INTRODUCTION
Understanding the relationships between gene products is fundamental to biology. Measuring genetic interactions (GIs), the extent to which the function of one gene depends on a second, is an unbiased way of determining functional relationships and has proven to be a powerful technique for discovering gene function, grouping genes into complexes, and organizing them into pathways (Tong et al, 2004; Schuldiner et al., 2005; Ooi et al., 2006; Roguev et al., 2008; Costanzo et al., 2010; Horn et al., 2011). The development of high-density, quantitative assays for GI mapping in the budding yeast S. cerevisiae (Sc) led to numerous findings. For example, maps of ER and mitochondrial genes led to the discovery of the complex responsible for very-long-chain fatty acid biosynthesis (Denic and Weissman, 2007), identification of the GET complex and other factors responsible for tail-anchored membrane protein insertion (Schuldiner et al., 2008, 2005; Costanzo et al., 2010; Jonikas et al., 2009), discovery of the SPOTS complex as a regulator of sphingolipid homeostasis (Breslow et al., 2010), and identification of MitOS as a determinant of mitochondrial morphology (Hoppins et al. 2011). GI maps of other pathways have led to a range of insights, including discovery of novel mechanisms of epigenetic control (Collins et al., 2007; Costanzo et al., 2010; Dai et al., 2008).
These discoveries speak to the power of GI analysis to group genes into complexes and to chart connections between pathways independently of a priori knowledge. But how plastic are genetic pathways over the course of evolution? In addition to searching for novel factors and pathways in the fission yeast Schizosaccharomyces pombe (Sp), we sought to determine systematically the extent to which conserved genes have adapted to serve in different roles with different partners. It is a fundamental consequence of evolution that conserved genes encode macromolecules with conserved biochemical properties. Yet gene-to-phenotype relationships are not as predictable. For example, hypoxanthine-guanine phosphoribosyltransferase (HGPRT) catalyzes purine monophosphate generation in every organism, but mutations in yeast lead to abnormal mitochondrial genome maintenance and cisplatin resistance (Kowalski et al., 2008), while mutations in humans lead to the neuro-psychiatric signs of Lesch-Nyhan syndrome. Developmental biologists have noted that orthologous genes have been repurposed to control the morphologies of distinct body parts in highly divergent organisms (Niwa et al. 2010). Furthermore, point mutations in the active sites of metabolic enzymes can change substrate specificity or electron transfer steps with profound phenotypic consequences (e.g. IDH1/2, Dang et al., 2009). Finally, genetic relationships have been reported to change markedly when cells are challenged with a stress like DNA damage (Bandyopadhyay et al., 2010).
Efforts to study functional repurposing have been limited by a lack of global comparisons. Work in Sp now makes large scale comparisons to Sc possible. Fission yeasts diverged from budding yeasts ~500 million years ago and their genomes show no synteny (Rhind et al., 2011). Efforts to curate the genomes of Sp and Sc have identified a shared subset of ~4450 apparent orthologs (Aslett and Wood, 2006). In light of extensive GI data generated in Sc, availability of a Sp deletion collection (Kim, et al. 2010), methods for generating Sp double mutants in high-throughput (Roguev, et al. 2007, Dixon, et al. 2008), and annotation of ortholog tables between these organisms, we saw a unique opportunity to assess how often conserved genes acquire new functions and partners over the course of evolution.
In addition to an evolutionary analysis, functional mapping in Sp is valuable for discovering new cell biology. Investigators from many disciplines have reported that certain aspects of Sp are better models of metazoan biology. For example, they possess: i) an RNAi pathway, ii) repetitive centromeres, iii) G2/M cell cycle control, iv) contractile ring-driven cytokinesis, and v) complex heterochromatin and splicing regulation (Sabatinos and Forsburg, 2010; Rhind et al., 2011). Some of these processes have been studied extensively while others have received limited attention. We intended to characterize functional repurposing through evolution and – of equal value – to identify processes in which Sp genetics can predict the properties of mammalian cells.
We report that many genes displayed conserved patterns of GI and that core complexes or modules displayed highly correlated patterns of GI (Roguev et al., 2008; Dixon et al., 2008). However, an important subset of conserved genes manifested divergent genetic relationships. By comparing the functional profiles of annotated orthologs, we identified genes that have acquired new partners or were participating in different or additional pathways in Sp versus Sc. We chose three disparate cases of divergence for detailed investigation. Our findings reveal new organelle homeostasis mechanisms and mitosis control factors. Considered together, our findings suggest new applications for model organism-based research and impact our view of evolution.
RESULTS AND DISCUSSION
S. pombe Genetic Interaction Map
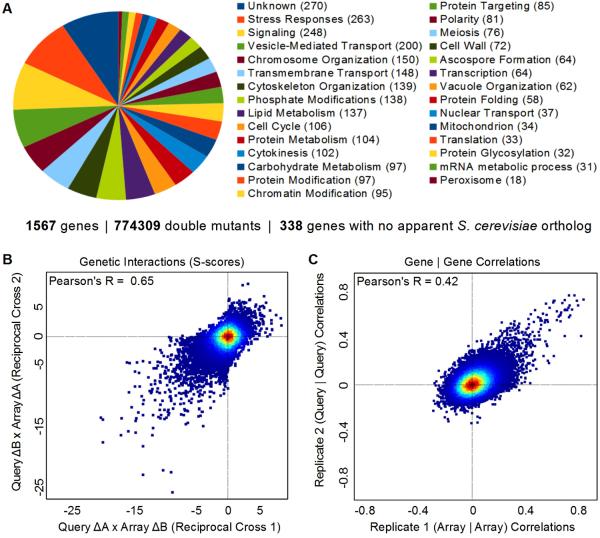
We evaluated pairwise GIs for 1,297 alleles in G418-marked “Array” strains crossed against 597 nourseothricin (NAT)-marked “Query” strains (Fig S1). Strains harboring 1,503 unique gene deletions and 64 unique hypomorphic (Degron-DAmP ; Schuldiner et al., 2005; Breslow et al., 2008) alleles of essential genes were used to generate 774,309 double mutants (expanding by ~eight-fold the number of GIs measured Sp Fig 1A). Interaction scores were determined by comparing the observed fitness of the double mutants with the typical fitness determined empirically from the expected penalty associated with each mutation (Fig S2, Collins et al., 2010). Our map consists of a matrix of GIs for 40% of the non-essential genome. Each row and column corresponds to the GI profile for one allele and is a phenotypic signature. We sorted the rows and columns of the matrix using hierarchical clustering such that neighbors have similar functional profiles. The resulting map is modular: correlated genes cluster together as a result of their shared GIs (Fig S3).
Figure 1. Dataset Overview and Reproducibility Benchmarks.
(A) Functional classification of genes in the map. (B) Scatter plot comparing scores from reciprocal crosses (C) Scatter plot of correlation coefficients between genetic interaction (GI) profiles for Query versus Array pairs of strains.
Multiple metrics for evaluating the dataset argued for its quality. First, biological triplicate measurements revealed that the scores are reproducible (overall correlation of ~ 0.78). Second, the same triplicate sets measured 6–12 months later remained highly correlated (~ 0.72). We also assessed the reproducibility of scores identified for reciprocal crosses: two scores that derived from independent measurements in which the antibiotic-resistance marker and the mating type of each strain were swapped (Query A x Array B versus Query B x Array A). Query strains and Array strains have different histories and are subjected to different storage and growth conditions during the assay and consequently may have differences in fitness. Despite these differences, the correlation of 0.65 for scores derived from reciprocal crosses is comparable to the highest quality Sc studies (Fig 1B) (Baryshnikova et al., 2010; Collins et al., 2010; Hoppins et al., 2011). From the score matrix, we computed pairwise correlations for all pairs of alleles from cells of the same mating type background and the reproducibility of the correlations observed between profiles (Fig 1C). Irreproducible correlations between profiles do occur as the majority of scores are between unrelated genes, but the diagonal is enriched with gene-to-gene correlations that were reproducible whether the strains being compared were h- or h+ pairs.
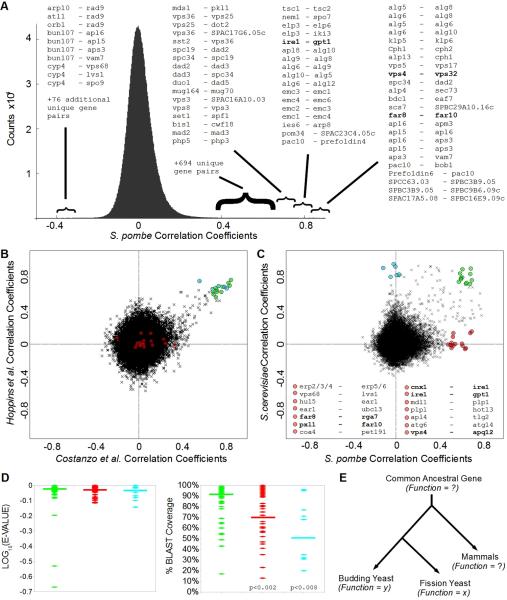
Our Sp map identified >700 high-confidence gene-to-gene correlations indicative of genes with related functions. Many of these are internal validations since they are known to be related (Fig 2A, Fig S3). Among the most notable clusters of genes, our analysis identified correctly the relationships between factors involved in the contractile ring, glycosylation, autophagy, retromer and ESCRT pathways, protein folding and quality control, the peroxisome, the G2/M transition, spindle and kinetochore assembly, lipid biosynthesis, hypoxia responses, clathrin adaptors and SNARE complexes, prefoldin, ubiquitin ligases and substrate adapters, mannosyltransferase and N-acetyltransferase complexes, mitochondria import and export, G-protein coupled receptor signaling, the Elongator complex, mRNA splicing, histone deacetylases, and the relationship between the alternative translocon and ER Membrane Protein Complex (Supplemental datasets, Fig 2A, Fig S3).
Figure 2. Functional Conservation versus Functional Repurposing.
(A) Distribution of correlation coefficients between GI profiles, with extreme cases of pairwise correlation and anti-correlation annotated as gene a - gene b. (B) Scatter plot of correlation coefficients comparing Sc data reported by Hoppins et al versus Costanzo et al. See Table S1 for a list of highly correlated profiles and highly reproducible (green) correlation relationships. (C) Scatter plot of correlation coefficients comparing GI profiles for Sp versus Sc. Highlighted examples of pairwise relationships that are correlated in Sp but not Sc are listed below the scatter plot. Bold indicates functional relationships explored in this study. See Table S1 for additional examples of correlated pairwise relationships conserved between Sp and Sc (green), and pairwise relationships that are correlated in Sc but not Sp (cyan) or Sp but not Sc (red). (D) Amino acid sequence comparison-based statistics for Sp versus Sc orthologs highlighted in 2B,C. Horizontal bars = median values. See Table S1 for BLAST scores, E-VALUEs, percent identities and overlap. (E) Functional Repurposing: the functions of ancestral genes are unknown, but for the factors studied here the apparent gene-to-gene and gene-to-phenotype relationships in Sp are divergent from those in Sc.
How reliable are such pairwise correlations for identifying bona fide functional partners? In addition to the reproducibility within a single dataset as shown in Fig 1C, the adoption of high-throughput GI assays by multiple groups makes it possible to compare inter-lab reproducibility. We compared the Sc gene-to-gene correlations reported by Costanzo et al. with those reported by Hoppins et al. from a shared subset of interactions that overlapped partially with orthologous interactions sampled in our Sp study (~500 interaction scores per profile). Gene-to-gene correlations that exceed ~0.4 between labs or within a dataset (Fig 1C versus Fig 2B) are highly likely to be reproducible, true positives that are robust to differences in data collection or analysis.
Functional Conservation versus Functional Repurposing
Using this same subset of orthologous interactions, we compared the gene-to-gene correlations observed in our Sp dataset with those observed in Sc (Fig 2C). As reported, (Roguev et al., 2008; Dixon et al., 2008), there is widespread functional conservation of gene-to-gene relationships. This is true especially for known complexes and pathways that appear to have descended from the last common ancestor unmodified (green, Table S1). However, our systematic view also revealed subsets of genes whose correlations in Sc were not observed in Sp and another subset whose correlations in Sp were not observed in Sc (Fig 2B–D, Table S1). We considered these to be plausible cases of functional repurposing: the adaption of conserved factors to serve additional or different roles in one organism versus the other (Fig 2E). We computed amino acid sequence comparison-based statistics for each case of highly correlated pairwise relationships conserved between Sp and Sc (green), versus relationships that are correlated in Sc but not Sp (cyan) or Sp but not Sc (red). Lower amino acid similarity did not correlate with repurposing (Fig 2D, left), but lower percentage coverage (i.e. additional motifs or domains present in only one of the orthologs) did correlate with apparent repurposing (Fig 2D, right). At the same time, these genes appear to be unique descendants of the same ancestral gene and to have adapted within the system of one organism versus the other to serve alternative functions.
Another explanation for some cases of divergent genetic relationships is the relative degree of redundancy within a pathway. For example, in Sp there are only two genes of the GOLD-domain family of COP-II coat components (SPAC17A5.08 and SPBC16E9.09). In Sc, there are three homologs of SPAC17A5.08 (ERP2, ERP3, and ERP4) and two homologs of SPBC16E9.09 (ERP5 and ERP6). SPAC17A5.08 and SPBC16E9.09 share virtually all of the same interactions in Sp, while none of the pairwise comparisons of ERP2/3/4 versus ERP5/6 profiles shared significant overlap in Sc (Fig 2C). This is expected but it highlights the value of Sp as a model eukaryote: it has few paralogs and thus there is an increased probability of detecting relationships between non-redundant factors (Aslett and Wood, 2006). A third explanation for divergent genetic correlations is simply that these organisms have different dependencies on a given process under the conditions of the assay (“organismal emphasis”). For example, autophagy genes were identified in Sc, but this analysis derived from growth on rich media where autophagy genes display few robust interactions in Sc. In Sp, autophagy pathway genes displayed strong interactions under the conditions used in our protocol (Supplemental Datasets).
As we were interested in functional repurposing, we selected three disparate cases of divergence that we could not explain by environmental dependencies or redundancy (Fig 2D, Table S1). The divergence was evident when comparing correlations from the subset of interactions used for Fig 2 and was clear when comparing the entire datasets. Each case represents a distinct pathway: the Unfolded Protein Response (UPR), spindle pole body (SPB) duplication, and mitosis. The genes involved have strong GIs and robust—but different—correlated partners in Sc versus Sp. Validation assays in all cases indicated that these genes have evolved different genetic relationships and serve in new or additional roles in fission versus budding yeast.
The Unfolded Protein Response Requires Gpt1 and Ire1
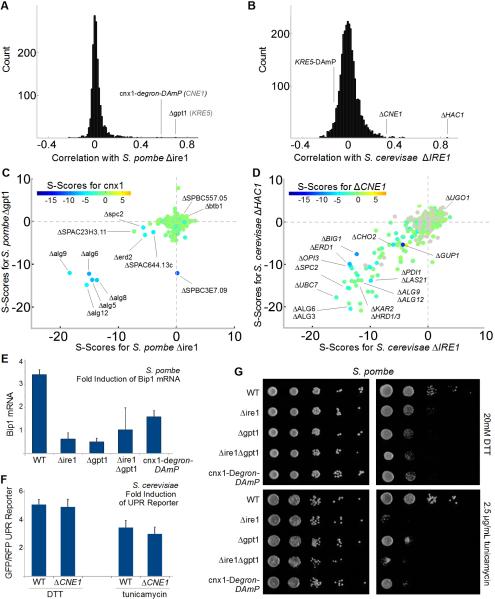
Ire1 is a conserved transmembrane kinase and nuclease that serves as the central sensor for misfolding stress in the ER. In Sc, Ire1 senses ER stress via its lumenal domains leading to oligomerization and activation of its nuclease to catalyze the unconventional splicing and activation of Ire1's direct and only substrate – the transcription factor Hac1. Active Hac1 then induces the UPR transcriptional program (Walter and Ron, 2011). This pathway is conserved though in metazoans the IRE1 ortholog has additional outputs and substrates. For example, in addition to splicing the Hac1 ortholog XBP1, metazoan IRE1 degrades ER-localized mRNAs (Hollien and Weissman, 2006; Hollien et al., 2009) thereby decreasing the ER folding burden in a pathway termed Regulated Ire1-Dependent Decay (RIDD). Conservation of the UPR in fission yeast remains unclear: Sp possesses Ire1 but does not possess an apparent Hac1/XBP1 ortholog.
In Sc, the functional relationship between IRE1 and HAC1 is among the most compelling cases of an unbranched, linear pathway. Accordingly, the GI profiles for IRE1 and HAC1 are highly correlated (Fig 3B). The correlation between these genes in Sc reflects their shared aggravating interactions with genes involved in lipid biosynthesis, protein folding, glycosylation, and quality control – indicating that budding yeast are dependent equally on both genes to survive ER stress (Fig 3D). The profile for ire1 in Sp revealed many of the same aggravating interactions (Fig 3C). Consistent with these GIs and its presumed role in inducing the UPR, Δire1 cells are sensitive to the ER protein folding stressors DTT and tunicamycin (Fig 3C,E,G). However, instead of a transcription factor like Hac1, ire1's most highly correlated partners in Sp were the UDP-Glucose-glycoprotein glucosylTransferase (UGT) (gpt1) and a calnexin ortholog (cnx1) (Fig 3A,C).
Figure 3. the UPR Depends on Gpt1/Cnx1 and Ire1.
(A) Distribution of correlation coefficients for Sp Δire1 compared with (B) distribution of correlation coefficients for Sc ΔIRE1 (when different, names for Sc orthologs in gray text). (C) 3D scatter plot for Sp scores comparing Δire1 with Δgpt1 on the x and y axes, respectively, and the calnexin ortholog, cnx1-degron-DAmP, color coded according to the inset scale. (D) 3D scatter plot for Sc scores comparing ΔIRE1 with ΔHAC1 on the x and y axes, respectively, and the non-essential calnexin ortholog, ΔCNE1, color coded according to the inset scale. (E) Fold induction of normalized bip1 mRNA levels by qPCR in ER stress-inducing conditions. Each bar is the mean of three biological and three technical replicates per strain per condition. (F) Fold induction of GFP/RFP ratios in cells harboring a reporter system in which a Hac1p-responsive promoter drives green fluorescent protein corrected for nonspecific expression changes by comparing GFP to co-expressed RFP from a constitutive (TEF2) promoter (Jonikas et al 2009). Error bars = S.D. (G) Growth sensitivity of the indicated Sp strains to 20mM DTT or 2.5μg/mL tunicamycin.
This is a remarkable and unexpected finding, as in Sp and mammalian cells, Gpt1 and Cnx1 are core components of a lectin-chaperone system for glycoprotein folding (Ellgaard and Helenius, 2003). Misfolded proteins are recognized by Gpt1, which then appends terminal glucose residues to the core oligosaccaharide. The calnexin ortholog Cnx1 recognizes the terminal glucose modification made by Gpt1 and binds to glucosylated substrates to facilitate folding (Sousa and Parodi, 1995; Fanchiotti et al., 1998). The strength and reproducibility of the correlations between ire1, gpt1 and cnx1 were among the most robust relationships in the entire Sp dataset (Fig 2A, 3A). In Sc, the putative UGT is the essential enzyme KRE5. The GI profiles for temperature-sensitive and constitutive hypomorphic alleles of this enzyme are consistent with its enzymatic role – including significant correlations with CWH41 and the calnexin ortholog CNE1– but neither its profile nor CNE1's show strong similarity to the profiles for IRE1 or HAC1.
The correlations between gpt1, cnx1 and ire1 in Sp imply a fundamental functional connection between these distinct sensors of misfolding – a connection which despite extensive studies was not apparent in Sc. We probed this putative functional connection by challenging Δire1, Δgpt1, cnx1-Degron-DAmP single and double mutant cells with DTT and tunicamycin, drugs that specifically disrupt ER protein folding. As predicted by their overlapping genetic signatures, Δire1 and Δgpt1 cells have the same sensitivities and transcriptional UPR defects, while the double mutant phenotypes are no stronger than the single mutant phenotypes (Fig 3E,G). By contrast, budding yeast ΔCNE1 cells display robust Ire1-dependent activation of Hac1 (Fig 3F). Moreover, KRE5 hypomorphic cells are insensitive to DTT and tunicamycin (Breslow et al., 2008). Finally, KRE5 hypomorphs display no GI with IRE1 or HAC1 (Costanzo et al., 2010). These results imply that, in comparison with Sc, the conserved enzymes Gpt1 and Ire1 have been repurposed. The unanticipated connection between these stress sensors raises many questions about how misfolded proteins are sensed and how stress signals are transduced into differential outputs (e.g. transcription factor splicing or RIDD) in fission yeast and mammalian cells.
ESCRT-III and Vps4 Proteins Regulate SPB Duplication
Studies in Sc led to the discovery and characterization of the Endosomal Sorting Complex Required for Transport (ESCRT) genes in endosomal maturation (Hurley and Emr, 2006). Subsequent work in mammals confirmed the role of ESCRTs in multi-vesicular body formation, but also revealed that ESCRTs act as membrane fission factors during enveloped virus budding (Raiborg and Stenmark, 2009; von Schwedler et al., 2003). Further work in archaea (Samson et al., 2008; Lindås et al., 2008) and in mammalian cells demonstrated that ESCRTs mediate the final abscission step of cytokinesis (Carlton and Martin-Serrano, 2007; Morita et al., 2007). Finally, depletion of ESCRT-III and VPS4 proteins was reported to produce multi-polar spindles, suggesting these factors are required for centrosome dynamics (Morita et al., 2010). The centrosomal defects in cells depleted of ESCRT-III/VPS4 were profound: up to ~80% of depleted HeLa cells exhibit five or more centrosomes during the first mitosis after siRNA treatment. Thus ESCRT genes serve in a diversity of cellular pathways but this diversity was not apparent in pioneering Sc studies.
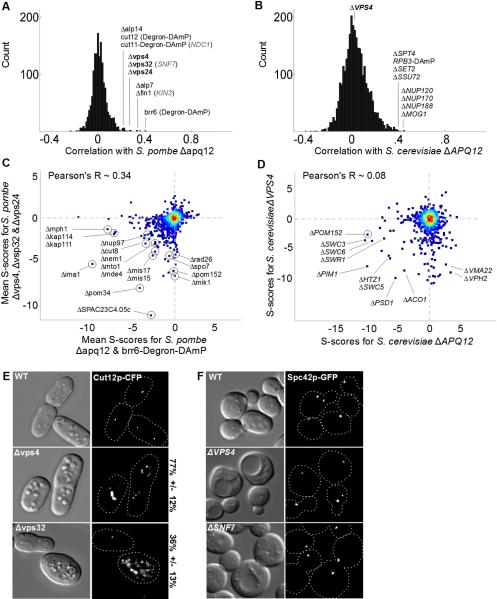
The GI profiles between ESCRTs and the rest of the endo-lysosomal system were among the most robust in our study. In addition to the expected interactions, vps32, vps24 and vps4 also displayed significant albeit weaker degrees of correlation with two nuclear membrane proteins, apq12 and brr6, which are determinants of spindle-pole body (SPB) duplication (Fig 4A, Tamm et al., 2011). In fungi, the SPB has a bulky cytoplasmic MTOC that is separated from the nuclear MTOC by the nuclear envelope. Duplication of the cytoplasmic MTOC precedes insertion of the structure into the nuclear membrane (Jaspersen and Winey, 2004). Brr6 and Apq12 are recruited to SPBs and are required for SPB insertion and nuclear envelope integrity during SPB insertion (Tamm et al., 2011). In Sp, apq12 and brr6 are most correlated with each other. In addition they display moderate correlations with components of the TACC/TOG complex (alp7/alp14) which regulates spindle formation (Sato and Toda, 2007), the NIMA kinase (fin1) which regulates SPB duplication (Grallert et al., 2004), core SPB components (cut11 and cut12) (West et al., 1998; Tallada et al., 2009), and—unlike Sc—the ESCRTs vps4, vps32 and vps24 (Fig 4A,B).
Figure 4. ESCRT-III Proteins and Vps4 Regulate Spindle Pole Body Duplication.
(A) Distribution of correlation coefficients for Sp apq12 compared with (B) distribution of correlation coefficients for ScAPQ12 (when different, names for Sc orthologs shown in gray text). (C) Scatter plot of mean scores for Sp apq12 and brr6 versus mean scores for vps32, vps24 and vps4. The following Sc orthologs have different names: SPAC23C4.05c = MSC1, mik1 = SWE1, rad26 = LCD1, mis15 = CHL4, mde4 = LRS4, cut8 = STS1, kap111 = KAP122, nup97 = NIC96, mph1 = MPS1. (D) Scatter plot of mean scores for Sc APQ12 versus VPS4. (E,F) DIC and fluorescence micrographs of the indicated cells expressing constitutive spindle-pole body markers (Cut12-CFP in Sp, Spc42p-GFP in Sc). For the fragmentation/duplication phenotypes observed in Sp the percent penetrance +/− standard deviation is noted. In addition to Spc42p, an outer plaque (Cnm67p-GFP), inner plaque (Spc110p-GFP), and gamma-tubulin (Tub4p-GFP) marked strains were scored (see Fig S4).
The correlation between brr6/apq12 and late ESCRTs reflects their shared aggravating interactions with nuclear membrane-proteins implicated in SPB duplication, nuclear morphology and pore biogenesis (SPAC23C4.05c, pom34, nup97, ima1, nem1/spo7 complex, Fig 4C). These genes also share moderate aggravating interactions with regulators of mitosis, spindle formation and kintechore components (mis17, mis15, mde4, rad26 and cut8, Fig 4C). By contrast, APQ12 and VPS4 are uncorrelated in Sc (Fig 4B,D). The few shared synthetic sick interactions include the Swr1 nucleosome remodeling complex, which shows synthetic interactions with many functionally unrelated genes (Fig 4D). With the exception of POM152 interactions with genes implicated in the SPB, nuclear pore, spindle or kinetochore were not observed in Sc (Fig 4D).
These GI profiles suggested that, despite the extensive differences between yeast SPBs and mammalian centrosomes, ESCRTs in Sp serve in an analogous role during the duplication of MTOCs (Morita et al., 2010). This possibility is also suggested by the report that deletion of the Sp ESCRT-II subunit, dot2, leads to over-amplification of SPBs in meiosis (Jin et al., 2005). We examined mitotic SPB phenotypes directly in ESCRT mutants using an integrated, constitutive marker of the SPB: Cut12-CFP (Toya et al., 2007). As predicted, both Δvps4 and Δvps32 cells displayed an over-amplification of Cut12-CFP labeled structures (Fig 4E). Of these, Δvps4 led to the more penetrant phenotype and was associated with large cytoplasmic bodies (Fig 4E). We also noted that for both Δvps4 and Δvps32 the severity and penetrance of the phenotype decreased over time, perhaps explaining why this phenotype was only observed in Δdot2 cells during meiotic divisions.
We next explored the possibility that ESCRT factors regulate SPB dynamics in Sc in detail. We examined four different SPB proteins: the core component Spc42p, outer plaque (Cnm67p-GFP) and inner plaque (Spc110p-GFP) components, and gamma-tubulin (Tub4p-GFP) (Fig 4F and Fig S4) but were unable to observe any indication of SPB duplication or fragmentation errors. Thus, both the global GI analysis and focused studies indicate that the role of the ESCRTs in regulating SPB duplication represents a novel activity not seen in Sc. In addition to identifying another example of functional repurposing, these studies indicate that fission yeast will become an important model for further study of ESCRT factors and their role at centrosomes.
The FAR Complex Regulates Mitosis in Fission Yeast
In Sc mating pheromones initiate signaling cascades that lead to cell cycle arrest (Elion, 2000). Multiple genes implicated in this phenomenon have been named FAR (Factor Arrest) genes, including a six-member complex composed of FAR3, FAR7, FAR8, FAR9/VPS64, FAR10 and FAR11. Initial characterizations indicated that mutation of this complex did not prevent pheromone-induced cell cycle arrest but rather premature resumption of budding (Kemp and Sprague Jr, 2003). Sc studies have found that the functional profiles of the FAR complex correlate with one another and other factors (Fig 5B, Hoppins et al., 2011, Costanzo et al., 2009). Among the most salient, a moderate degree of correlation between the FAR complex and protein phosphatase type 2A (PP2A, PPG1 subunit) and notable anti-correlation with a well-characterized factor arrest gene, FAR1 were observed (Fig 5B). The shared interactions between components of the FAR complex include the TORC2 kinase complex, lipid synthesis and ERMES complex genes (Fig 5D). These finding are consistent with the idea that the FAR complex regulates PP2A but suggest it has pleiotropic roles.
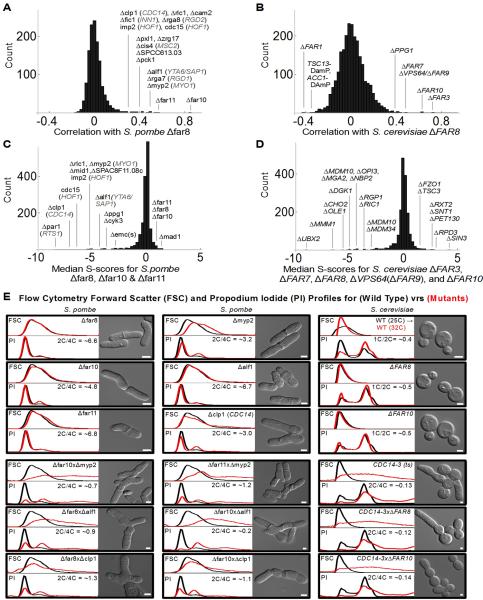
Figure 5. the FAR Complex Regulates the Cell Cycle in Fission Yeast.
(A) Distribution of correlation coefficients for Sp far8 compared with (B) distribution of correlation coefficients for ScFAR8 (when different, names for Sc orthologs shown in gray text). Distribution of aggravating and alleviating GIs shared by the FAR complex genes in (C) Sp and (D) Sc. (E) Flow cytometry analysis showing forward scatter (FSC, top panel) and DNA content histograms (PI, bottom panel) for wild type (black) and mutant cells (red). Traces represent mean values from triplicate experiments next to corresponding DIC images of representative cells. Bars 3 μm.
The FAR complex is an intriguing candidate for functional repurposing in that some of its constituents are unique to budding yeast. FAR3 and FAR7 are only found in a restricted set of budding yeasts (Fig S5) with no apparent orthologs in metazoa or Sp. Recently, an IP-MS study of human PP2A complexes identified a homologous complex: the Striatin-Interacting Phosphatase and Kinase (STRIPAK) complex (Goudreault et al., 2009). STRIPAK contains the PP2A catalytic (PP2Ac) and scaffolding (PP2A A) subunits, the Striatins (FAR8 homologs which possess PP2A regulatory B”' domains), the transmembrane STRiatin-Interacting Proteins (STRIP1 and STRIP2, FAR11 homologs), and the tail-anchored membrane protein SarcoLemmal Membrane-Associated Protein (SLMAP, a FAR10 homolog).
In addition to the above, STRIPAK contains a homolog of the yeast protein Mob1 (Moreno et al., 2001; Goudreault et al., 2009)—named MOBKL3 in human cells—which is a critical component of the Septation Initiation Network (SIN) in fission yeast (McCollum and Gould, 2001). Multiple Mob1 homologs exist in mammals and their function as kinase activators appears to be conserved (Hergovich et al., 2006). STRIPAK assemblies also contain Ste20-family kinases (Goudreault et al., 2009). The absence of FAR3 and FAR7 from Sp and metazoa and the presence of additional proteins not found in the Sc complex suggests that the cellular roles of STRIPAK complexes have ramified. Recent studies have identified STRIPAK as a regulator of the Hippo pathway (Ribeiro et al., 2010), as a modulator of Ras-MAPK signaling (Horn et al., 2011), and as a regulator of SIN in Sp (Sing et al. 2011).
The GI profiles for Sp far8 (SPBC1773.01), far10 (SPBC3H7.13) and far11 (SPBC27B12.04c) are correlated with each other and—in contrast to Sc—with multiple genes involved in cytokinesis and mitosis, including components of the actomyosin contractile ring (rlc1, cam2, fic1, rga8, imp2, ccd15, pxl1, rga7, myp2), the CDC14 ortholog clp1, the kinase pck1, and Golgi proteins zrg17, cis4, and SPCC613.03 (Fig 5A). This pattern suggested that the FAR complex plays a role in mitosis control in Sp. The correlations between FAR complex genes in Sp reflects strong aggravating interactions with a PP2A regulatory subunit (par1), and aggravating interactions with the mitotic exit phosphatase (clp1 /CDC14), core components of the contractile ring, and the catalytic PP2A subunit (SPAC22H10.04, Fig 5C). Shared alleviating interactions include interactions between different FAR subunits, a spindle attachment factor (mad1), and a phosphatase (stp1) implicated in the G2/M transition (Fig 5C).
These relationships suggest that the FAR complex regulates PP2A-mediated mitotic transitions. Moreover, the strong aggravating interaction between FAR complex genes and par1 suggests that the regulatory specificity conferred on PP2A by the FAR complex can be compensated for in Sp by this alternative B subunit. Single mutants of far8, far10 and far11 do not have striking phenotypes as assayed by flow cytometry for size and DNA-content, or by DIC imaging (Fig 5E). However, as predicted by their GIs, double mutants of FAR complex genes with genes functioning in mitotic signaling, cytokinesis or abscission have profound phenotypes. Double mutations with the type II myosin heavy chain myp2, a AAA ATPase we have named ATPase Like Fidgetin-1 (alf1), and the CDC14-related protein phosphatase, clp1, have abnormal morphologies and enhanced ploidy as measured by flow cytometry and DIC microscopy (Fig 5E). The aggravating interactions seen in Sp were not observed in Sc. ΔFAR8 and ΔFAR10 cells do not have cell cycle defects and double mutants with a ts allele of CDC14 have the same maximum permissive temperature as the single mutant CDC14-3 (32°C). After two hours at 32°C, single mutant CDC14-3 cells manifest clear shifts in ploidy and bud hyper-elongation. Double mutant ΔFAR8/CDC14-3 and ΔFAR10/CDC14-3 cells are indistinguishable from CDC14-3 single mutants (Fig 5E). Thus, as indicated by the systematic GI data, the role of the FAR complex in directing late mitotic events is not found in Sc.
STRIPAK Signaling Complexes Bridge the Golgi, the Centrosome, and the Nuclear Membrane
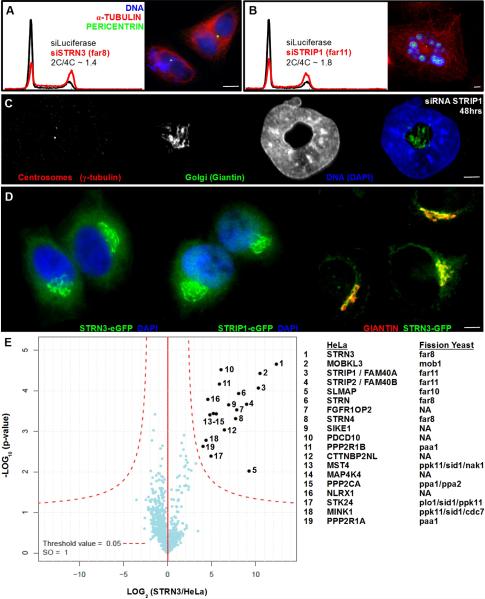
We sought to determine whether the gene-to-phenotype relationships observed for the Sp FAR complex were predictive of human STRIPAK complexes. The mitocheck consortium reported that silencing of STRN (Far8) resulted in bi-nuclear cells and cell death (Neumann, B. et al 2010). We found that siRNA-mediated depletions of core STRIPAK components, including STRN3 (Far8) and STRIP1 (Far11) in HeLa cells resulted in strong shifts from 2C to 4C DNA content (Fig 6A). Microscopy of silenced cells corroborated the increase in DNA content and revealed a range from bi-nuclear to horseshoe- and torus-shaped nuclei or fragmented nuclear remnants. Most remarkable, we often observed that centrosomes and intact Golgi stacks were found within the cavity of horseshoe- or torus-shaped nuclei (Fig 6C, Fig S6B–C).
Figure 6. Striatins and STRIPs form a Complex at the Golgi that Regulates Mitosis.
(A,B) Flow cytometry analysis showing DNA content histograms of HeLa cells treated with control siRNA targeting Luciferase (black) or siRNAs to deplete STRIPAK complex subunits (red). HeLa cell depletions revealing multinucleate cells and fragmented nuclei. (C) HeLa cell depleted of STRIP1 (Far11) for 48 hours. Fluorescence staining shows centrosomes (red), Golgi (green) and nuclei (blue). (D) HeLa cells harboring bacterial artificial chromosomes for eGFP-tagged STRN3 (Far8) or STRIP1 (Far11), demonstrating Golgi-like morphology and colocalization with the Golgi resident protein GIANTIN (red). See Fig S6 for additional colocalizations after siRNA depletion. (E) Volcano plot representation of STRN3-interacting proteins. For each protein identified by IP-MS, the ratio of the intensities in the STRN3 IPs over the control was calculated and plotted against the p-value of a t-test calculated from triplicates. The red curve is a cutoff calculated from false discovery rate estimation.
Given that STRIPs (Far11) and SLMAP (Far10) are membrane proteins and depletion of STRN3 or STRIP1 led to Golgi ribbons surrounded by dysmorphic nuclei, we sought to determine in which organelle this complex resides. We generated HeLa lines expressing STRN3-eGFP and STRIP1-eGFP from single-copy bacterial artificial chromosomes (Poser et al., 2008) under control of native promoters and untranslated sequences. Co-localization microscopy indicated that these proteins exist in the Golgi (Fig 6D). To evaluate the functionality of the tagged proteins we purified them for analysis by mass spectrometry and recovered the STRIPAK constituents reported previously, including SLMAP, indicating the Golgi-localized GFP fusion proteins form functional complexes (Fig 6E).
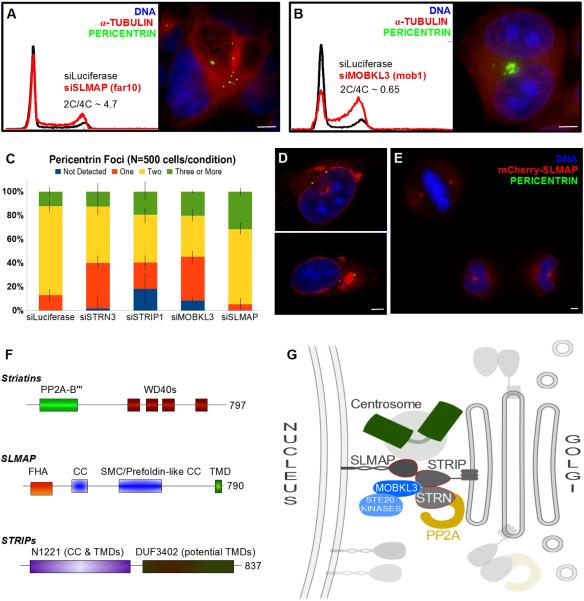
Despite the co-purification of SLMAP with Striatin and STRIP proteins, siRNA deletion of SLMAP produced a different phenotype and SLMAP did not localize to the Golgi (Fig 7, Fig S6). Depletion of SLMAP produced a subtle increase in 4C DNA and S-phase cells while microscopy revealed an significant increase in the number of pericentrin foci observed in interphase cells (Fig 7A,C). This suggests that SLMAP serves as a physical and signaling connection between the Golgi and centrosomes that is important for SPB duplication or spindle assembly. Early studies of SLMAP truncations revealed that it can localize to centrosomes via the FHA domain and that over-expression of SLMAP truncations induced mitotic arrest (Guzzo et al., 2004). C-terminal truncations of SLMAP localized to centrosomes—however, the tail-anchored and full-length membrane protein tagged at the N-terminus localized to the outer nuclear envelope during interphase (Fig 7D). During mitosis, full-length SLMAP localized clearly to centrosomes and the membranous material surrounding the mitotic spindle after nuclear envelope breakdown (Fig 7E).
Figure 7. STRIPAK Signaling Complexes Bridge the Nuclear Envelope, Centrosomes and Golgi.
(A,B) Flow cytometry analysis showing DNA content histograms of HeLa cells treated with control siRNA targeting Luciferase (black) or siRNAs to deplete SLMAP (Far10) or MOBKL3 (Mob1). HeLa cells depleted of the designated proteins and labled for immuno-fluorescence. See Fig S6 for additional images of siMOBKL3 phenotypes. (C) Fraction of interphase cells with abnormal numbers of pericentrin foci following siRNA treatment. Errors Bars = S.E.M. (D,E) HeLa cells transiently expressing mCherry-SLMAP and labeled for immuno-fluorescence. See Fig S7 for the lack of co-localization between mCherry-SLMAP and the Golgi. (F) Domain architecture of the FAR/STRIPAK components. (G) STRIPAK Model: Striatin and STRIPs reside at the Golgi. Striatins serve as regulatory B”' subunits of a PP2A trimer. Striatin/STRIP recruit MOBKL3, which directs Ste20/Germinal Center kinases. STRIPs interact with SLMAP in the outer nuclear membrane, bridging the Golgi, the centrosome, and the nuclear envelope. These interactions are likely restricted to specific cell cycle phases and the interaction between SLMAP and centrosomes predominates during mitosis. Disruption of STRIPAK leads to diverse failures during mitosis: including centrosome duplication errors, spindle assembly errors and cytokinesis failure.
The presence of the Striatins and STRIPs in the Golgi, the presence of SLMAP in the outer nuclear envelope, the association between SLMAP and centrosomes, and the co-purification of MOBKL3 and PP2A with the complex are important clues to its function in human cells. Mob1-like proteins activate mitotic kinases after being recruited to the spindle poles (Wurzenberger and Gerlich, 2011). Disruption of these signals results in mitotic failures in yeast. Accordingly, when we depleted MOBKL3 in HeLa cells we observed nearly universal spindle failures followed by cell death (Fig 7B, Fig S6D). Integrating these observations, we propose that human STRIPAK complexes serve to direct mitotic signaling events (Fig 7G). STRIPAK complexes appear positioned to regulate the activity of Mob/kinase complexes and to form a unique PP2A holoenzyme directed toward mitotic substrates. Furthermore, the fact that two components of this complex reside in the Golgi and a third resides in the outer nuclear envelope suggests STRIPAK complexes participate in the tethering of centrosomes to the Golgi, centrosome duplication signaling, Golgi fragmentation at the G2/M transition, or targeting Golgi fragments to spindle poles during mitosis (Fig 7G). Given the increasing evidence that the Golgi and spindles have functional interactions throughout the cell cycle (Sütterlin and Colanzi, 2010), our observations suggest the STRIPAK complex mediates communication between these organelles. The functional repurposing of the STRIPAK complex in Sp and mammals, in contrast to the distinctly different complex formed in Sc, correlates with the evolution of major differences between these organisms in cytokinesis, cell cycle phasing, and Golgi morphology.
PERPSECTIVE
Jacques Monod's expression of biological unity, “Anything found to be true of E. coli must also be true of elephants,” can be answered with a nuanced view in light of the dramatic increase in functional genomic information. We analyzed genes conserved in budding yeast, fission yeast and mammals with a focus on functional divergence. In an important subset of genes we found evidence of functional repurposing: the use of conserved machines in different pathways with different inputs or outputs. Monod's reductionist view describes the depth of conservation between structure and function: folds and key residues confer durable properties through evolution. Protein complexes tend to be conserved but not as deeply as structure, while connections between complexes or pathways can be quite plastic.
The unique opportunity to conduct a functional comparison between two divergent eukaryotes with comprehensive ortholog mapping provided us with an unparalleled view of repurposing. Having this view enabled us to document an unanticipated degree of malleability in function and functional connections. Our Sp map led to several mechanistic insights that are relevant to understanding mammalian cells. It also yielded a rich resource for other investigators as we have described only a fraction of the connections in our data. To aid these efforts we have created a website for the community to navigate this dataset (www.YeastQuantitativeGenetics.org). Future studies will enhance our understanding of which components and connections are invariant across evolution versus those that are adaptable. Moreover, it should be possible to connect repurposing events to changes in cellular physiology (e.g. switching from fission to budding cell division). Such insights may prove to be useful in medicine in that malignancy and infection are both problems of rapid evolution. Our ability to design “conditionally lethal” therapies will also depend on understanding which functional relationships can be repurposed.
EXPERIMENTAL PROCEDURES
Strains and Genetic Crosses
Array G418-resistant haploid single deletion mutants were isogenic to SP286 (h+ ade6-M210;ura4-D18;leu1–32) selected from the BIONEER collection (Kim et al., 2010). The nourseothricin-resistant h- query strains were made in the PEM2 strain (Roguev et al., 2007). Targeting cassettes were built using a 2-step, fusion PCR protocol in which long (~3kb) cassettes were amplified after annealing mediated by non-palindromic and unique GC-rich overlapping sequences (Fig S1). Integration of the resistance markers into the target locus was verified by PCR. Query strains harboring constitutive hypomorphic Degron-DAmP alleles were made via the same strategy except a degron sequence (Ravid and Hochstrasser, 2008) was fused in place of the stop codon, followed by a selectable maker in place of the 3'UTR. Mating, selection and propagation of the double mutants were carried on a Singer RoToR pinning robot using the PEM2 procedure (Roguev et al., 2008, 2007). For directed assays in Sc single and double mutations were generated in W303 diploids followed by sporulation and tetrad dissection.
Genetic Interaction Score Acquisition and Analysis
Double mutant plates were scanned on a flat-bed scanner (EPSON PhotoPerfection 350, Fig S2) and integrated colony intensities extracted using a custom algorithm (scripts available upon request) executed in MATLAB (The Mathworks, Natcik, MA). Fitness analysis was performed by a strategy modified from Collins et al. (2010), including normalization of plate surface artifacts, row/column normalization artifacts, and batch artifacts. Linkage biases due to the reduced frequency of recombination between linked loci (manifested by a reduced number of spores and a spurious negative score) were used to identify contaminated or mis-annotated strains (Fig S2D).
Supplementary Material
Acknowledgments
We thank A Roguev, N Krogan, C Ryan, P Espenshade, W Sundquist, D Ward and T Formosa for helpful discussions, reagents and protocols; N Elde, J. Kaplan, L. Colf and M. Karen for critical discussions of the manuscript; J. Rutter and J. Shaw for microscope access; This work was support by funds from HHMI (JSW), NIH MCRTP 5T32 CA93247-9 (ME), the German National Genome Research Network grant (01GS0861 AH, MM) and the Huntsman Cancer Institute Center Grant (P30 CA042014, AF). OB and SC are HHMI fellows of the Helen Hay Whitney Foundation. JW was supported by a long term fellowship from the International Human Frontier Science Program (LT 00821/2007-L). AF was a HHMI Fellow of the Life Science Research Foundation and is supported by the University of Utah.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
See the supplemental material for a full description of the materials and methods.
Citations
- Aslett M, Wood V. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung M-K, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of Genetic Networks in Response to DNA Damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn J-Y, Ou J, San Luis B-J, Bandyopadhyay S, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Meth. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 2008;5:711. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Meth. Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome biology. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing Nucleosome Function: A Highly Versatile Library of Synthetic Histone H3 and H4 Mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130:663–677. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JLY, Prasad TSK, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe K-L, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- Fanchiotti S, Fernández F, D'Alessio C, Parodi AJ. The UDP-Glc:Glycoprotein glucosyltransferase is essential for Schizosaccharomyces pombe viability under conditions of extreme endoplasmic reticulum stress. J. Cell Biol. 1998;143:625–635. doi: 10.1083/jcb.143.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreault M, D'Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Molecular & Cellular Proteomics. 2009;8:157. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Krapp A, Bagley S, Simanis V, Hagan IM. Recruitment of NIMA kinase shows that maturation of the S. pombe spindle-pole body occurs over consecutive cell cycles and reveals a role for NIMA in modulating SIN activity. Genes Dev. 2004;18:1007–1021. doi: 10.1101/gad.296204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo RM, Sevinc S, Salih M, Tuana BS. A novel isoform of sarcolemmal membrane-associated protein (SLMAP) is a component of the microtubule organizing centre. Journal of Cell Science. 2004;117:2271–2281. doi: 10.1242/jcs.01079. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 2011;95(2):323. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Fischer B, Axelsson E, Huber W, Boutros M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat. Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Jin Y, Mancuso JJ, Uzawa S, Cronembold D, Cande WZ. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev. Cell. 2005;9:63–73. doi: 10.1016/j.devcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp HA, Sprague GF., Jr Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Molecular and cellular biology. 2003;23:1750. doi: 10.1128/MCB.23.5.1750-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, Yoo H-S, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D, Pendyala L, Daignan-Fornier B, Howell SB, Huang R-Y. Dysregulation of purine nucleotide biosynthesis pathways modulates cisplatin cytotoxicity in Saccharomyces cerevisiae. Mol. Pharmacol. 2008;74:1092–1100. doi: 10.1124/mol.108.048256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindås A-C, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- Moreno CS, Lane WS, Pallas DC. A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 2001;276:24253–24260. doi: 10.1074/jbc.M102398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. The EMBO journal. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, et al. Phenotypinc profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464(7289):721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa N, Akimoto-Kato A, Niimi T, Tojo K, Machida R, Hayashi S. Evolutionary origin of the insect wing via integration of two developmental modules. Evol. Dev. 2010;12:168–176. doi: 10.1111/j.1525-142X.2010.00402.x. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Pan X, Peyser BD, Ye P, Meluh PB, Yuan DS, Irizarry RA, Bader JS, Spencer FA, Boeke JD. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Poser I, Sarov M, Hutchins JRA, Hériché J-K, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Josué F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Molecular Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nature methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Meth. Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NS, Shao N, McLean JR, Sevugan M, Ren L, Chew TG, Bimbo A, Sharma R, Tang X, Gould KL, et al. SIN-inhibitory phosphatase complex promotes Cdc11p dephosphorylation and propagates SIN asymmetry in fission yeast. Curr. Biol. 2011;21:1968–1978. doi: 10.1016/j.cub.2011.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm T, Grallert A, Grossan EPS, Alvarez-Tabares I, Stevens FE, Hagan IM. Brr6 drives the S. pombe spindle pole body nuclear envelope insertion/extrusion cycle. J. Cell Biol. 2011;195:467–484. doi: 10.1083/jcb.201106076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung H-Y, Morita E, Wang HE, Davis T, He G-P, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Sousa M, Parodi AJ. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc: glycoprotein glucosyltransferase. The EMBO Journal. 1995;14:4196. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J. Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallada VA, Tanaka K, Yanagida M, Hagan IM. The S. pombe mitotic regulator Cut12 promotes spindle pole body activation and integration into the nuclear envelope. J. Cell Biol. 2009;185:875–888. doi: 10.1083/jcb.200812108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303 doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Toya M, Sato M, Haselmann U, Asakawa K, Brunner D, Antony C, Toda T. Gamma-tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat. Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Sceince. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell. 1998;9:2839–2855. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 2011;12:469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.