomes may actively mediate tumor growth and dissemination. Therefore, we next examined the effect of normal and MM BM-MSC–derived exosomes on in vitro and in vivo MM cell growth.
The MM.1S and RPMI.8226 MM cell lines were exposed to increasing concentrations of conditioned medium–derived exosomes isolated from both primary normal and MM BM-MSCs. Exosomes were tested for their functional effects at concentrations previously reported (19, 28, 29). Normal BM-MSC–derived exosomes were able to significantly reduce MM cell proliferation compared with MM BM-MSC–derived exosomes, which slightly increased MM cell growth in vitro (P < 0.05; Supplemental Figure 3A). Similar findings were obtained using 4 MM, 2 monoclonal gammopathy of undetermined significance (MGUS), 2 smoldering MM, and 4 normal BM-MSC–derived exosome samples (Figure 2, A and B). It was previously reported that the whole BM stromal cell population promotes MM cell proliferation, due the presence of either autocrine or paracrine circuits of growth that support MM cell proliferation (30–35). This was previously established using primary MM BM-MSCs and HS-5 cells as a model of normal BM-MSCs (36). We therefore compared the effect of the whole BM-MSC compartment using normal BM-MSCs, MM BM-MSCs, or HS-5 cells — and their related exosomal counterparts — on MM cell proliferation. We found that primary MM BM-MSCs clearly increased MM cell proliferation (36%–38%), and both HS-5 cells and normal BM-MSCs exerted similar effects (15%–32% and 9%–16%, respectively), thus recapitulating previous findings (30–36). Importantly, MM BM-MSC–derived pure exosomes exerted a proproliferative effect of about 22%–30% on MM cells, whereas HS-5 cells and normal BM-MSC–derived pure exosomes inhibited MM cell proliferation (28%–31% and 30%–40%, respectively; Supplemental Figure 3B).
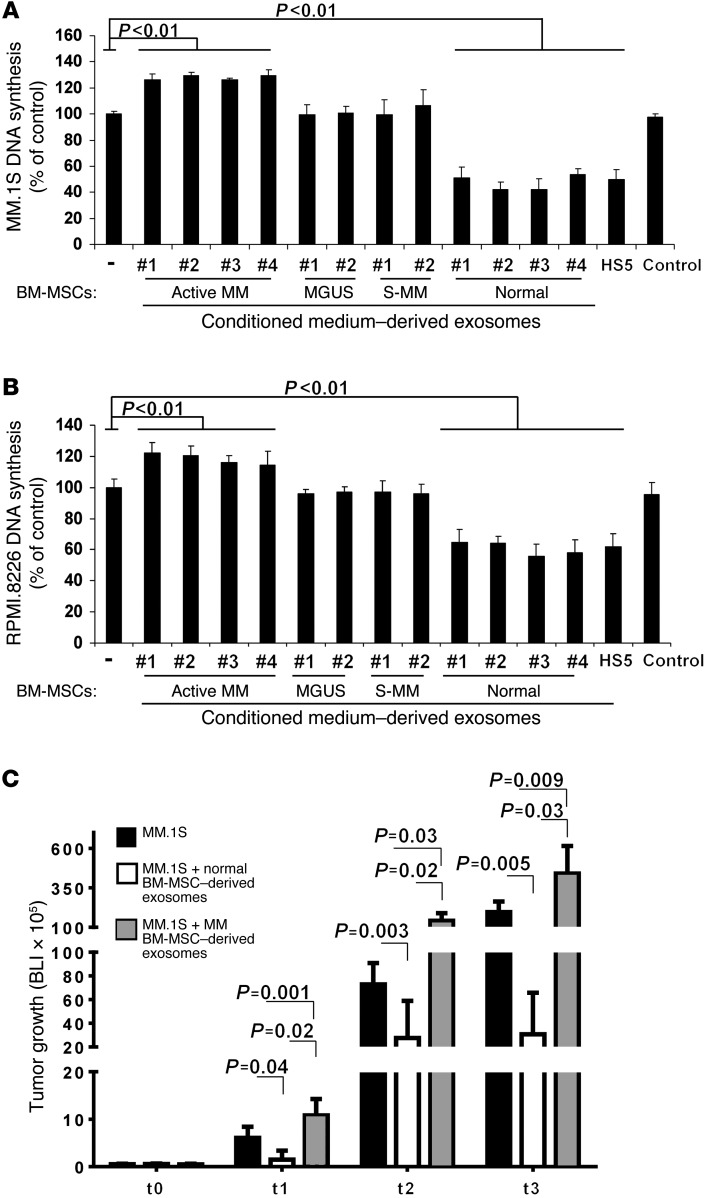
Figure 2. Normal and MM BM-MSC–derived exosomes differentially affect MM cell proliferation in vitro and in vivo.
(A and B) MM cell lines MM.1S (A) and RPMI.8226 (B) (30,000 cells/well; RPMI medium plus 10% exosome-depleted FBS) were cultured in the absence or presence of MM (n = 4), MGUS (n = 2), smoldering MM (S-MM; n = 2), or normal (n = 4) BM-MSC–derived exosomes (200 μg/ml; 48 hours). Loaded exosomes are expressed as μg of protein-containing exosomes. Cell proliferation was assessed using [3H]-thymidine uptake. Cell-conditioned media absent cells and processed as in all samples tested served as control. Average of 3 independent experiments is shown. P values were generated using ANOVA. MM and normal BM-MSC–derived exosomes showed a differential impact on MM cell growth in vitro. (C) TEBs were loaded with GFP+Luc+ MM.1S cells alone or with primary MM or normal BM-MSC–derived exosomes (3 × 106 cells/TEB; 1 μg exosomes) and implanted subcutaneously in SCID mice. Exosomes (1 μg) were also injected in situ every 4 days until the end of the studies. Tumor growth was determined by measuring bioluminescence imaging (BLI) intensity at baseline (t0) and days 7 (t1), 10 (t2), and 14 (t3) (n = 5 per group). MM and normal BM-MSC–derived exosomes showed a differential impact on MM cell growth in vivo.
We next investigated the ability of normal and MM BM-MSC–derived exosomes to modulate MM cell growth and dissemination in vivo. Tissue-engineered bones (TEBs) were loaded with equal numbers of GFP+Luc+ MM.1S cells in the presence of either primary MM BM-MSC–derived exosomes or primary normal MSC–derived exosomes (37). TEBs loaded with GFP+Luc+ MM.1S cells were used as a control. TEBs were then injected subcutaneously in SCID mice. Tumor burden was evaluated using bioluminescence in vivo imaging. Mice had similar tumor penetrance at baseline, whereas at days 7, 10, and 14 after implantation, recipients of MM BM-MSC–derived exosomes had a significantly higher tumor growth rate than did recipients of MM cells only or of normal BM-MSC–derived exosomes (Figure 2C and Supplemental Figure 3, C and D).
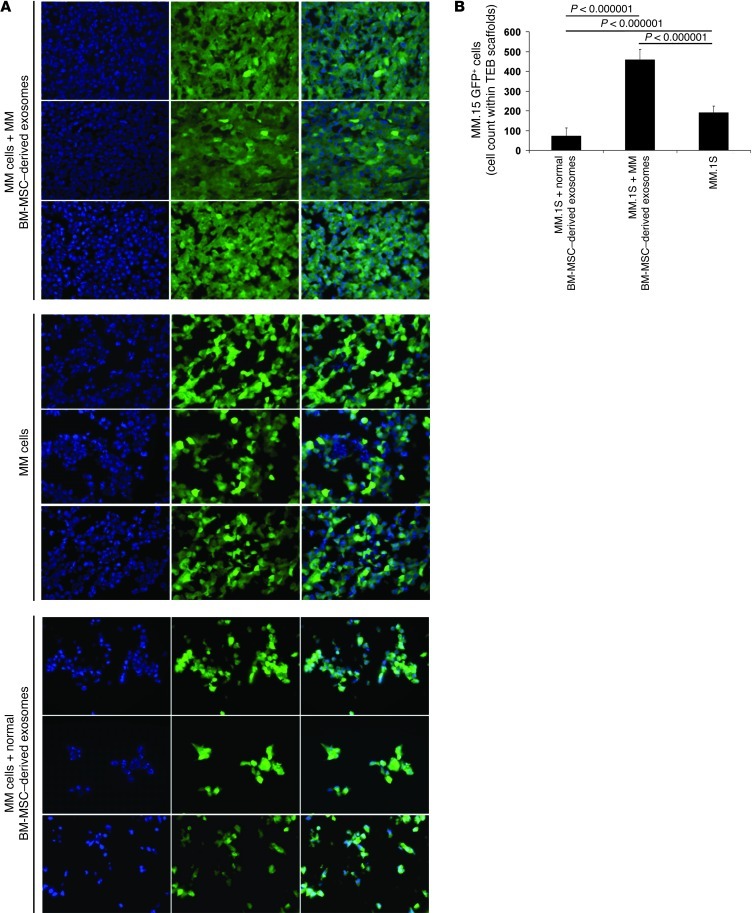
These findings were further confirmed by immunofluorescence on TEB scaffolds ex vivo (Figure 3A). A significantly higher number of MM cells was found in TEBs loaded with MM BM-MSC–derived exosomes versus those loaded with normal BM-MSC–derived exosomes (Figure 3B), which indicated that the presence of normal and MM BM-MSC–derived exosomes inhibited and induced tumor growth, respectively, compared with control MM cells alone.
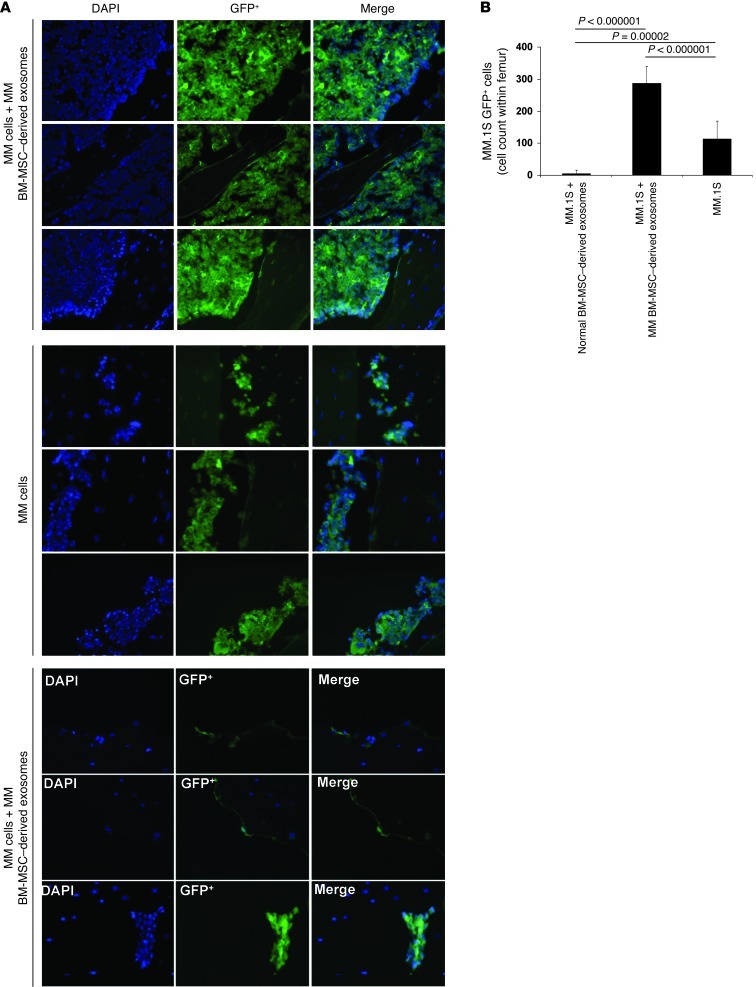
Figure 3. Visualization and quantification of MM cells ex vivo on TEB scaffolds.
(A) Immunofluorescence detection of GFP+ MM.1S cells ex vivo on TEB scaffolds. Nuclei were stained using DAPI. 1 representative image per group is shown. Original magnification, ×40. (B) GFP+ MM.1S cells were counted in 4 different regions per TEB scaffold per mouse. Average ± SD count is shown.
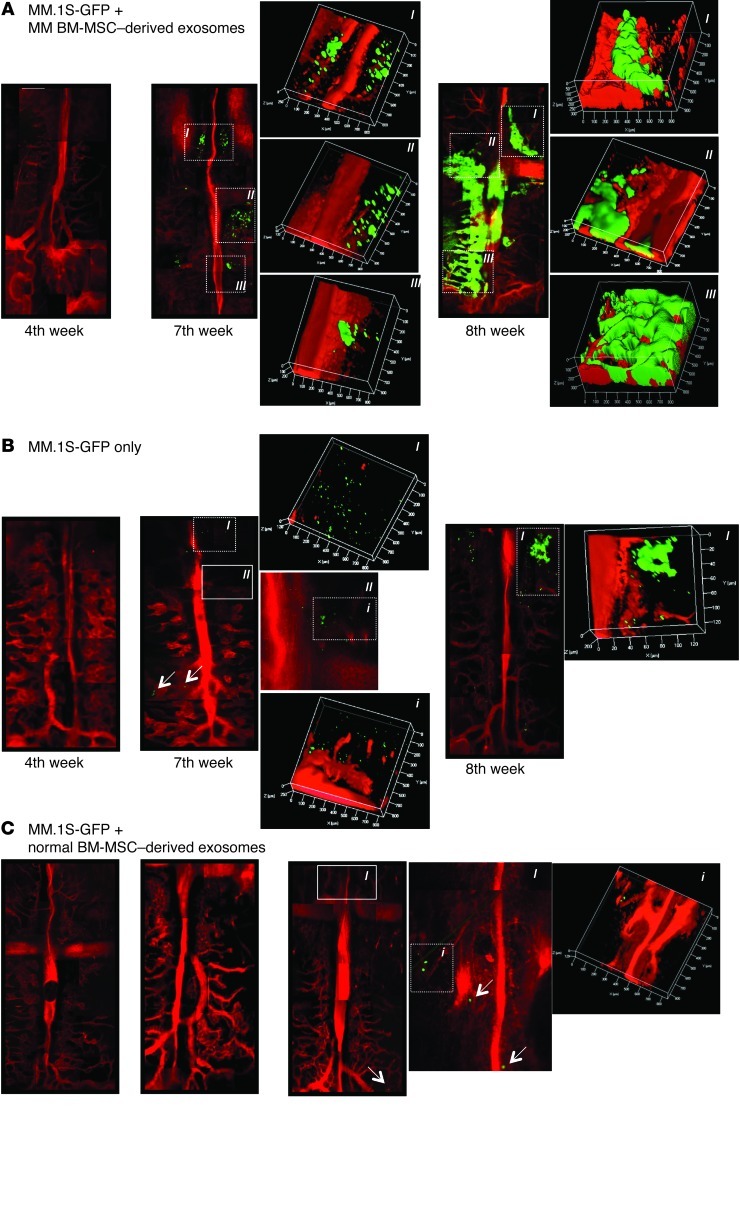
To further corroborate the differential impact of exosomes in regulating MM progression, we next examined whether MM BM-MSC–derived exosomes also induce cell dissemination and metastasis to distant BM niches, which is a hallmark of MM. Using in vivo confocal imaging, we were able to visualize the BM vasculature and the possible presence of GFP+ MM cells (38). Mice were imaged 4 weeks after subcutaneous implantation of the TEBs; this time point was considered the baseline across the 3 cohorts of animals, in which no detection of GFP+ MM cells was observed. At 7 and 8 weeks after implantation, imaging showed clear differences in the ability of MM cells to disseminate to the distant BM niches in vivo. At week 7, a higher tumor burden was observed in recipients of TEB implants loaded with MM cells exposed to MM BM-MSC–derived exosomes compared with those loaded with MM cells only (Figure 4, A and B). In contrast, in recipients of TEBs loaded with MM cells mixed with normal BM-MSC–derived exosomes, only a weak GFP signal was detected (Figure 4C). These results were further corroborated by femur BM immunofluorescence ex vivo (Figure 5, A and B). These findings suggest that BM-MSCs may play an oncogenic role in the progression and widespread dissemination of this disease.
Figure 4. Normal and MM BM-MSC–derived exosomes differentially affect MM cell homing and growth in vivo.
Detection of MM cell homing to the BM was performed by in vivo confocal microscopy (original magnification, ×5). Green, GFP+ MM cells (denoted by arrows); red, Evans Blue–positive blood vessels. Specific BM niches are shown in boxed regions with dotted lines; relative tridimensional reconstruction is shown for each panel (scale expressed in μm). In B, specific BM niches were obtained by changing the focal plane, moving toward the skull of the mouse (boxed region with solid line).
Figure 5. Visualization and quantification of MM cells ex vivo on femur BM.
(A) Immunofluorescence detection of GFP+ MM.1S cells ex vivo on bone tissues. Nuclei were stained using DAPI. 1 representative image per mouse per group is shown. Original magnification, ×40. (B) GFP+ MM.1S cells were counted in 4 different regions per femur per mouse. Average ± SD count is shown.
Exosomal miR and protein content differs between normal and MM BM-MSCs.
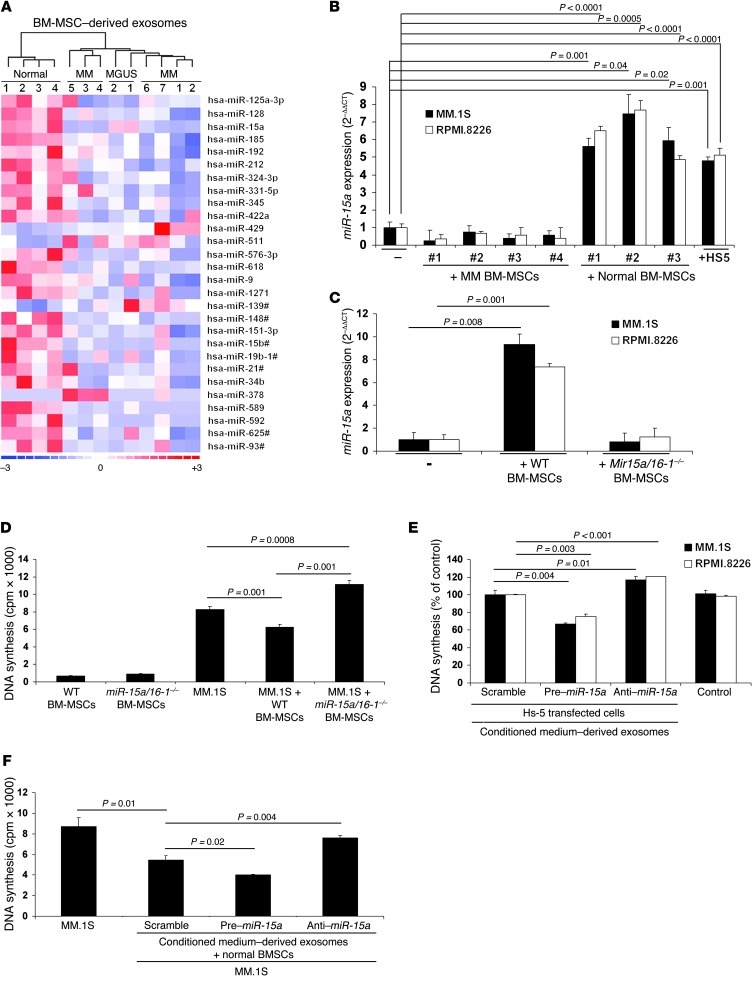
Large amounts of small RNAs have recently been reported in exosomes, which suggests that they may contain miRs. Indeed, the presence of miRs in exosomes was recently documented in mast cells (18). We therefore hypothesized that BM-MSC–derived exosomes may differ in their miR content, leading to epigenetic transfer of this information to the tumor clone. We next performed miR expression profiling on normal, MGUS, and MM BM-MSC–derived exosomes. Supervised clustering analysis comparing normal versus MM and MGUS BM-MSC–derived exosomes showed statistically significant differences between the 2 cohorts of samples: specifically, reduced expression of 16 miRs and increased expression of 2 miRs was observed in MM BM-MSC–derived exosomes compared with normal and MGUS BM-MSC–derived exosomes, which presented with similar patterns (P < 0.05; Figure 6A). In particular, we found miR-15a to be one of the downmodulated miRs in MM BM-MSC–isolated exosomes. We next validated and confirmed the lower miR-15a levels in 5 additional MM BM-MSC–derived exosome samples, compared with 2 normal BM-MSC–derived exosome samples (Supplemental Figure 4A).
Figure 6. miR-15a expression differs between normal and MM BM-MSCs, and miR-15a–containing exosomes are transferred into MM cells.
(A) miR expression profiling on total RNA isolated from normal (n = 4), MM (n = 7), and MGUS (n = 2) BM-MSC–derived exosomes. A heatmap was generated after supervised hierarchical cluster analysis. Differential miR expression is shown by red (upregulation) versus blue (downregulation) intensity (d-Chip software; normal versus MM, 1.5-fold change, P < 0.05). (B) MM.1S and RPMI.8226 MM cells were cultured in the absence or presence of primary MM (n = 4) or normal (n = 3) BM-MSCs or the HS-5 cell line for 48 hours. miR-15a expression was determined by qRT-PCR in MM cells (2–ΔΔCt method, normalized to RNU6B miR as reference). Results are average ± SD of 3 independent experiments. miR-15a was upregulated in MM cells when in contact with normal BM-MSCs. (C) Murine exosomes were isolated from BM of WT or miR-15a/16-1–/– mice and subsequently added to MM cells for 48 hours. miR-15a levels were determined by qRT-PCR in human MM cells (2–ΔΔCt method, normalized to C. elegans miR-39 reference, used as spiked control). Bars represent SD. (D) MM cells were cultured in the presence or absence of murine WT or miR-15a/16-1–/– BM-MSCs for 48 hours, and cell proliferation was assessed as [3H]-thymidine uptake. Bars indicate SD. (E and F) HS-5 cells (E) or primary BM-MSCs (F) were transfected with scramble, pre–miR-15a, or anti–miR-15a probe. Cells were then exposed to the indicated exosomes for 48 hours. Cell-conditioned media absent cells and processed as in all samples tested served as control. Cell proliferation was assessed using [3H]-thymidine uptake assay. Bars indicate SD.
The presence of miRs in exosomes led us to hypothesize that exosomes may represent an active vehicle between the BM microenvironment and MM cells, thus functionally modulating the biological behavior of MM cells. We previously reported that primary MM cells present with reduced expression of miR-15a compared with their normal cellular counterparts, and functional studies have corroborated the role of miR-15a as a tumor suppressor miR in this disease (32). However, the finding that miR-15a was also deficient in MM BM-MSCs, not only in the tumor clone, led us to hypothesize that the lack of this miR in exosome transfer from MM BM-MSCs to the tumor clone mediates oncogenesis in MM. We first evaluated the expression of miR-15a in BM-MSCs and found that primary normal BM-MSCs had higher miR-15a expression than did primary MM BM-MSCs. In addition, miR-15a levels were comparable in HS-5 cells and normal primary BM-MSCs (Supplemental Figure 4B). We next evaluated the expression of miR-15a in MM cells cultured either alone or in the presence of BM-MSCs; miR-15a was significantly upregulated in MM cells cocultured with normal BM-MSCs or HS-5 cells, but not with MM BM-MSCs (Figure 6B), suggestive of miR-15a transfer from BM-MSCs to MM plasma cells. To further confirm that miR-15a is actively transferred to the tumor cells, we used WT and miR-15a/16-1–/– C57BL/6 mice. Murine BM-MSCs were used at the fourth passage and presented with a multipotent MSC phenotype (Supplemental Figure 4C), as previously described (39, 40).
Exosomes were isolated from BM-MSCs of WT or miR-15a/16-1–/– mice. Levels of miR-15a and miR-16-1 were confirmed by quantitative RT-PCR (qRT-PCR) on both BM-MSCs and BM-MSC–derived exosomes from miR-15a/16-1–/– and WT mice (Supplemental Figure 4, D and E). After coculture of MM cells with murine WT BM-MSC–derived exosomes, miR-15a expression increased in MM cells, providing further evidence of transfer of MSC-derived exosomes containing miR-15a into the plasma cells. No changes were observed in MM cells exposed to miR-15a/16-1–/– BM-MSC–derived exosomes (Figure 6C). Similar findings were obtained for miR-16-1 (Supplemental Figure 4F). We next sought to determine whether murine miR-15a/16-1–/– BM-MSCs can functionally target MM cells. MM cells were cultured in the presence or absence of BM-MSCs isolated from WT or miR-15a/16-1–/– mice. miR-15a/16-1–/– BM-MSCs were found to significantly induce MM.1S cell proliferation, and similar results were obtained using RPMI.8226 cells (Figure 6D and Supplemental Figure 4G).
To further confirm that miR-15a plays a critical role in MSC-mediated tumor progression, we performed loss- and gain-of-function studies. HS-5 cells were transfected with either pre– or anti–miR-15a, and transfection efficiency was tested by qRT-PCR (Supplemental Figure 5A). MM cells were exposed to exosomes isolated from transfected HS-5 cells for 48 hours; importantly, exosomes isolated from pre–miR-15a–transfected HS-5 cells significantly inhibited MM cell proliferation (Figure 6E). To further confirm the ability of miR-15a–containing exosomes to inhibit MM cell proliferation, we collected exosomes from primary BM-MSCs transfected with pre– or anti–miR-15a. Transfection efficiency was tested by qRT-PCR (Supplemental Figure 5B). We observed a lower proliferation rate of MM.1S cells exposed to pre–miR-15a–transfected BM-MSC–derived exosomes, and similar results were obtained using RPMI.8226 cells (Figure 6F and Supplemental Figure 5C). Together, these findings confirmed that miR-15a acts as a tumor suppressor miR that is present in normal BM-MSCs but absent in MM BM-MSCs and that the lack of exosome-mediated miR-15a transfer to malignant plasma cells is permissive for the growth and dissemination of clonogenic MM cells.
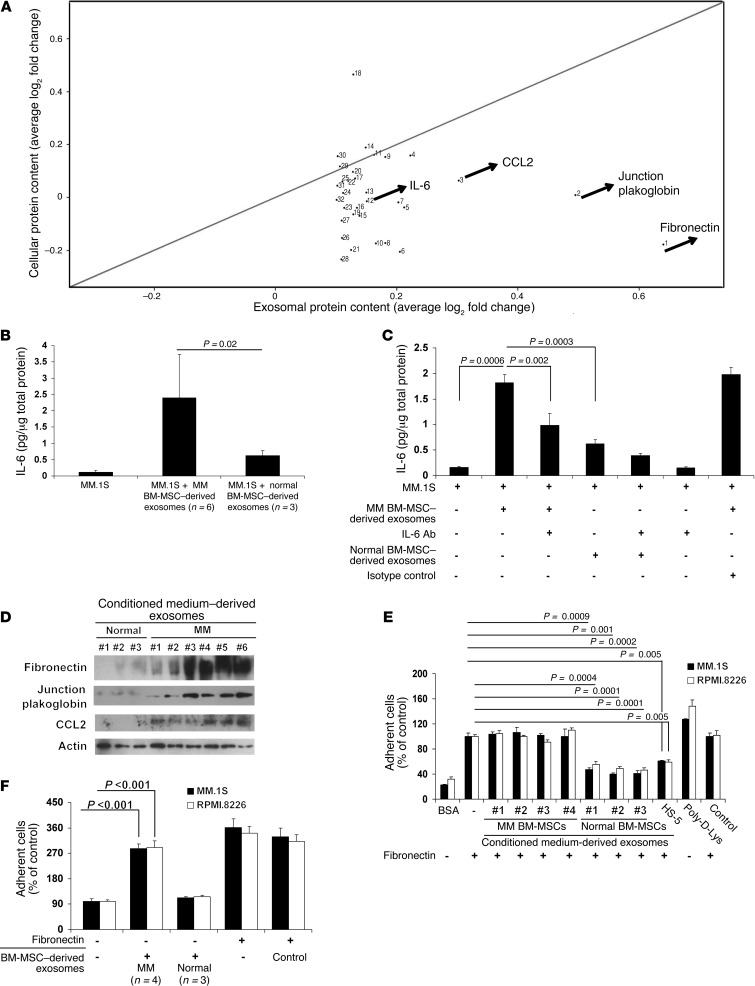
However, the lack of transfer of tumor suppressor miRs could not explain all of the changes in MM cells in response to MM BM-MSC–derived exosomes. Therefore, we next sought to determine whether exosomes may selectively transfer certain proteins from BM-MSCs to recipient tumor plasma cells. A high-throughput antibody-based protein array was performed on exosomes isolated from primary normal and MM BM-MSCs, as well as on the whole population of normal and MM BM-MSCs. BM-MSC–derived exosomal proteins differed from BM-MSC–derived cellular proteins (Supplemental Table 2), which suggests that MM BM-MSC–derived exosomes present specifically higher content levels of IL-6, CCL2 (also known as MCP1), junction plakoglobin (also known as γ-catenin), or fibronectin that does not reflect the protein content of the cells of origin. Indeed, IL-6, fibronectin, CCL2, and junction plakoglobin demonstrated minimal changes between normal and MM BM-MSCs (Supplemental Tables 2 and 3). These findings suggest that exosomes may selectively transfer certain proteins to the recipients cells, behaving as vesicles that selectively transport specific proteins to cells they interact with (Figure 7A). It was previously reported that CCL2 plays a crucial role in MM pathogenesis and disease progression, demonstrated both in vitro and in vivo (41, 42). Our present findings also demonstrated that MM BM-MSC–derived exosomes contain higher levels of CCL2 than do normal BM-MSC–derived exosomes, confirming the role of this protein in supporting MM biology.
Figure 7. Characterization of exosomal protein content and functional sequelae.
(A) Scatter plot showing exosomal and cellular protein content of MM and normal BM-MSC–derived samples. (B) MM.1S cells were exposed to exosomes isolated from MM (n = 6) or normal (n = 3) BM-MSCs for 24 hours, and IL-6 concentration was measured on conditioned media using human IL-6 ELISA. Bars indicate SD. (C) MM.1S cells were exposed to exosomes isolated from MM or normal BM-MSCs for 24 hours, and IL-6 concentration was measured in conditioned media in the presence or absence of an IL-6–blocking antibody (0.2 μg/ml) using human IL-6 ELISA. Mouse IgG2B served as isotype control. Bars indicate SD. (D) Western blot on MM (n = 6) and normal (n = 3) BM-MSC–derived exosomes using anti-fibronectin, anti–junction plakoglobin, anti-CCL2, and anti-actin antibodies. (E and F) Adhesion of MM cells to BSA (negative control), poly-d-lysine (positive control), and fibronectin-coated wells, exposed or not to MM (n = 4) or normal (n = 3) BM-MSC–derived exosomes (200 μg/ml; 6 hours). Length of adhesion was 2 hours. All data are mean ± SD of triplicate experiments. Cell-conditioned media absent cells and processed as in all samples tested served as control.
We next sought to determine whether the differentially expressed exosomal protein content between MM and normal BM-MSCs could functionally affect MM cells. MM cells were exposed to MM or normal BM-MSC–derived exosomes, and IL-6 levels were found to be increased in the conditioned media of MM cells exposed to MM compared with normal BM-MSC–derived exosomes (Figure 7B and Supplemental Figure 6A). A recent study on the proteomic content of exosomes and their function indicates that the proteins are not restricted to the interior of the exosome (43). Exosomal proteins may therefore be released from the intraexosomal space and interact with extracellular proteins via as-yet undefined mechanisms. Our present data are similar in showing that MM BM-MSC–derived exosomes increased IL-6 levels in the conditioned medium. We next demonstrated that MM BM-MSC–derived exosome-dependent induction of IL-6 release from MM cells was significantly abolished when cells were exposed to IL-6 blocking antibody (Figure 7C and Supplemental Figure 6B), which indicates that IL-6 released by exosomes is functionally important in MM cell growth.
Detection of fibronectin, CCL2, and junction plakoglobin was further confirmed by Western blot (Figure 7D). Because we observed that exosomes from MM BM-MSCs presented with higher fibronectin content, and based on accelerated dissemination of MM cells to distant BM niches, we next examined whether exosomes regulate adhesion properties of MM cells to the surrounding BM milieu. MM cell adhesion to fibronectin was reduced when cells were cultured in the presence of primary normal BM-MSC–derived exosomes, whereas MM BM-MSC–derived exosomes did not affect the cells’ ability to adhere to fibronectin (Figure 7E). These findings indicate that BM-MSC–derived exosomes may differentially impact MM cell adhesion. To further corroborate that this is indeed mediated by different fibronectin contents, we tested adhesion of MM cells to non–fibronectin-coated wells and found that MM cells increased their adhesion abilities when exposed to MM versus normal BM-MSC–derived exosomes (Figure 7F).
Additionally, downmodulated phosphorylation of the adhesion protein FAK was observed when MM cells were treated with normal versus MM BM-MSC–derived exosomes, and inactivation of cofilin was found in MM cells exposed to MM BM-MSC–derived exosomes, compared with normal MSC–derived exosomes, in which increased phosphorylation of cofilin was observed (Supplemental Figure 6C).
Discussion
It has long been believed that BM-MSCs support MM cell growth, thus resulting in MM progression (1–3). Although many investigations have explored the role of the BM microenvironment in releasing cytokines and growth factors that may support MM pathogenesis and therefore disease progression, to date, little is known of the transfer of genetic and protein information from BM-MSCs to clonal plasma cells via exosomes. Our present study revealed a novel mechanism by which BM-MSCs play an oncogenic role in MM.
We first confirmed that BM-MSCs promoted release and transfer of exosomes to MM cells, and next demonstrated differences in miR profiling of normal versus MM BM-MSC–derived exosomes. Indeed, we observed lower miR-15a expression in MM versus normal BM-MSC–derived exosomes. We previously showed that primary MM plasma cells isolated from patients with relapsed/refractory disease have lower expression of miR-15a than do their normal cellular counterparts and that miR-15a displayed tumor-suppressive properties, as shown by inhibition of cell proliferation in miR-15a–overexpressing MM cells, both in vitro and in vivo (32). Here we found that primary MM BM-MSCs shared similarities with MM tumor cells, including lower miR-15a levels. We therefore sought to examine whether transfer of genetic information (in the form of oncogenic miRs) or, alternatively, lack of transfer of tumor suppressor miRs (such as miR-15a) could lead to significant changes in tumor growth and dissemination in MM. We found that the lack of transfer of the tumor suppressor miR-15a from MM BM-MSCs to MM cells via exosomes could partially explain the observed increased tumor burden together with enhanced tumor dissemination to distant BM niches in vivo. In contrast, the observed inhibition of tumor growth and MM cell dissemination may be explained, at least in part, by the effect of miR-15a–expressing BM-MSC–derived exosomes. The finding that both tumor cells (32) and BM-MSCs had low levels of miR-15a is intriguing and indicates that both cells share similar epigenetic information. Importantly, miR-15a may represent a potential tumor suppressor miR in MM, not only because of its absence in the plasma cell clone (32), but also because of its absence in MM BM-MSCs. Future studies will examine whether regulation of miR-15a originates in BM-MSCs and becomes transferred to MM cells during the early stages of oncogenesis, or whether both cell types acquire the same epigenetic event concurrently.
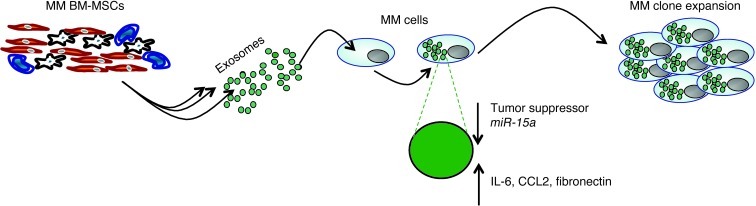
However, the data related to miR tumor suppressor regulation does not fully explain the positive regulation of tumor progression induced by MM BM-MSC–derived exosomes. Therefore, we further examined the protein content of these exosomes and found that they selectively carried a number of oncogenic proteins, as well as cytokines or proteins known to be regulators of adhesion and migration. These findings support the hypothesis that exosomes may act as active vesicles responsible for molding the microenvironment surrounding MM cells, leading to MM growth, dissemination, and subsequent disease progression (Figure 8).
Figure 8. BM-MSC–derived exosomes support MM clone expansion.
In the present study, we did not examine transfer of exosomes and miRs from the tumor clone to BM-MSCs. We believe that this phenomenon is also likely to occur (which would indicate that epigenetic transfer is a bidirectional phenomenon) and that the BM microenvironment not only plays a supportive role in tumor cell growth, but also acts as a conduit of epigenetic information leading to behavioral changes in the tumor clone. Collectively, our findings suggest that transfer of exosomes from BM-MSCs to clonal plasma cells represents a previously undescribed mechanism that may highlight the active contribution of BM-MSCs to MM disease progression.
Methods
Cells and patient characteristics.
Primary BM-MSCs obtained from normal healthy subjects, relapsed/refractory MM patients, MGUS patients, and smoldering MM patients were cultured and selected in plastic flasks and used at the third to fourth passage. In keeping with previous reports and International Society for Cellular Therapy recommendations (24, 25), combinations of monoclonal antibodies were used to define the growing cells as MSCs, including FITC-conjugated anti-CD34, anti-CD19, and anti-CD138; PerCP-Cy5.5–conjugated anti-CD105, anti-CD90, and anti-CD14; allophycocyanin-conjugated anti-CD106 and anti-CD73; and PE-conjugated anti-CD45 antibodies (BD Biosciences). Appropriate isotype controls were used. The MM cell lines MM.1S, RPMI.8226, and U266 and the stromal cell line HS-5 were purchased from ATCC and used for in vitro studies. GFP+Luc+ MM.1S cell lines were generated by retroviral transduction with the pGC-GFP/Luc vector (gift of A. Kung, Dana-Farber Cancer Institute). Patients presented with median age of 69 years (range, 60–72 years) and were diagnosed with either MM, MGUS, or smoldering MM based on the International Myeloma Working Group criteria (44). All MM patients presented with active MM, all with relapsed/refractory disease.
Exosome purification and fluorescent labeling.
Exosomes were purified from cell culture supernatant of BM-MSCs. FBS used in culture media for exosome isolation was precleared by ultracentrifugation at 100,000 g for 3 hours at 4°C. Supernatant fractions collected from 48-hour BM-MSC cultures were filtered using filtration on 0.22-μm pore filters, as previously described (18, 22), followed by ultracentrifugation at 20,000 g for 20 minutes and incubation with an exosome precipitation solution (ExoQuick; System Biosciences) (23). Exosomes were then harvested by ultracentrifugation at 100,000 g for 70 minutes, as previously described (45). Exosomes were also isolated following the same procedure as described above, but without including the ExoQuick incubation step, as previously described (18, 22, 45, 46). For functional assays where exosomes were used, the concentration of total proteins contained in each exosome pellet was quantified using the Bradford assay (BioRad), as described previously (7, 22); exosome quantities are therefore expressed as micrograms of containing proteins. Exosomes were labeled with the green fluorescent linker PKH67 (Sigma-Aldrich), as described previously (47). Exosomes isolated using the 2 procedures (i.e., with and without ExoQuick solution) showed similar results at the ultrastructural level, as shown by electron microscopy and immunogold labeling.
Electron microscopy.
Exosomes were adsorbed for 1 minute to a carbon-coated grid rendered hydrophilic by a 30-second exposure to a glow discharge. Excess liquid was removed with a filter paper (Whatman no. 1), and samples were stained with 1% uranyl acetate for 30 seconds. After excess uranyl formate was removed with a filter paper, grids were examined in a TecnaiGβ Spirit BioTWIN, and images were recorded with an AMT 2k CCD camera. For immunogold labeling, pelleted exosomes were fixed with 2% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4), then processed for ultrathin sectioning and immunogold labeling using anti-CD63 and anti-CD81 antibodies and protein A coupled with 10- or 15-nm gold particles as described previously (18, 22). Sections were observed at 80 kV with a TecnaiGβ Spirit BioTWIN Transmission electron microscope (FEI), and images were recorded with an AMT 2k CCD camera.
Immunoblotting.
Exosomes were lysed using lysis buffer (Cell Signaling Technology) reconstituted with 5 mM NaF, 2 mM Na3VO4, 1 mM PMSF, 5 μg/ml leupeptine, and 5 μg/ml aprotinin. Whole-exosomal lysates (100 μg/lane) were subjected to SDS-PAGE and transferred to a PVDF membrane (BioRad Laboratories). Anti-CD63, anti-CD81, anti–p-cofillin, anti–p-FAK, anti-fibronectin, anti–junction plakoglobin, anti-CXCL2, anti-tubulin, and anti-actin antibodies were used.
ELISA.
IL-6 levels were detected using IL-6 ELISA (human IL-6 immunoassay; R&D Systems) according to the manufacturer’s protocol. Anti–IL-6 neutralizing antibody and the related isotype control were purchased from R&D Systems.
Exosomal RNA isolation and miR profiling.
RNA was isolated using RNeasy mini kit (Qiagen), according to manufacturer’s protocol and as previously described (18). miR profiling was assessed using TaqMan human miR profiling. C. elegans miR-39 was chosen as an internal spiked control because of a lack of sequence homology to human miRs and absence of empiric hybridization to human miR probes on miR microarrays, as previously described (48). Mean miR expression was used for miR qRT-PCR data normalization, as previously described (49). Comparison among normal (n = 4), MM (n = 7), and MGUS (n = 2) BM-MSC–derived exosomes was performed using dChip software (1.5-fold change, P < 0.05).
qRT-PCR.
qRT-PCR for miRs of interest (TaqMan MicroRNA Assays; Applied Biosystems) was performed on an Applied Biosystems AB7500 Real Time PCR system. All PCR reactions were run in triplicate, and miR expression — relative to RNU6B or C. elegans miR-39 — was calculated using the 2–ΔΔCt method (50).
miR transfection.
HS-5 and primary normal BM-MSCs were transfected with precursor (pre–miR-15a), anti–miR-15a, or a scrambled probe used as control (Exiqon), at a final concentration of 40 nM, as previously described (32), using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Efficiency of transfection was validated by qRT-PCR for detection of miR-15a levels.
Confocal microscopy.
Exosomes and MM cells were fluorescently labeled using PKH67 and FITC-conjugated anti-tubulin antibody. Cellular nuclei were stained using DAPI. Imaging of exosome uptake was performed using a Yokogawa spinning disk confocal system (Andor Technology) with a ×100 1.4NA Plan-Apo chromatic objective on a Nikon Ti-E microscope equipped with a Hamamatsu OrcaER CCD camera. Laser illumination, shutters, and filter wheels were controlled by Andor iQ software (Andor Technology). GFP+ MM.1S cells were incubated with DiD-labeled exosomes for 30 minutes. Laser excitation of GFP and DiD was performed sequentially using 488- and 640-nm lasers. Images were acquired using a ×100 Plan-Apo objective lens with a Hamamatsu OrcaER camera. Acquisition parameters, shutters, filter positions, and focus were controlled by Andor iQ software.
In vivo studies.
MM cell homing to the BM was imaged in vivo using a Zeiss 710 confocal system (Carl Zeiss Microimaging) on an upright Examiner stand with a custom stage. A skin flap was made in the mouse scalp to expose the underlying dorsal skull surface. Images of the tumors were captured in approximately 1-hour sessions. High-resolution images with cellular detail were obtained through the intact mouse skull at depths of up to 250 μm from the surface of the skull using a ×5 0.45 NA Plan-Apo objective (Carl Zeiss Microimaging). Multiple imaging depths were acquired, and a maximum intensity z projection was performed in Image J to merge the images. GFP was excited with the 488-nm line on an Argon laser. Blood vessels were imaged using Evans Blue (Sigma-Aldrich) excited with a 633-nm laser. Emission signals were collected using Zeiss internal confocal Quasar detectors. In vivo tumor growth was assessed using in vivo bioluminescence imaging. Mice were injected with 75 mg/kg luciferase (Xenogen), and tumor growth was detected by bioluminescence using a Xenogen In Vivo Imaging System (Caliper Life Sciences). 5-week-old female SCID-beige mice were obtained from Charles River Labs. miR-15a/16-1–/– mice (C57BL/6 background) were a gift of R. Dalla-Favera (Columbia University, New York, New York, USA). Murine BM-MSCs were isolated as previously described (39, 40), used at the fourth passage, and characterized by flow cytometry using FITC-conjugated anti-CD34 and anti-CD44, PE-conjugated anti-CD45 and anti-CD105, and allophycocyanin-Cy7–conjugated anti-CD45R antibodies (BD Biosciences).
TEB.
Silk scaffolds were created and differentiated into TEBs as previously reported (30). TEBs were loaded with MM cells (3 × 106 cells) alone or in the presence of normal or MM BM-MSC–derived exosomes (1 μg). MM cells were previously cultured in the presence or absence of exosomes for 48 hours and subsequently implanted s.c. Exosomes (1 μg) were injected in situ, every 4 days, until the end of the studies. TEB scaffold loaded with MM cells and materials recovered after Exoquick precipitation of medium that had not been conditioned by any cells was used as control in vivo.
In vitro studies.
DNA synthesis was measured by [3H]-thymidine ([3H]-TdR; Perkin Elmer) uptake, as previously described (32). Adhesion was evaluated using an in vitro adhesion assay to fibronectin, following the manufacturer’s recommendations (EMD Biosciences). Calcein AM was used to measure adherent cells, and the degree of fluorescence was measured using a spectrophotometer (485–520 nm). BSA-coated wells served as a negative control. 48-hour coculture of MM.1S and RPMI.8226 MM cells with primary normal BM-MSCs or HS-5 cells were performed, and MM cell proliferation rate was evaluated by [3H]-thymidine uptake, as previously described (32). Exosomes were tested for their functional effects at concentrations previously reported (19, 28, 29).
Tissue immunofluorescence.
Tissue immunofluorescence was performed on both TEBs and femurs harvested from mice as described previously (51). GFP+ MM.1S cells were considered cells of interest. DAPI nuclear stain was added to each slide. Slides were analyzed using a fluorescence microscope (Nikon TE2000-E; objective ×40 plan fluor 0.75 NA). GFP+ MM.1S cells were counted from 4 separate fields per slide. Images were taken using a Hamamatsu OrcaER camera and NIS-Element software. Image J software was used to merge the 2 channels.
Protein array.
Exosomal and cellular protein contents were evaluated using antibody microarray (BD Biosciences — Clontech) as described previously (52).
Accession number. miR expression profiling data were deposited in GEO (series accession no.
GSE39571).
Statistics.
P values described in the in vitro assays were based on t tests (2-tailed; α 0.05) or ANOVA. P values less than 0.05 were considered significant. Exact P values are provided in the figures. In the protein arrays, prior to analysis of the protein expression, the data were background corrected (53), normalized within the array using loess normalization of the M values, and normalized across arrays using quantile normalization to the A values (54). Linear model with adjustment for correlation between duplicate spots was then applied using the FDR approach. The limma package in R was used for the analysis (55).
Study approval.
Approval for these studies was obtained from the Dana-Farber Cancer Institute Institutional Review Board. Informed consent was obtained from all patients and healthy volunteers in accordance with the Declaration of Helsinki. All mice were treated, monitored, and sacrificed in accordance with an approved protocol of the Dana-Farber Cancer Institute Animal Care and Use Committee.
Supplementary Material
Acknowledgments
The authors thank Lisa Cameron (Confocal and Light Microscopy Core Facility, Dana-Farber Cancer Institute) and Maria Ericcson (Electron Microscopy Facility, Harvard Medical School) for technical help; Riccardo Dalla-Favera for providing C57BL/6 miR-15a/16-1–/– mice; and Siobhan Glavey for editing the manuscript. This work was supported by NIH grants R01CA154648, R01CA133799, and R01CA125690 and by Accademia Nazionale dei Lincei (Rome, Italy).
Footnotes
Conflict of interest: Irene M. Ghobrial serves on advisory boards for Bristol-Myers Squibb, Millennium, Novartis, and Noxxon and receives research support from Noxxon and Bristol-Myers Squibb. Kenneth C. Anderson serves on the board of directors and/or advisory committees for Celgene, Millennium, Bristol-Myers Squibb, and Onyx and is a scientific founder of Acetylon and Oncopep. David T. Scadden is a stock owner and consultant for Fate Therapeutics, serves as a consultant for GlaxoSmithKline and Bone Therapeutics, and receives research support from GlaxoSmithKline.
Citation for this article: J Clin Invest. 2013;123(4):1542–1555. doi:10.1172/JCI66517.
References
- 1.Chauhan D, et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood. 1996;87(3):1104–1112. [PubMed] [Google Scholar]
- 2.Gupta D, et al. Adherence of multiple myeloma cells to bone marrow stromal cells upregulates vascular endothelial growth factor secretion: therapeutic applications. Leukemia. 2001;15(12):1950–1961. doi: 10.1038/sj.leu.2402295. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S, et al. Bone marrow angiogenic ability and expression of angiogenic cytokines in myeloma: evidence favoring loss of marrow angiogenesis inhibitory activity with disease progression. Blood. 2004;104(4):1159–1165. doi: 10.1182/blood-2003-11-3811. [DOI] [PubMed] [Google Scholar]
- 4.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raposo G, Tenza D, Mecheri S, Peronet R, Bonnerot C, Desaymard C. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol Biol Cell. 1997;8(12):2631–2645. doi: 10.1091/mbc.8.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Théry C, et al. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147(3):599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Niel G, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121(2):337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 9.Blanchard N, et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol. 2002;168(7):3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 10.Mears R, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4(12):4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 11.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 12.Giri PK, Schorey JS. Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLoS One. 2008;3(6):e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 14.Andreola G, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195(10):1303–1316. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11(3):1010–1020. [PubMed] [Google Scholar]
- 16.Liu C, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176(3):1375–1385. doi: 10.4049/jimmunol.176.3.1375. [DOI] [PubMed] [Google Scholar]
- 17.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180(11):7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 18.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 19.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 21.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 22.Ostrowski M, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 23. Karolina DS, et al. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS One. 2011. 6 8 e22839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominici M, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 25.Garayoa M, et al. Mesenchymal stem cells from multiple myeloma patients display distinct genomic profile as compared with those from normal donors. Leukemia. 2009;23(8):1515–1527. doi: 10.1038/leu.2009.65. [DOI] [PubMed] [Google Scholar]
- 26.Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009;4(1):e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsiades CS, Mitsiades NS, Munshi NC, Richardson PG, Anderson KC. The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: interplay of growth factors, their receptors and stromal interactions. Eur J Cancer. 2006;42(11):1564–1573. doi: 10.1016/j.ejca.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Montecalvo A, et al. Exosomes as a short-range mechanism to spread alloantigen between dendritic cells during T cell allorecognition. J Immunol. 2008;180(5):3081–3090. doi: 10.4049/jimmunol.180.5.3081. [DOI] [PubMed] [Google Scholar]
- 29.Yu S, et al. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178(11):6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 30. Chauhan D, et al. Targeting mitochondrial factor Smac/DIABLO as therapy for multiple myeloma (MM). Blood. 2007;109(3):1220–1227. doi: 10.1182/blood-2006-04-015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podar K, et al. Targeting PKC in multiple myeloma: in vitro and in vivo effects of the novel, orally available small-molecule inhibitor enzastaurin (LY317615.HCl). Blood. 2007;109(4):1669–1677. doi: 10.1182/blood-2006-08-042747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roccaro AM, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113(26):6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsiades CS, et al. Aplidin, a marine organism-derived compound with potent antimyeloma activity in vitro and in vivo. Cancer Res. 2008;68(13):5216–5225. doi: 10.1158/0008-5472.CAN-07-5725. [DOI] [PubMed] [Google Scholar]
- 34.Hideshima T, et al. Biologic sequelae of I{kappa}B kinase (IKK) inhibition in multiple myeloma: therapeutic implications. Blood. 2009;113(21):5228–5236. doi: 10.1182/blood-2008-06-161505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitsiades CS, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010. 16 4 483 -9. 10.1038/nm.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mhyre AJ, Marcondes AM, Spaulding EY, Deeg HJ. Stroma-dependent apoptosis in clonal hematopoietic precursors correlates with expression of PYCARD. Blood. 2009;113(3):649–658. doi: 10.1182/blood-2008-04-152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldstein RH, Reagan MR, Anderson K, Kaplan DL, Rosenblatt M. Human bone marrow-derived MSCs can home to orthotopic breast cancer tumors and promote bone metastasis. Cancer Res. 2010;70(24):10044–10050. doi: 10.1158/0008-5472.CAN-10-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sipkins DA, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435(7044):969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soleimani M, Nadri S. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat Protoc. 2009;4(1):102–106. doi: 10.1038/nprot.2008.221. [DOI] [PubMed] [Google Scholar]
- 40.Maggini J, et al. Mouse bone marrow-derived mesenchymal stromal cells turn activated macrophages into a regulatory-like profile. PLoS One. 2010;5(2):e9252. doi: 10.1371/journal.pone.0009252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arendt BK, et al. Interleukin 6 induces monocyte chemoattractant protein-1 expression in myeloma cells. Leukemia. 2002;16(10):2142–2147. doi: 10.1038/sj.leu.2402714. [DOI] [PubMed] [Google Scholar]
- 42.Jöhrer K, Janke K, Krugmann J, Fiegl M, Greil R. Transendothelial migration of myeloma cells is increased by tumor necrosis factor (TNF)-alpha via TNF receptor 2 and autocrine up-regulation of MCP-1. Clin Cancer Res. 2004;10(6):1901–1910. doi: 10.1158/1078-0432.CCR-1053-03. [DOI] [PubMed] [Google Scholar]
- 43.Khan S, Jutzy JM, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16(1):1–12. doi: 10.1007/s10495-010-0534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The International Myeloma Working Group Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121(5):749–757. doi: 10.1046/j.1365-2141.2003.04355.x. [DOI] [PubMed] [Google Scholar]
- 45.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells towards a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012. 109 31 E2110 –E2116 10.1073/pnas.1209414109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang C, Kim SH, Bianco NR, Robbins PD. Tumor-derived exosomes confer antigen-specific immunosuppression in a murine delayed-type hypersensitivity model. PLoS One. 2011. 6 8 e22517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mestdagh P, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–60. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghobrial IM, et al. Proteomic analysis of mantle-cell lymphoma by protein microarray. Blood. 2005;105(9):3722–3730. doi: 10.1182/blood-2004-10-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie ME, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23(20):2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 54.Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 55.Smyth GK. Bioinformatics and Computational Biology Solutions using R and Bioconductor, R. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit R, Irizarry W, eds. New York, New York, USA: Springer; 2005:397–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.