Abstract
A transgenic primate model for Huntington's Disease (HD) first reported by our group that (HD monkeys) carry the mutant Huntingtin (HTT) gene with expanded polyglutamine (CAG) repeats and, develop chorea, dystonia, and other involuntary motor deficiencies similar to HD [1]. More recently, we have found that longitudinal magnetic resonance imaging of the HD monkey brain revealed significant atrophy in regions associated with cognitive deficits symptomatic in HD patients, providing the first animal model which replicates clinical phenotypes of diagnosed humans. Here we report germline transmission of the pathogenic mutant HTT in HD monkey by the production of embryos and subsequent derivation of HD monkey embryonic stem cells (rHD-ESCs) using HD monkey sperm. rHD-ESCs inherit mutant HTT and green fluorescent protein (GFP) genes through the gametes of HD monkey. rHD-ESCs express mutant HTT and form intranuclear inclusion, a classical cellular feature of HD. Notably, mosaicism of the pathogenic polyQ region in the sperm as well as derived ESCs were also observed, consistent with intraindividual and intergenerational reports of mosaic CAG repeats [2,3]and CAG expansion in HD patients [4–7]. The confirmation of transgene inheritability and development of pathogenic HD phenotype in derived rHD-ESCs reported in this study is a milestone in the pursuit of a transgenic primate model with inherited mutant HTT for development of novel disease biomarkers and therapeutics.
Introduction
Huntington's Disease (HD) is a devastating neurodegenerative disorder that leads to motor disability and cognitive deterioration throughout the course of the disease, with a duration of approximately 15 years after clinical symptoms appear [8,9]. HD is a genetic disorder caused by the expansion of the polyQ at the exon 1 of the IT15 gene that encodes the HTT protein. CAG repeat lengths over 39 in humans result in pathological HD. A negative correlation has been shown between repeat length and age of onset [6,7,10,11] and lifespan [12]. Patients with the long CAG repeats exhibit severe symptoms of HD in adolescence [6,7,11]. Although recent advancement in induced pluripotent stem cell technology has opened a new gateway for modeling human diseases using patients own pluripotent stem cells [13–19], the rationale in the derivation of rHD-embryonic stem cells (ESCs) from a transgenic HD monkey are (1) confirming germline transmission of transgenic HD monkeys and (2) predicting expression pattern of the mutant HTT transgene in second generation HD monkeys. Nonhuman primates are a unique model for studying human diseases due to the high similarity in physiology and genetics with humans [1,20,21]. A transgenic nonhuman primate which carries a genetic defect leading to a human disease, such as HD will not only recapitulate human conditions but also the pathogenic events that are critical for investigating disease pathogenesis and the development of novel biomarkers and treatments [1,20,21].
Materials and Methods
Generation of transgenic HD monkey [1,21,22]
We developed a transgenic HD rhesus macaque, rHD1, born and raised at the Yerkes National Primate Center of Emory University following the IACUC protocols for animal care. In brief, lentiviruses carrying exon 1 of the HTT gene containing 84 CAGs under the control of human polyubiquitin C (UBC) promoter was used to infect metaphase II arrested rhesus monkey oocytes followed by fertilization and embryo transfer into surrogate females.
Regimen of follicular stimulation [21,23]
Female rhesus monkeys exhibiting regular menstrual cycles were induced with exogenous gonadotropins. On day 1 or 2 of menses, subcutaneous human follicle-stimulating hormone [r-FSH: Organon Inc. 30 IU, intramuscular injection (IM)] was injected twice daily for 6 days. On day 7–10, daily injection of Gonadotropin-releasing hormone (GnRH) antagonist (Acyline 75 mg/kg; NICHD), r-FSH (20 IU, IM, twice daily) and recombinant human-luteinizing hormone (r-hLH; Ares Serono; 30 IU each, IM, twice daily) were administered. Ultrasonography was performed on day 7 of the stimulation to confirm follicular responses. An IM injection of 1,000 IU recombinant human chorionic gonadotropin (r-hCG; Ares Serono) was administered for ovulation at approximately 37 h before oocyte retrieval.
In vitro maturation [21,23]
Oocytes were matured in modified CMRL-1066 containing 10% heat-inactivated fetal bovine serum (FBS; Hyclone Laboratories, Inc.) supplemented with 40 μg/mL Sodium pyruvate, 150 μg/mL Glutamine, 550 μg/mL Calcium lactate, 100 ng/mL estradiol, and 3 μg/mL of Progesterone for up to 36 h in 35-μL drops of medium under mineral oil at 37°C with 5% CO2, 5% O2, and 90% N2.
Production of transgenic HD monkey embryos and establishment of ESCs [21,23–25]
MII arrested oocytes were retrieved from hormone-stimulated female rhesus macaques and used for in vitro fertilization by intracytoplasmic sperm injection (ICSI) followed by in vitro culture using HECM-9 [24]. HD and WT-Control monkeys were chair trained for semen collection using IACUC approved electroejaculation protocol [20,21,23]. Expanded blastocysts were selected for inner cell mass (ICM) isolation by using XYClone laser (Hamilton Throne, Inc.). The isolated ICMs were cultured until attached onto mouse fetal fibroblasts (MFFs; ICR mice) to form an outgrowth. Outgrowths with prominent stem cell morphology were mechanically passaged and continued in culture. Monkey ESCs were cultured in medium composed of knockout-Dulbecco's modified Eagle's medium (KO-DMEM) supplemented with 20% Knock-out Serum Replacement (KSR; Invitrogen), 1 mM glutamine, 1% nonessential amino acids, and supplemented with 4 ng/mL of human basic fibroblast growth factor (bFGF; Chemicon).
Immunostaining of stem cell markers [24,25]
rHD-ESC culture was fixed in 4% paraformaldehyde (PFA), permeabilized by 1% Triton-X (excluded for cell surface markers), blocked with 2% BSA and 130 mM glycine in a phosphate buffer saline (PBS). After overnight incubation with primary antibodies, Nanog (Santa Cruz Biotechnology Cat# sc-30329 Conc: 1:200), Oct4 (Santa Cruz Biotechnology Cat# sc-5279 Conc: 1:500), SSEA-4 (Chemicon Cat# MB4303 Conc: 1:250), TRA-1-60 (Chemicon MAB4360 1:200) followed by thorough washes, a secondary antibody conjugated with Alexa Red (Molecular Probe Cat# A21203 Conc: 1:1000) was used for detection of the primary antibodies. DNA was counterstained with Hoechst 33342 (5 μg/mL Molecularprobe Conc 1:10000). The specimen was examined using epifluorescent microscope. Alkaline phosphatase assay was performed following the manufacturer's instruction (Vector Lab Cat# SK-5300).
Immunostaining of mutant HTT [24,25]
Differentiated rHD-ESCs were fixed using 4% PFA for 15 min, permeabilized, and blocked. The sample was then incubated with primary antibody mEM48 (1:50) at 4°C overnight. After washing with the PBS, the samples were processed with avidin/biotinylated enzyme complex using the Vectastain Elite ABC kit (Vector Laboratories, Cat# PK-6102), and immediately stained with DAB (Vector Laboratories, Cat# SK-4100) for 30–40 s. Cell samples were examined and captured by CellSens software (Olympus, Inc.). For fluorescent imaging, a secondary antibody conjugated with Alexa Red (Molecular Probe) was used for detection of the primary antibody. DNA was counterstained with Hoechst 33342 (5 μg/mL).
Cytogenetic analysis/G-banding analysis [24,25]
rHD-ESCs were treated with KaryoMax® colcemid (Invitrogen) for 20 min, dislodged with 0.05% Trypsin-EDTA, centrifuged, and resuspended in hypotonic 0.075 M KCl solution for 20 min. After centrifugation, the cells were fixed thrice in a 3:1 ratio of methanol to glacial acetic acid. The cell pellet was resuspended in 1 mL of fixative and stored at 4°C. For GTL-Banding, the fixed cell suspension was dropped on wet slides, air dried, and baked at 90°C for 1 h. Slides were immersed in 0.5× Trypsin-EDTA (Invitrogen) with two drops of 67 mM Na2HPO4 for 20 to 30 s, rinsed in distilled water and stained with Leishman Stain (Sigma) for 90 s. Twenty metaphases were analyzed for numerical and structural chromosome abnormalities using an Olympus BX-40 microscope. Images were captured and at least two cells were karyotyped using the CytoVysion® digital imaging system (Applied Imaging). Karyotyping service provided by Cell Line Genetics, LLC.
In vitro differentiation to neuronal lineage [24,25]
rHD-ESC clumps were cultured in suspension for 7 days for the formation of embryoid bodies (EBs) and then allowed to attach onto a gelatin-coated plate. EBs were cultured in N1 medium (selection of neural progenitor cells; KO-DMEM [Invitrogen Cat# 10829-018]) supplemented with minimum essential amino acid (Invitrogen 10370-021), 200 mM of L-glutamine (Invitrogen Cat# 25030-081) and N2 supplement (Invitrogen Cat# 17502048) for 7 days, in N2 medium (neural progenitor cells expansion; N1 medium supplemented with 20 ng/mL bFGF [R&D Systems 233-FB-025]) for 14 days and in N3 medium (maturation of neuronal cells; KO-DMEM supplemented with 1% FBS [Hyclone]) and B27 supplement (Invitrogen 17504-044) for 7 days to allow differentiation into mature neuronal cell types. Successful differentiation of neuronal cell types was confirmed by the expression of Nestin (Chemicon Cat# MAB353,CONC 1:500), microtubule-associated protein (MAP2 Millipore Cat# AB5266 Conc: 1:200), neuron specific βIII tubulin(Chemicon Cat# MAB1637 Conc: 1:500), glial fibrillary acidic protein (GFAP Chemicon Cat# MAB360 Conc:1:500), and choline acetyltransferase (CHAT; Millipore Cat# AB143,Conc 1:200) by using specific antibodies [24,25].
Formation of teratoma in SCID mice [24,25]
Undifferentiated rHD-ESC clumps were recovered mechanically and implanted under the kidney capsule of severely compromised immune deficient (SCID) mice for 6 to 8 weeks. Animals were then euthanized followed by recovering of the teratoma and histological analysis. All animal procedures were approved by the IACUC at Emory University.
Genomic DNA isolation and genotyping PCR [1]
Genomic DNA (gDNA) was extracted from a cell pellet generated from all cell lines using the Promega Wizard Kit. Additionally, gDNA was extracted from sperm with phenol-chloroform extraction and precipitation with isopropanol. For genotyping of mutant HTT and green fluorescent protein (GFP) transgenes, 50 ng of gDNA was amplified by PCR at an annealing temperature of 68° for 40 cycles. For mutant HTT, primer set of HD32-F 5′-CTACGAGTCCCTCAAGTCCTTCCAGC-3′ and MD177-R 5′-GACGCAGCAGCGGCTGTGCCTG-3′ were used with expected amplicon size with 29 CAGs of 185 bp. For GFP, primer set of Ubiquitin-F1 5′-GAGGCGTCAGTTTCTTTGGTC-3′ and EGFP-R1 5′-CTGCTGCCCGACAACCACTA-3′ were used with expected amplicon size of 869 bp. All products were electrophoresed on a 1.5% agarose gel and visualized by ethidium bromide.
Sequencing of polyQ [1]
For polyQ sequencing from the transgenic HTT transcripts of all HD cell lines, 500 ng of total RNA was extracted from the cells and reverse-transcribed to cDNA using the High Capacity Reverse Transcription Kit (Applied Biosystems). The HTT transcript was amplified from the cDNA by PCR at an annealing temperature of 67° for 40 cycles with the following primers; HD32-Forward 5′-CTACGAGTCCCTCAAGTCCTTCCAGC-3′ and MD177-Reverse 5′-GACGCAGCAGCGGCTGTGCCTG-3′. All PCR products were electrophoresed on a 1.5% agarose gel and target bands were gel purified, cloned into the pGEM-T easy vector (Promega), and subsequently sequenced at Genewiz Corporation with T7 and SP6 primers. Both the expanded polyQ mHTT transgene and the endogenous HTT gene were cloned into the pGEM-T easy vector and sequenced.
Real-time PCR quantitation of HTT mRNA expression [1]
To quantitate the expression of HTT mRNA from all cell lines, 500 ng of total RNA from all cells was reverse transcribed to cDNA. Quantitation of HTT mRNA expression was performed using custom-designed gene-specific Taqman assays (forward sequence - 5′GCCGCTGCTGCCTCA4 ′3; reverse sequence–5′TGCAGCGGCTCCTCAG′3; probe sequence- 5′CCGCCGCCCCCGCC′3) in a 1× final reaction of Taqman Gene Expression Master Mix (Applied Biosystems). All real-time PCR (qPCR) results were first normalized with the geometric mean of 2 endogenous controls, beta-actin (ACTB) and glyceraldehyde-3-phosphate dehydrogenase and (GAPDH) (custom Taqman assay for ACTB forward sequence- 5′GCCGTCTTCCCCTCCAT3′; reverse sequence–5′CTGACCCATGCCCACCAT′3; probe sequence–5′CACGCCCTGGTGCCTG′3; GAPDH measured with commercially available rhesus Taqman assay from ABI). The ACTB-GAPDH normalized qPCR results were then further normalized to expression of the endogenous HTT transcript via a Taqman assay designed to exon 10–11 of HTT (forward sequence–5′AGCCCTGTCCTTTCAAGAAAACAA′3; reverse sequence–5′CATCCTCCAAGGCTTCTTCTTCT′3; probe sequence - 5′CCTAAGAGCACTTTGCCTTT′3).
Results
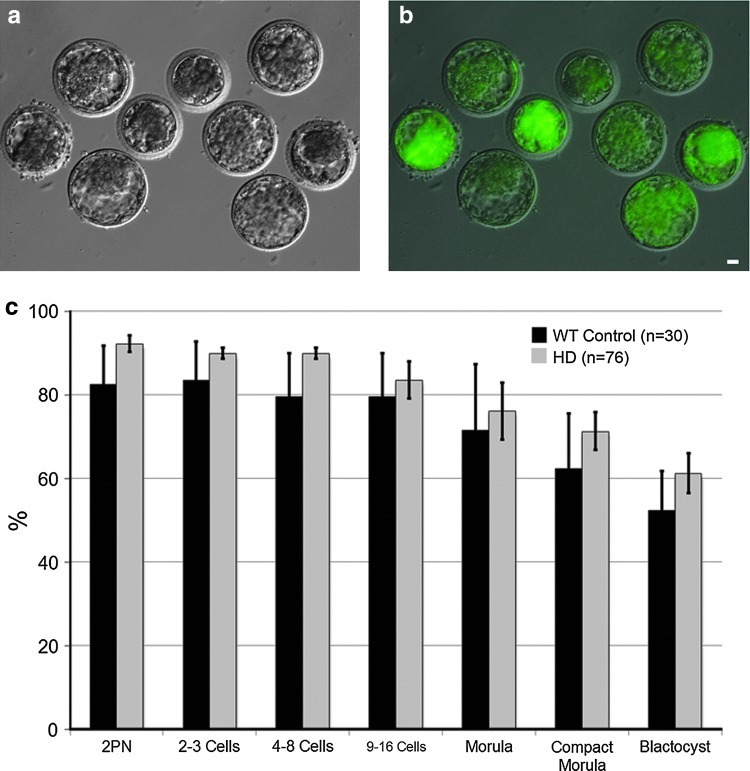
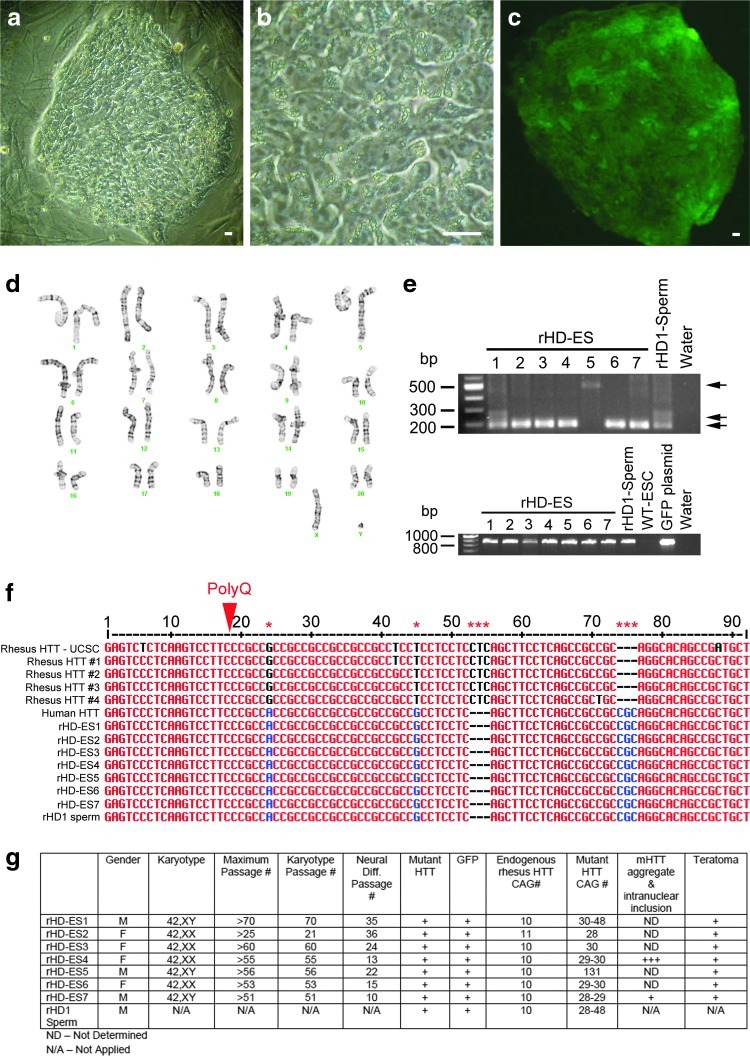
A male transgenic HD monkey, rHD1, carrying exon 1 of the human HTT (hHTT) gene with 29 CAG repeats and a GFP gene under the regulation of human polyubiquitin C (UBC) promoter was generated by lentiviral transfection of a mature oocyte followed by in vitro fertilization, culture, and embryo transfer into a surrogate female as described previously [1]. To determine successful germline transmission of the mHTT and GFP transgenes, we performed ICSI using sperm of rHD1 followed by in vitro production of embryos (Fig. 1) and the derivation of ESCs from the ICM of the resulting blastocyst stage embryos (Fig. 2a–c). Development of embryos fertilized with the sperm of rHD1 was similar to that of wild-type nontransgenic rhesus macaque embryos (WT-Control; Fig. 1c). Fertilization rate of rHD1 embryos was 92.5%±2% and 61.3%±4.8% for blastocyst, compared to WT-control group of 82.5%±9.2% and 52.3%±9.4%, respectively (Fig. 1c). A total of seven ESC lines were established from rHD1 derived blastocysts. Three of the rHD-ESC lines are male and four are female with normal karyotype based on a total of 20 cells analyzed in each cell line (Fig. 2d, g). All rHD-ESC lines carry mutant HTT and GFP transgenes and were confirmed by PCR (Fig. 2e). GFP expression was observed in all cell lines with the various intensity of green fluorescent (Fig. 1b), while all rHD-ESCs carry mutant HTT gene (Fig. 2e). Sperm from rHD1 carry 28 to 48 CAG repeats and rHD-ESC lines carry 28–131 CAG repeats suggesting possible germline expansion of polyQ (Fig. 2g). To further confirm if rHD-ESCs carry the human mutant HTT transgene, sequence analysis of the PCR amplicon flanking the polyQ region was aligned and compared with the following genes; HTT sequences of four control rhesus macaque, rhesus HTT sequence from the University of California at Santa Cruz (UCSC) genome database, human HTT, and human mutant HTT transgene from the 7 rHD-ESC cell lines (Fig. 2f). All rHD-ESC lines carry the human specific HTT sequence with variations in the number of pathogenic CAG repeats (Fig. 2e–g).
FIG. 1.
Development of transgenic Huntington's Disease (HD) monkey embryos. (a) Transmission light image of HD monkey blastocyst; (b) epifluorescent image of HD monkey blastocyst expressing green fluorescent protein (GFP); (c) development rate of HD monkey embryos. Bar=20 μm.
FIG. 2.
Establishment and characterization of rHD-embryonic stem cells (ESCs). (a) Transmission light image of rHD-ESC colony (low magnification); (b) transmission light image of rHD-ESC colony (high magnification); (c) epifluorescent image of rHD-ESC colony expressing GFP; (d) karyotype of rHD-ESC; (e) genotyping of rHD-ESCs by PCR analysis. Top panel-genotype of mutant HTT transgene. Lower panel-genotype of GFP transgene; (f) alignment of flanking regions of the CAG repeats of PCR amplicon of human HTT, rhesus macaque HTT, rHD1 sperm, and rHD-ESC mutant HTT genes; (g) summary of rHD-ESCs. Bar=10 μm.
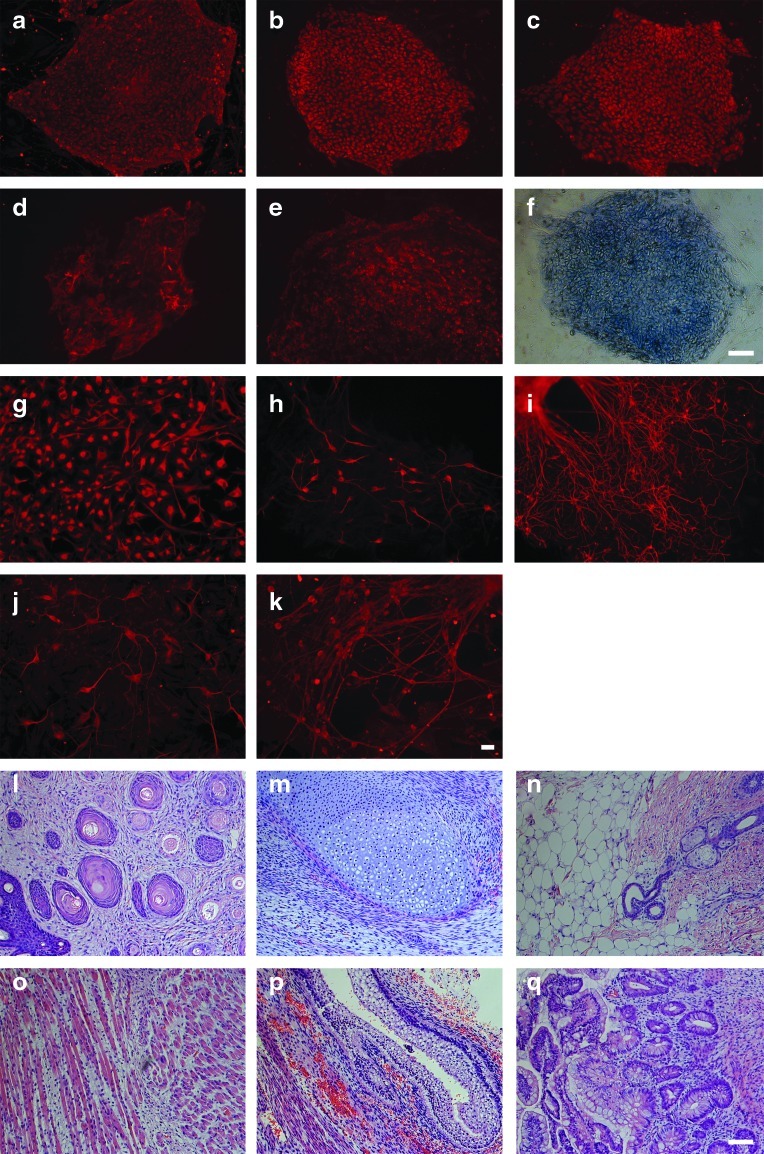
Each of the rHD-ESCs expresses stem cell markers, including Nanog, Oct4, Sox2, SSEA4, TRA-1-60, and alkaline phosphatase by immunocytochemistry (Fig. 3a–f). In addition to the expression of stem cell markers, rHD-ESC lines can differentiate into neural cells in vitro and were confirmed by immunocytochemistry using specific antibodies that recognize Nestin (NES), microtubule-associated protein (MAP2), neuron specific βIII tubulin (TUBB3), GFAP, and CHAT (Fig. 3g–k). Teratoma assay in immune compromised mice further demonstrated the pluripotency of rHD-ESC lines and their capability to differentiate into different lineages (Fig. 2g and Fig. 3l–q).
FIG. 3.
Expression of stem cell and differentiation markers, and the formation of teratoma. (a–e) Immunostaining of undifferentiated rHD-ESCs with antibodies which recognize stem cell specific proteins (A-Nanog, B-Oct4, C-Sox2, D-SSEA4 and E-TRA-1-60); (f) alkaline phosphatase staining; (g–k) immunostaining of in vitro neural-differentiated rHD-ESCs with antibodies which recognize neural-specific proteins (G-Nestin, H-Map2, I-βIII tubulin, J-GFAP, and K-CHAT). (l–q) Teratoma derived from rHD-ESC grafts under the kidney capsule of immune compromised mice (l: ectoderm; m–o: mesoderm; p–q: endoderm). Teratoma composed of tissues originated from different germlayers. Bar (a–f)=50 μm; Bar (g–k)=10 μm; Bar (l–q)=50 μm.
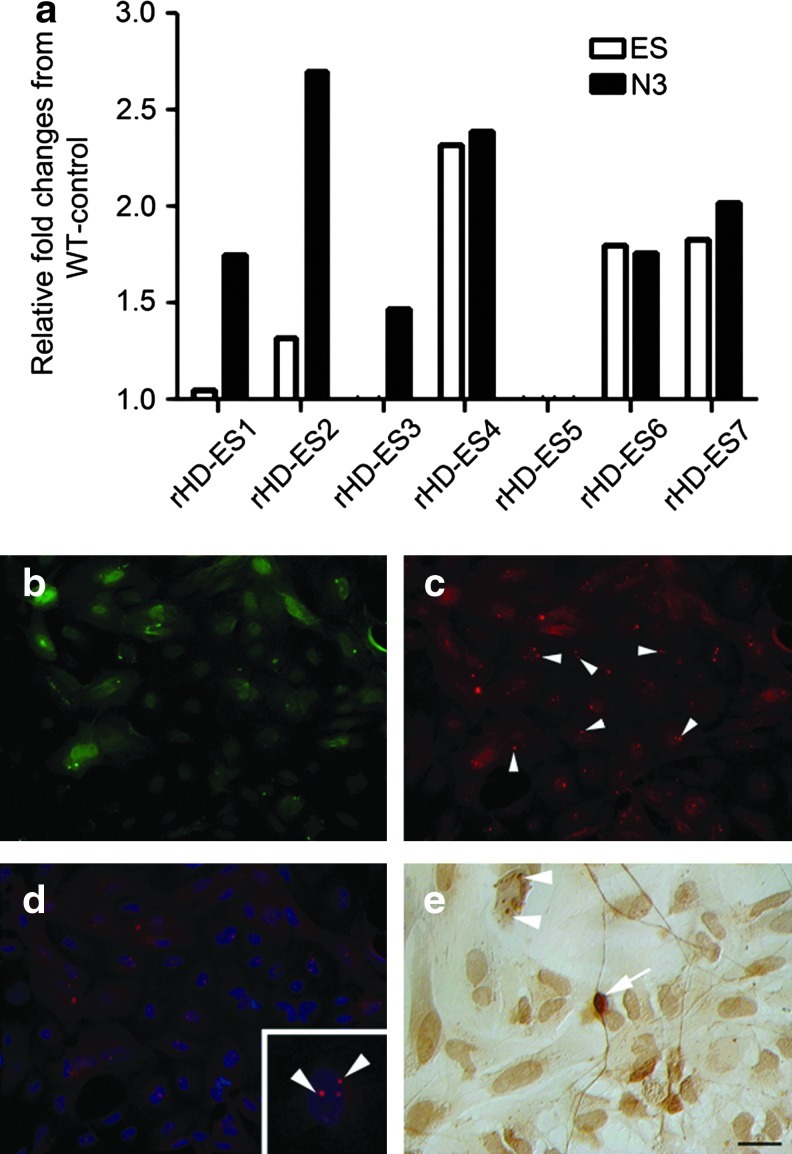
Expression of the GFP transgene was observed in embryos and rHD-ESCs (Fig. 1b and Fig. 2c) by fluorescent microscopy. The expression level of mutant HTT in the rHD-ESC lines was evaluated by Q-PCR analysis (Fig. 4a) and immunostaining using an antibody specifically recognizing mutant HTT (Fig. 4b–e). The expression of mutant HTT was detected in undifferentiated rHD-ESCs with increased expression as neural differentiation progresses in culture of rHD-ES1, 2, 3, and 7 when compared with endogenous HTT and WT-control (Fig. 4a). The expression level of HTT in rHD-ES5 was not upregulated compared to control ESC levels (Fig. 2e). It is not clear whether the expression of mutant HTT in rHD-ES5 is impacted by the expanded polyQ (131Q), or if aberrant gene rearrangement occurred. While the protein level of aggregated mutant HTT was too low to be detected by western blot analysis, aggregated mutant HTT and intranuclear inclusions in the neural-differentiated rHD-ESCs was detected by immunostaining in rHD-ES4 and rHD-ES7 (Fig. 4b–e).
FIG. 4.
Expression of mutant HTT. (a) Expression level of mHTT in rHD-ESCs was determined by Q-PCR and was normalized to endogenous HTT and WT-control monkey HTT expression. Undifferentiated rHD-ESCs (ES) and in vitro neural-differentiated rHD-ESCs (N3) were collected, followed by RNA extraction and real-time PCR (qPCR) analysis with HTT specific Taqman assays (Applied Biosystems). All qPCR results were normalized with the geometric mean of two endogenous controls, beta-actin (ACTB) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and then normalized to WT-Control; (b) epifluorescent image of GFP expression in neural-differentiated rHD-ESCs; (c) immunstaining of in vitro neural-differentiated rHD-ESCs using antibody specific to mutant HTT aggregates; (d) a montage of epifluorescent images of DNA staining (blue) and (c); (e) immunohistochemistry of mutant HTT aggregates using antibody specific to mutant HTT aggregates and detected by avidin/biotinylated enzyme complex followed by staining with DAB (arrow-cytoplasmic mutant HTT aggregate; arrowhead-nuclear inclusion). Bar=10 μm.
Discussion
One of the key rationales in developing a transgenic animal model is the inheritance of the transgene through the germ cells which mimics the inheritable pattern of human genetic disorders [1,20,22,26,27]. Unlike most of the laboratory animal species, rhesus macaque reaches pubertal age at around 4 years old. It is a relatively long physiological event compared to most laboratory species, including small nonhuman primates, such as marmosets [28]. While there is no perfect animal model to embody humans, it is important to identify appropriate model systems to address specific underlying pathogenic mechanisms of inherited neurological disorders, such as HD, that are influenced by aging events [29,30]. A nonhuman primate model is particularly important in modeling diseases, such as HD since various systems in the body are impacted during the course of the disease, which include motor functions, psychiatric, and metabolic disturbance [9,21,31]. To capture the systemic impact of disease, such as HD, nonhuman primates can effectively model the progression of HD through longitudinal analysis with similar clinical measurements evaluated in human patients, such as magnetic resonance imaging. Rhesus macaques also share a similar motor repertoire that allows the evaluation of fine movement control that most of the currently available model systems cannot offer [29]. Most importantly, psychiatric impact can be evaluated with a sophisticated battery of cognitive behavioral tests developed for macaques that cannot be examined in other species [32–34].
rHD1, one of the first transgenic HD monkeys, has reached pubertal age and germline transmission was confirmed by fertilization of mature macaque oocytes with HD monkey sperm followed by the production of transgenic embryos (Fig. 1a, b) and subsequent derivation of rHD-ESCs (Fig. 2). Our result suggests that sperms carrying mutant HTT and GFP transgene are not impacted, and the development competence of the resulted embryos was similar to wild-type nontransgenic monkey embryos. rHD-ESCs inherit and express both the mutant HTT and GFP transgenes (Fig. 2c, e, f), and develop unique HD cellular features, including intranuclear inclusions, as they differentiate toward neuronal lineage (Fig. 4b–e). It is interesting that all rHD-ESC lines carry both mutant HTT and GFP transgenes. rHD1 carry a single copy of mutant HTT transgene and at least six copies of the GFP transgenes [1]. The two transgenes are expected to segregate independently because independent lentiviral integration event is expected [35]. As a result, each rHD-ESC line may carry different copy of GFP transgene which may result in the variation in GFP expression level (Fig. 1b). Furthermore, we found instability of the pathogenic CAG repeat in the gametes of the HD monkey as well in the ESCs, which is an underlying feature of not only HD [6,7,11] but other triplet nucleotide repeat diseases as well, including spinocerebellar ataxia type 1 [36,37] and dentatorubral-pallidoluysian atrophy [38]. Notably, this is the first report, to the best of our knowledge, of CAG mosaicism in a primate animal model for HD, and could, therefore, serve as a unique tool for further evaluation of this pathogenic mechanism. While rHD-ESCs could be a useful stem cell model for studying HD, the confirmation of germline transmission through the gametes is an important and critical step in the development of transgenic primate models of human inherited genetic diseases. Overall, the successful inheritability of the mutant HTT transgene in our nonhuman primate model can now lead to the establishment of second generation HD monkeys for studies of HD pathogenic trajectories and much needed development of novel treatments.
Acknowledgments
All protocols involving animal care and handling were approved by Emory University's IACUC. We thank Tim Chi and Adam Neumann for assisted reproduction in rhesus macaque and Dr. X.J.Li for providing mEM48 antibody. Acyline was provided by NICHD. We thank Yerkes National Primate Research Center (YNPRC) veterinarian and animal care staff for providing outstanding services. KP and RP are supported by the Royal Golden Jubilee PhD program of Thailand Research Fund. YNPRC is supported by the National Center for Research Resources P51RR165 and is currently supported by the Office of Research and Infrastructure Program (ORIP)/OD P51OD11132. This study is supported by a grant awarded by the ORIP/NIH (RR018827) to AWSC.
Author Disclosure Statement
The authors have no commercial association that might create a conflict of interest in connection with the submitted manuscript.
References
- 1.Yang SH. Cheng PH. Banta H. Piotrowska-Nitsche K. Yang JJ. Cheng EC. Snyder B. Larkin K. Liu J, et al. Towards a transgenic model of Huntington's disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telenius H. Kremer B. Goldberg YP. Theilmann J. Andrew SE. Zeisler J. Adam S. Greenberg C. Ives EJ. Clarke LA. Hayden MR. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 3.Telenius H. Kremer HP. Theilmann J. Andrew SE. Almqvist E. Anvret M. Greenberg C. Greenberg J. Lucotte G, et al. Molecular analysis of juvenile Huntington disease: the major influence on (CAG)n repeat length is the sex of the affected parent. Hum Mol Genet. 1993;2:1535–1540. doi: 10.1093/hmg/2.10.1535. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas G. Devys D. Goldenberg A. Maltete D. Herve C. Hannequin D. Guyant-Marechal L. Juvenile Huntington disease in an 18-month-old boy revealed by global developmental delay and reduced cerebellar volume. Am J Med Genet. 2011;155A:815–818. doi: 10.1002/ajmg.a.33911. [DOI] [PubMed] [Google Scholar]
- 5.Ribai P. Nguyen K. Hahn-Barma V. Gourfinkel-An I. Vidailhet M. Legout A. Dode C. Brice A. Durr A. Psychiatric and cognitive difficulties as indicators of juvenile huntington disease onset in 29 patients. Arch Neurol. 2007;64:813–819. doi: 10.1001/archneur.64.6.813. [DOI] [PubMed] [Google Scholar]
- 6.Langbehn DR. Hayden MR. Paulsen JS. CAG-repeat length and the age of onset in Huntington disease (HD): a review and validation study of statistical approaches. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:397–408. doi: 10.1002/ajmg.b.30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JM. Ramos EM. Lee JH. Gillis T. Mysore JS. Hayden MR. Warby SC. Morrison P. Nance M, et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78:690–695. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scahill RI. Hobbs NZ. Say MJ. Bechtel N. Henley SM. Hyare H. Langbehn DR. Jones R. Leavitt BR, et al. Clinical impairment in premanifest and early Huntington's disease is associated with regionally specific atrophy. Hum Brain Mapp. 2011 doi: 10.1002/hbm.21449. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabrizi SJ. Reilmann R. Roos RA. Durr A. Leavitt B. Owen G. Jones R. Johnson H. Craufurd D, et al. Potential endpoints for clinical trials in premanifest and early Huntington's disease in the TRACK-HD study: analysis of 24 month observational data. Lancet Neurol. 2012;11:42–53. doi: 10.1016/S1474-4422(11)70263-0. [DOI] [PubMed] [Google Scholar]
- 10.Snell RG. MacMillan JC. Cheadle JP. Fenton I. Lazarou LP. Davies P. MacDonald ME. Gusella JF. Harper PS. Shaw D DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 11.Geevasinga N. Richards FH. Jones KJ. Ryan MM. Juvenile Huntington disease. J Paediatr Child Health. 2006;42:552–554. doi: 10.1111/j.1440-1754.2006.00921.x. [DOI] [PubMed] [Google Scholar]
- 12.Andrew SE. Goldberg YP. Kremer B. Telenius H. Theilmann J. Adam S. Starr E. Squitieri F. Lin B, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 13.HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An MC. Zhang N. Scott G. Montoro D. Wittkop T. Mooney S. Melov S. Ellerby LM. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimos JT. Rodolfa KT. Niakan KK. Weisenthal LM. Mitsumoto H. Chung W. Croft GF. Saphier G. Leibel R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 16.Ma L. Hu B. Liu Y. Vermilyea SC. Liu H. Gao L. Sun Y. Zhang X. Zhang SC. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park IH. Arora N. Huo H. Maherali N. Ahfeldt T. Shimamura A. Lensch MW. Cowan C. Hochedlinger K. Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrier A. Peschanski M. How can human pluripotent stem cells help decipher and cure Huntington's disease? Cell Stem Cell. 2012;11:153–161. doi: 10.1016/j.stem.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Carter RL. Chan AW. Pluripotent stem cells models for Huntington's disease: prospects and challenges. J Genet Genomics. 2012;39:253–259. doi: 10.1016/j.jgg.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan AWS. Transgenic nonhuman primates of neurodegenerative diseases. Reprod Biol Endocrinol. 2004;2:39. doi: 10.1186/1477-7827-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang SH. Chan AW. Transgenic animal models of Huntington's disease. Curr Top Behav Neurosci. 2011;7:61–85. doi: 10.1007/7854_2010_105. [DOI] [PubMed] [Google Scholar]
- 22.Chan AW. Chong KY. Martinovich C. Simerly C. Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291:309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- 23.Chan AWS. Chong KY. Schatten G. Production of transgenic primates. In: Pinkert CA, editor. Transgenic Animal Technology. Academic Press; San Diego: 2002. pp. 359–394. [Google Scholar]
- 24.Chan AW. Cheng PH. Neumann A. Yang JJ. Reprogramming Huntington monkey skin cells into pluripotent stem cell. Cell Reprogram. 2010;12:509–517. doi: 10.1089/cell.2010.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laowtammathron C. Cheng EC. Cheng PH. Snyder BR. Yang SH. Johnson Z. Lorthongpanich C. Kuo HC. Parnpai R. Chan AW. Monkey hybrid stem cells develop cellular features of Huntington's disease. BMC Cell Biol. 2010;11:12. doi: 10.1186/1471-2121-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates GP. Mangiarini L. Davies SW. Transgenic mice in the study of polyglutamine repeat expansion diseases. Brain Pathol. 1998;8:699–714. doi: 10.1111/j.1750-3639.1998.tb00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schilling G. Becher MW. Sharp AH. Jinnah HA. Duan K. Kotzuk JA. Slunt HH. Ratovitski T. Cooper JK, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki E. Suemizu H. Shimada A. Hanazawa K. Oiwa R. Kamioka M. Tomioka I. Sotomaru Y. Hirakawa R, et al. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 29.Courtine G. Bunge MB. Fawcett JW. Grossman RG. Kaas JH. Lemon R. Maier I. Martin J. Nudo RJ, et al. Can experiments in nonhuman primates expedite the translation of treatments for spinal cord injury in humans? Nat Med. 2007;13:561–566. doi: 10.1038/nm1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice J. Animal models: Not close enough. Nature. 2012;484:S9–S9. doi: 10.1038/nature11102. [DOI] [PubMed] [Google Scholar]
- 31.Tabrizi SJ. Scahill RI. Durr A. Roos RA. Leavitt BR. Jones R. Landwehrmeyer GB. Fox NC. Johnson H, et al. Biological and clinical changes in premanifest and early stage Huntington's disease in the TRACK-HD study: the 12-month longitudinal analysis. Lancet Neurol. 2011;10:31–42. doi: 10.1016/S1474-4422(10)70276-3. [DOI] [PubMed] [Google Scholar]
- 32.Bachevalier J. Machado CJ. Kazama A. Behavioral outcomes of late-onset or early-onset orbital frontal cortex (areas 11/13) lesions in rhesus monkeys. Ann N Y Acad Sci. 2011;1239:71–86. doi: 10.1111/j.1749-6632.2011.06211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachevalier J. Malkova L. Mishkin M. Effects of selective neonatal temporal lobe lesions on socioemotional behavior in infant rhesus monkeys (Macaca mulatta) Behav Neurosci. 2001;115:545–559. doi: 10.1037//0735-7044.115.3.545. [DOI] [PubMed] [Google Scholar]
- 34.Ewing-Cobbs L. Prasad MR. Swank P. Kramer L. Mendez D. Treble A. Payne C. Bachevalier J. Social communication in young children with traumatic brain injury: relations with corpus callosum morphometry. Int J Dev Neurosci. 2012;30:247–254. doi: 10.1016/j.ijdevneu.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang SH. Cheng PH. Sullivan RT. Thomas JW. Chan AW. Lentiviral integration preferences in transgenic mice. Genesis. 2008;46:711–718. doi: 10.1002/dvg.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung MY. Ranum LP. Duvick LA. Servadio A. Zoghbi HY. Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 37.Orr HT. Chung MY. Banfi S. Kwiatkowski TJ., Jr. Servadio A. Beaudet AL. McCall AE. Duvick LA. Ranum LP. Zoghbi HY. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 38.Takano H. Onodera O. Takahashi H. Igarashi S. Yamada M. Oyake M. Ikeuchi T. Koide R. Tanaka H. Iwabuchi K. Tsuji S. Somatic mosaicism of expanded CAG repeats in brains of patients with dentatorubral-pallidoluysian atrophy: cellular population-dependent dynamics of mitotic instability. Am J Hum Genet. 1996;58:1212–1222. [PMC free article] [PubMed] [Google Scholar]