SUMMARY
MicroRNAs (miRNAs) control gene expression by promoting degradation or repressing translation of target mRNAs. The components of the miRNA pathway are subject to diverse modifications that can modulate the abundance and function of miRNAs. Iron is essential for fundamental metabolic processes, and its homeostasis is tightly regulated. Here we identified iron chelators as a class of activator of the miRNA pathway that could promote the processing of miRNA precursors. We show that cytosolic iron could regulate the activity of the miRNA pathway through poly(C)-binding protein 2 (PCBP2). PCBP2 is associated with Dicer and promotes the processing of miRNA precursors. Cytosolic iron could modulate the association between PCBP2 and Dicer, as well as the multimerization of PCBP2 and its ability to bind to miRNA precursors, which can alter the processing of miRNA precursors. Our findings reveal a role of iron homeostasis in the regulation of miRNA biogenesis.
INTRODUCTION
MicroRNAs (miRNAs) constitute one of the most abundant classes of gene regulatory molecules in eukaryotic cells. Serving as guides in RNA-induced silencing complexes, miRNAs direct Argonaute proteins to specific target mRNAs to repress gene expression posttranscriptionally (Heo and Kim, 2009). miRNAs have been shown to shape diverse cellular pathways, from chromosome architecture, development, and growth control to apoptosis and stem cell maintenance (Czech et al., 2008; Ghildiyal and Zamore, 2009; Kawamura et al., 2008; Kim et al., 2009; Okamura et al., 2008; Okamura and Lai, 2008; Plasterk, 2006; Taft et al., 2009). Moreover, the miRNA pathway has been adopted by researchers for RNA interference (RNAi), which has broad utility in gene-function analysis, drug-target discovery and validation, and therapeutic development (Dykxhoorn and Lieberman, 2006). The endogenous mature miRNA is the product of multiple processing steps, which are tightly regulated to modulate miRNA functions (Heo and Kim, 2009). Chemical biology, in particular the use of diverse chemicals to interrogate molecular processes, provides a means of rapidly and effectively dissecting biological mechanisms and gene networks in ways not feasible with mutation-based genetic approaches (Hergenrother, 2006; Lipinski and Hopkins, 2004; Schreiber, 2005). To dissect the regulatory mechanisms of modulating the activity of the miRNA pathway, we previously developed a cell-based assay to monitor the activity of the miRNA pathway using a chemical biology approach (Shan et al., 2008).
Iron is the fundamental metal for life processes in an oxygen-rich atmosphere (Hentze et al., 2004; Lippard and Berg, 1994). It is a cofactor for the oxygen transporter hemoglobin and participates in numerous other enzymatic reactions. Iron is involved in many cellular processes, including respiration and many key metabolic reactions (Rouault, 2006; Theil and Goss, 2009). Both iron overload and iron deficiency can lead to cytotoxicity and cell growth arrest/apoptosis, respectively (Hentze et al., 2010).
Iron storage and transportation in most mammals are partially controlled by iron regulatory proteins (IRPs) in a posttranslational fashion (Rouault, 2006). The IRP system is the most extensively studied mode of coordinated regulation in iron homeostasis (Hentze et al., 2004). Trans-acting IRPs, through their metal availability sensors, interact with ferritin and transferrin receptor (TfR) iron-responsive elements (IREs), which are conserved hairpin structures found in messenger RNA (mRNA)-untranslated regions (UTRs), to control either mRNA stability or initiation of translation. Ferritins are the main iron storage proteins found in animals. The capacity to store iron in ferritin is essential for life in mammals, but the mechanism by which cytosolic iron is delivered to ferritin remained a mystery until recently, when poly(rC)-binding protein 1 (PCBP1) was found to bind to ferritin in vivo and facilitate the loading of iron into ferritin (Shi et al., 2008). Depletion of PCBP1 in human cells inhibits ferritin iron loading and increases cytosolic iron pools. Thus, PCBP1 can function as a cytosolic iron chaperone in the delivery of iron to ferritin. It was also noted that PCBP2, a paralog of PCBP1, could also potentially contribute to ferritin iron loading (Shi et al., 2008).
Here we describe the identification, via chemical screen, of iron chelators as a class of enhancer of the miRNA/RNAi pathway that could promote the processing of both short hairpin RNAs (shRNAs) and miRNA precursors. We show that cytosolic iron regulates the activity of the miRNA pathway through poly(C)-binding protein 2 (PCBP2). PCBP2 is associated with Dicer and promotes the processing of miRNA precursors. Cytosolic iron could modulate the association between PCBP2 and Dicer, as well as the multimerization of PCBP2 and its ability to bind to miRNA precursors, which can alter the processing of miRNA precursors. Our findings reveal a role of iron homeostasis in the regulation of miRNA biogenesis, and indicate that altered miRNA expression might contribute directly to the molecular pathogenesis of human diseases associated with disrupted iron homeostasis. Our results also suggest potential use of iron chelators to modulate the activity of the miRNA pathway.
RESULTS
Identification of Metal Chelators as a Class of shRNA-Mediated RNAi Enhancer
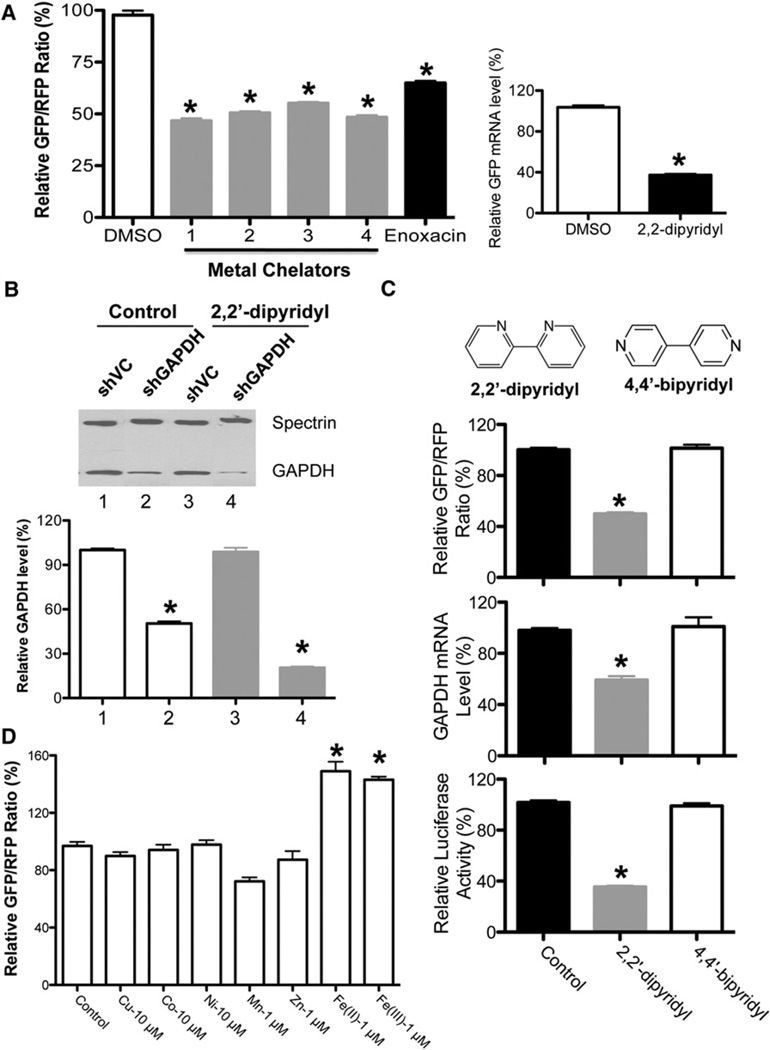
We previously developed a cell-based assay (RNAi-293-EGFP) to monitor the activity of the RNAi pathway using EGFP as a reporter (Shan et al., 2008). In this system, a HEK293-derived stable cell line expressing a GFP reporter gene (293-EGFP) was infected with a lentivirus expressing a shRNA that is processed into small interfering RNAs (siRNAs) specifically targeting GFP. Here we further optimized this system by introducing RFP into this reporter cell line as an internal control, so the GFP/RFP ratio would better reflect the activity of the miRNA/RNAi pathway. Using this modified system, we screened additional libraries and identified several structurally diverse small molecules with metal chelation activity that could enhance the activity of the RNAi pathway (Figure 1A and Figure S1A available online). The enhancement by metal chelators was further confirmed with additional shRNAs targeting both endogenous and additional reporter genes, indicating that the effect of these metal chelators on shRNA-mediated gene knockdown is universal (Figures 1B and S1B).
Figure 1. Identification of Iron Chelators as a Class of shRNA-Mediated RNAi Enhancer via Chemical Screen.
(A) Iron chelators enhance shRNA-EGFP-mediated gene silencing. DMSO was used as a negative control, whereas enoxacin, a previously identified RNAi enhancer, was used as a positive control. Values are mean ± SD for triplicate samples. *p < 0.001 when metal chelators were compared with control or enoxacin, and enoxacin was compared with control. The left panel shows the relative GFP/RFP ratios, while the right panel shows the GFP mRNA level determined by quantitative RT-PCR. Values are mean ± SD for triplicate samples.
(B) The metal chelator 2,2′-dipyridyl enhances shGAPDH-mediated gene silencing. The GAPDH protein levels were detected by western blots with anti-GAPDH antibody, with Spectrin used as a loading control. Metal chelator has no effect on GAPDH expression in shVC control cells. Values are mean ± SD for triplicate samples. *p < 0.001 when shGAPDH treated with 2,2′-dipyridyl was compared with shGAPDH control, and shGAPDH control was compared with shVC.
(C) The iron chelator 2,2′-dipyridyl, but not its inactive isomer 4,4′-bipyridyl, could enhance shRNA-mediated RNAi. Three reporter cell lines, shGFP, shLuciferase, and shGAPDH, were treated with 2,2′-dipyridyl and its isoform 4,4′-bipyridyl for 48 hr before analyses. Values are mean ± SD for triplicate samples. *p < 0.001 when 2,2′-dipyridyl was compared with control (DMSO) and 4,4′-bipyridyl.
(D) Iron specifically regulates the activity of the RNAi pathway. A metal substitution assay was performed with RNAi-293-EGFP/RFP reporter cells. The reporter cells were seeded in FBS-free DMEM supplemented with different metals at comparable toxicity levels for 24 hr before measuring relative fluorescence intensity. Values are mean ± SD for triplicate samples. *p < 0.001.
See also Figures S1 and S2.
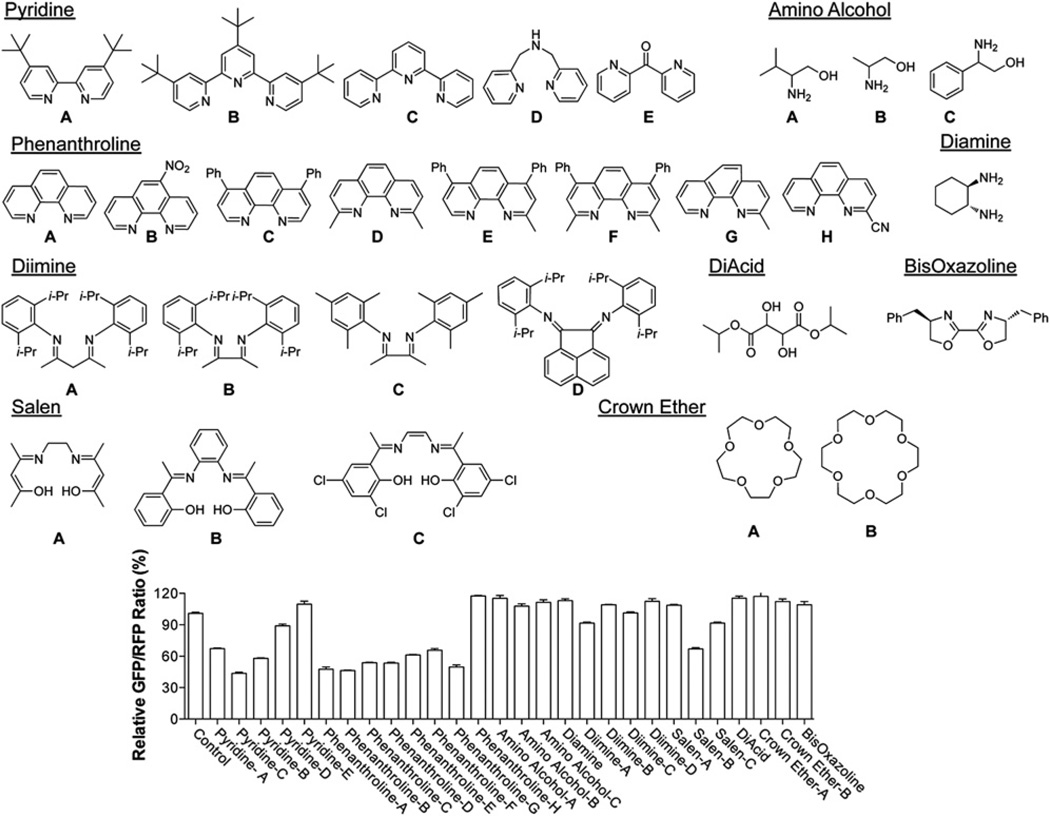
Through our initial screen we identified deferoxamine mesylate as a potent enhancer of the shRNA-mediated RNAi-enhancing activity. Deferoxamine mesylate is a well-known ligand for iron that exhibits iron chelation in vivo (Richardson and Ponka, 1998). We performed further tests and found that 1-(dipyridinylmethylene) thiosemicarbazide, another known iron chelator, also exhibits the same effect (Figures 1A and S1A). A broader screen of a variety of ligands indicated that neither diimines nor oxygen ligands, such as crown ethers, diacids, and amino alcohols, had any effect on shRNA-mediated RNAi, whereas iron chelators, such as pyridine-based ligands and some salen ligands, display significant RNAi-enhancing activity (Figure 2) (Whitnall et al., 2006). In addition, the common metal chelator 2,2′-dipyridyl, but not its inactive isomer 4,4′-bipyridyl, could enhance shRNA-mediated RNAi (Figures 1C and S2A) (Rosner et al., 2002). These results together strongly suggest that iron chelating activity is required for the RNAi-enhancing activity we observed. Among the small molecules we tested, we chose dipyridine for our follow-up studies, due to its small size as well as the availability of the inactive form of 4,4′-bipyridyl for control experiments. Similar RNAi-enhancing activity by iron chelator could also be observed in a hepatoma cell line (HepG2), which plays a critical role in regulating iron homeostasis (Figure S1C).
Figure 2. The RNAi-Enhancing Activity of Different Classes of Metal Chelators.
Chemical structures of small molecules tested are shown. DMSO was used as a negative control. The relative GFP/RFP ratio for each small molecule is shown on the bottom. See also Figure S2.
Cytosolic Iron Modulates the Activity of the RNAi Pathway
Since the metal chelators we tested above could potentially chelate multiple metals, we determined which metals specifically modulate the activity of the RNAi pathway by performing a metal substitution assay using our RNAi reporter system. In this assay, our reporter cell line was grown in serum-free medium that was supplemented with different metals, including manganese, zinc, copper, nickel, cobalt, and iron. The RNAi activity for each was determined by measuring the GFP/RFP ratio. Among the metals we tested at comparable toxicity levels, only iron could significantly increase the GFP/RFP ratio and inhibit RNAi activity, suggesting that intracellular iron can modulate the activity of the RNAi pathway (Figure 1D).
Cellular iron is well known to be tightly regulated and compartmentalized by nature to maximize controlled reactions with oxygen in cells (Hentze et al., 2004; Rouault, 2009). Once transported into the cells by transferrin or through other pathways, iron is distributed in three main compartments for different functions. In mitochondria, iron is mainly for biosynthesis of heme and Fe-S clusters. Surplus iron goes to ferritin for storage in cytoplasm. Heme serves as a prosthetic group for many cellular proteins with diverse biological functions (Rouault and Tong, 2008). Because iron chelators can also block heme biogenesis, we wanted to explore whether heme is involved in the RNAi pathway. Thus, our reporter cell line was cultured in serum-free medium supplemented with succinylacetone (SA) to deplete heme as described previously (Yin et al., 2007). Heme depletion was confirmed by heme quantification (Figure S2B). Furthermore, we also included a panel with both SA treatment and the addition of different concentrations of heme. We saw no alteration of RNAi activity with SA treatment in the presence or absence of heme, which suggests that heme is not involved in modulating the activity of the RNAi pathway (Figure S2C). In addition, we performed knockdown experiments using siRNAs against the heavy chain subunit of ferritin and transferrin receptor 1 (TFR1) and found that the reduction of ferritin or TFR1, which would alter the level of cytosolic iron, could modulate shRNA-mediated RNAi (Figures S2D–S2F). Finally, we altered the cytosolic iron level by manipulating the expression level of ferroportin, an iron exporter, and observed similar effects on the activity of the RNAi pathway (Figure S2G). These results together suggest that the cytosolic iron level regulates the activity of the RNAi pathway.
Iron Chelator Promotes the Processing of shRNAs and miRNA Precursors
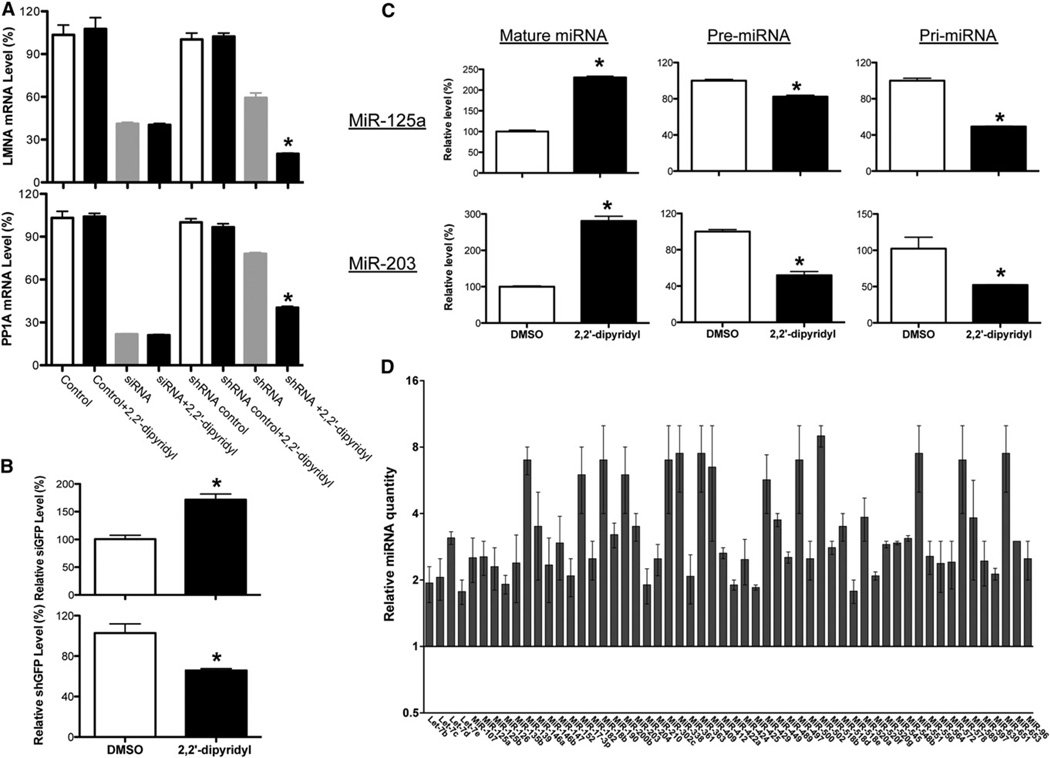
Since there are two approaches to induce the specific suppression of gene expression in cells, shRNAs and siRNA duplexes, we also tested siRNA duplexes and found that iron chelator has no effect on siRNA duplex-induced RNAi, while the shRNAs against the same genes could be enhanced by iron chelator (Figure 3A). We further examined the expression of mature small RNAs in cells expressing different shRNAs (Figures 3B and S3A). Although different promoters were used in these constructs, we observed consistent increases of processed siRNAs and corresponding decreases of shRNAs in cells treated with iron chelator (Figures 3B and S3A). Furthermore, we examined the levels of the mature form of miR-125a, pri-miR-125a, and pre-miR-125a in cells stably overexpressing the primary transcript of miR-125a (Pri-miR-125a) that we described previously by quantitative RT-PCR (Figure 3C) (Duan et al., 2007). The addition of iron chelator led to increases in the mature form of miR-125a and corresponding decreases in the levels of both pre-miRNA and pri-miRNA (Figure 3C). This suggests that iron chelator can promote the processing of miR-125a. Since the shGFP used in our initial reporter system mimics miRNA precursors and is therefore processed by Dicer, these data suggest that iron chelator may function at the level of Dicer-mediated precursor processing. To test this possibility, we used microRNA TaqMan assays to profile miRNA expression in the presence or absence of iron chelator. The majority of endogenous miRNAs (313/369) were not affected (data not shown). Among the miRNAs consistently altered by iron chelator, all showed increased expression of the mature form (Figures 3C, 3D, and S3B–S3E). Reminiscent of miR-125a, we also found decreased levels of the pri- and pre- forms of some of the miRNAs whose mature forms increased in the presence of metal chelator (Figure 3C and S4A). Interestingly, we noted that the precursor forms of the elevated endogenous miRNAs are generally abundant in untreated cells, while the endogenous miRNAs that are not significantly affected by iron chelator generally have very few or no detectable precursors in cells (Table S1). Furthermore, iron chelator treatment could increase the production of mature small RNAs from the miR-30a-based shRNAs (precursor-like RNAs) that were abundant in cells, whereas iron chelator had no effect on endogenous miR-30a, which had no detectable steady-state precursor form in cells (Figure S3). Similar enhancement of miRNA processing by iron chelator could be seen in HepG2 cells, as well (Figure S1D). These data together show that iron chelator can promote processing of both shRNA and miRNA precursors, which depends largely on the amount of precursor RNA in cells, rather than specific RNA sequences. The increased miRNA processing could impact the miRNA-mediated posttranscriptional regulation on the mRNA targets that were identified previously (Figure S4B and S4C) (Le et al., 2011; Le et al., 2009; Szulwach et al., 2010; Zhang et al., 2009).
Figure 3. Iron Chelator Promotes the Processing of shRNAs and miRNA Precursors.
(A) Metal chelator enhances only shRNA-mediated but not synthetic siRNA duplex-induced knockdown. Knockdown of each mRNA was graphed as the percentage of mock-treated samples in the presence or absence of metal chelators (2,2′-dipyridyl). Values are mean ± SD for triplicate samples. *p < 0.001. Control was transfected with control siRNA duplex.
(B) Quantitative RT-PCR was used to measure the levels of processed siRNA and shRNA in RNAi-293-EGFP/RFP reporter cells. 5S was used as an internal control. Values are mean ± SD for triplicate samples. *p < 0.001.
(C) Quantitative RT-PCR was used to measure the levels of pri-, pre-, and mature forms of miR-125a and miR-203 in mock- and iron chelator-treated HEK293 cells expressing the primary transcript of miR-125a. The relative expression levels as determined by DDCt analyses are shown. Values are mean ± SD for triplicate samples. *p < 0.001.
(D) Relative quantity of miRNA with consistent changes in expression shown for iron chelator-treated HEK293 cells (transfected HEK293 cells stably expressing pri-miR-125a were used), calibrated to mock-treated cells (mock-treated relative quantity = 1; the mean relative quantity from three pairs is plotted with error represented as a 95% confidence interval).
See also Figures S3 and S4 and Table S1.
Although our heme depletion assay suggests that heme is not involved in modulating the activity of the RNAi pathway (Figure S2C), a previous in vitro study suggested that heme is a cofactor of DGCR8, which is associated with Drosha and involved in pri-miRNA processing (Faller et al., 2007). To address this apparent discrepancy, we depleted heme in cells stably overexpressing the primary transcript of miR-125a (Pri-miR-125a) and examined the processing of miR-125a. Consistent with the results from our RNAi reporter assay, we saw no significant change in miR-125a processing (Figure S4D).
PCBP2 Mediates the Modulation of RNAi by Cytosolic Iron via Interactions with Dicer
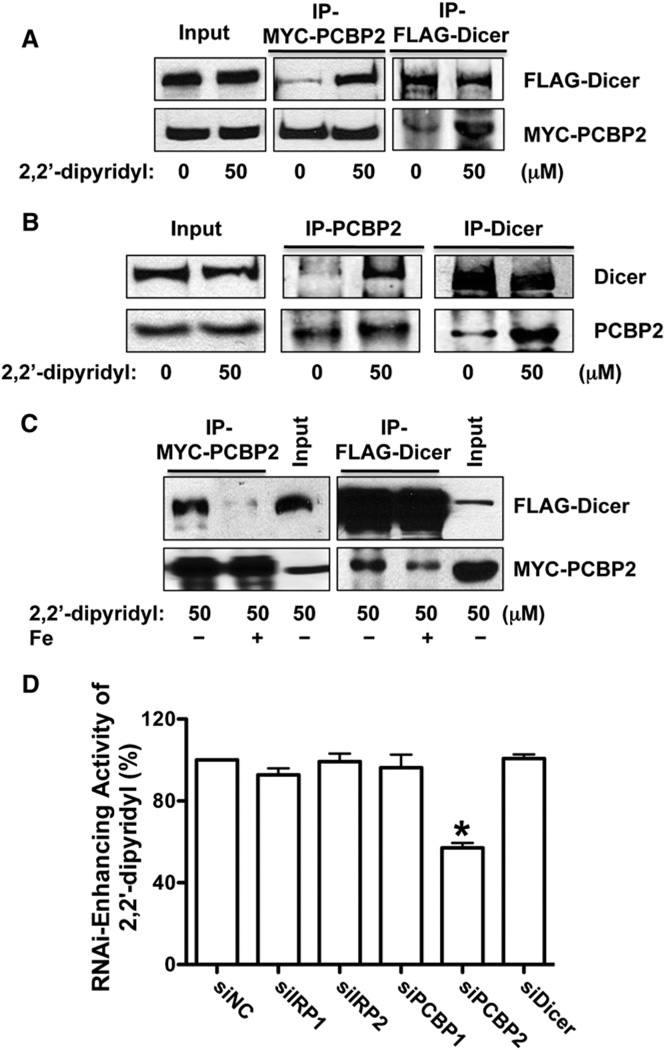
Iron regulatory proteins 1 and 2 (IRP1 and IRP2) are mammalian proteins that register cytosolic iron concentrations and posttranscriptionally regulate expression of iron metabolism genes to optimize cellular iron availability via interaction with IREs in mRNA (Rouault, 2006). Most recently, PCBP1 was identified as a cytosolic iron chaperone (Shi et al., 2008). It was also noted that PCBP2, a paralog of PCBP1, could potentially contribute to ferritin iron loading, as well (Shi et al., 2008). Because iron chelator could promote the processing of shRNAs and miRNA precursors, we next examined whether any of these proteins is associated with Dicer, the core component of miRNA precursor processing complex (Krol et al., 2010). We performed reciprocal coimmunoprecipitation experiments followed by western blot analyses using HEK293 cells transfected with FLAG-Dicer and IRP1, IRP2, PCBP1, or PCBP2 expression vectors (Paroo et al., 2009). We found that only PCBP2, but not other proteins, could be coimmunoprecipitated with Dicer. This association is RNA-independent, since the lysate used for immunoprecipitation was pretreated with RNase (Figure 4A and S5A). Interestingly, the association between PCBP2 and Dicer could be enhanced significantly by iron chelator (Figure 4A). These observations were further confirmed by the coimmunoprecipitation experiment using the antibodies recognizing the endogenous PCBP2 and Dicer proteins (Figure 4B). Furthermore, the addition of iron to the immunoprecipitation buffer could partially abolish the enhanced association between PCBP2 and Dicer by iron chelator (Figure 4C). We also used the siRNAs against each of these genes to knockdown their expression in our EGFP reporter cells and found again that only the knockdown of PCBP2 could block the RNAi-enhancing activity of iron chelator (Figure 4D andS5B). These results together demonstrate that PCBP2 mediates the modulation of the RNAi pathway by cytosolic iron via interaction with Dicer in an iron-dependent manner.
Figure 4. PCBP2 Mediates the Modulation of the RNAi Pathway by Cytosolic Iron via Interaction with Dicer.
(A) Iron modulates the association between exogenous PCBP2 and Dicer. The upper-middle panel shows the western blot to detect the coimmunoprecipitated FLAG-Dicer protein in the immunoprecipitated complex with anti-MYC antibody under different conditions. The lower-middle panel indicates the immunoprecipitated MYC-PCBP2. The upper-right panel shows the immunoprecipitated FLAG-Dicer, while the lower-right panel shows the western blot to detect the coimmunoprecipitated MYC-PCBP2 protein in the immunoprecipitated complex (IP) with anti-FLAG antibody under different conditions. The cells were treated with DMSO or 2,2′-dipyridyl for 48 hr before immunoprecipitation. Ten percent input and 50% IP were loaded.
(B) Iron modulates the association between endogenous PCBP2 and Dicer proteins. The upper panel shows the western blot to detect the Dicer protein in the immunoprecipitatedcomplex or input withanti-Dicerantibodyunder different conditions. The lower panel indicates the PCBP2 in the immunoprecipitated complex or input with anti-PCBP2 antibody under different conditions. The cells were treated with DMSO or 2,2′-dipyridyl for 48 hr before immunoprecipitation.
(C) The addition of iron to the IP buffer decreases the association between PCBP2 and Dicer. Immunoprecipitation experiments similar to (A) were performed with the addition of iron in the IP buffer.
(D) The RNAi-enhancing activity of iron chelators is PCBP2-dependent. The siRNAs against IRP1, IRP2, PCBP1, PCBP2, and Dicer were transfected into the RNAi-293-EGFP/RFP reporter cells. The cells were then treated with DMSO or 2,2′-dipyridyl for 48 hr before RNAi-enhancing activity was determined. Values are mean ± SD for triplicate samples. *p < 0.001.
See also Figures S4 and S5.
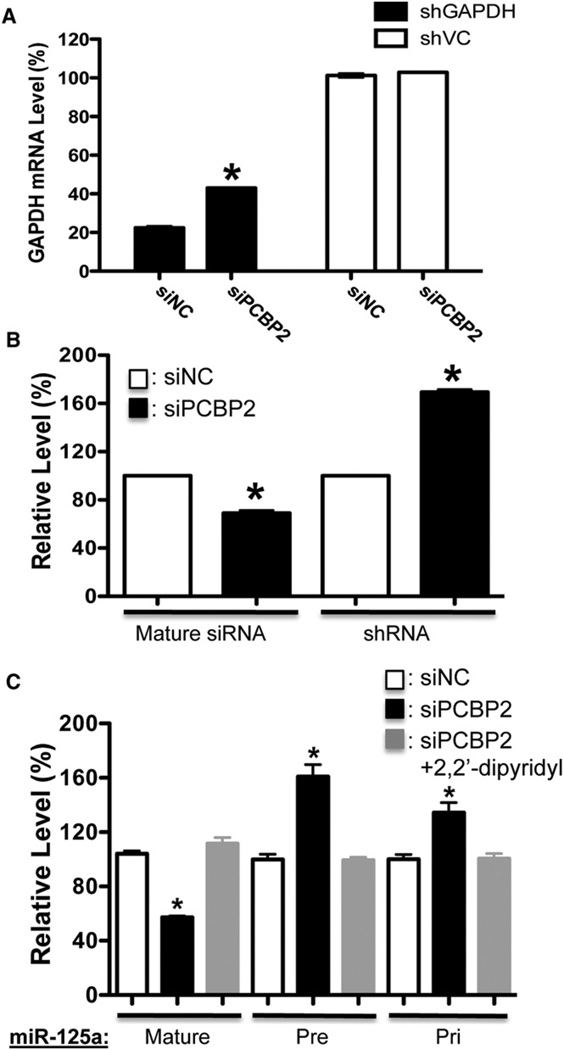
PCBP2 (or hnRNP-E2) is a member of a family of four homologous RNA-binding proteins belonging to the heterogeneous nuclear ribonucleoprotein K-homology domain superfamily and is widely expressed and highly conserved among mammals (Makeyev and Liebhaber, 2002). This group of proteins is believed to play roles in mRNA stabilization, translational activation, and translational silencing (Makeyev and Liebhaber, 2002). Intriguingly, miR-328 was recently shown to compete with other RNAs for PCBP2 binding (Eiring et al., 2010). To determine whether PCBP2 itself is involved in the miRNA/RNAi pathway, we knocked down the expression of PCBP2 in different shRNA-mediated RNAi reporter systems we have used. We found that the reduction of PCBP2 could consistently decrease the shRNA-mediated knockdown efficiency of these systems (Figures 5A and S5C). Furthermore, the knockdown of PCBP2 could inhibit the processing of both shRNA and miRNA precursors and block the enhanced miRNA processing mediated by iron chelator (Figures 5B, 5C, and S5D–S5E). These findings together suggest that PCBP2 is involved in the miRNA/RNAi pathway and can regulate the processing of miRNA precursors or shRNAs.
Figure 5. PCBP2 Modulates the Activity of the RNAi/MicroRNA Pathway.
(A) The knockdown of PCBP2 inhibits the shRNA-mediated RNAi. The cells expressing either shVC or shGAPDH were transfected with siRNA against PCBP2 or siNC (control siRNA). The GAPDH mRNA levels were determined by quantitative RT-PCR. Knockdown of PCBP2 has no effect on GAPDH expression in shVC control cells. Values are mean ± SD for triplicate samples. *: p < 0.001.
(B) The knockdown of PCBP2 blocks the processing of shRNA. Values are mean ± SD for triplicate samples. *p < 0.001.
(C) The knockdown of PCBP2 blocks the processing of miRNA precursors. The panel shows the processing of miR-125a. Values are mean ± SD for triplicate samples. *p < 0.001.
See also Figure S5.
Cytosolic Iron Modulates PCBP2 Multimerization and Its Association with Pre-miRNAs
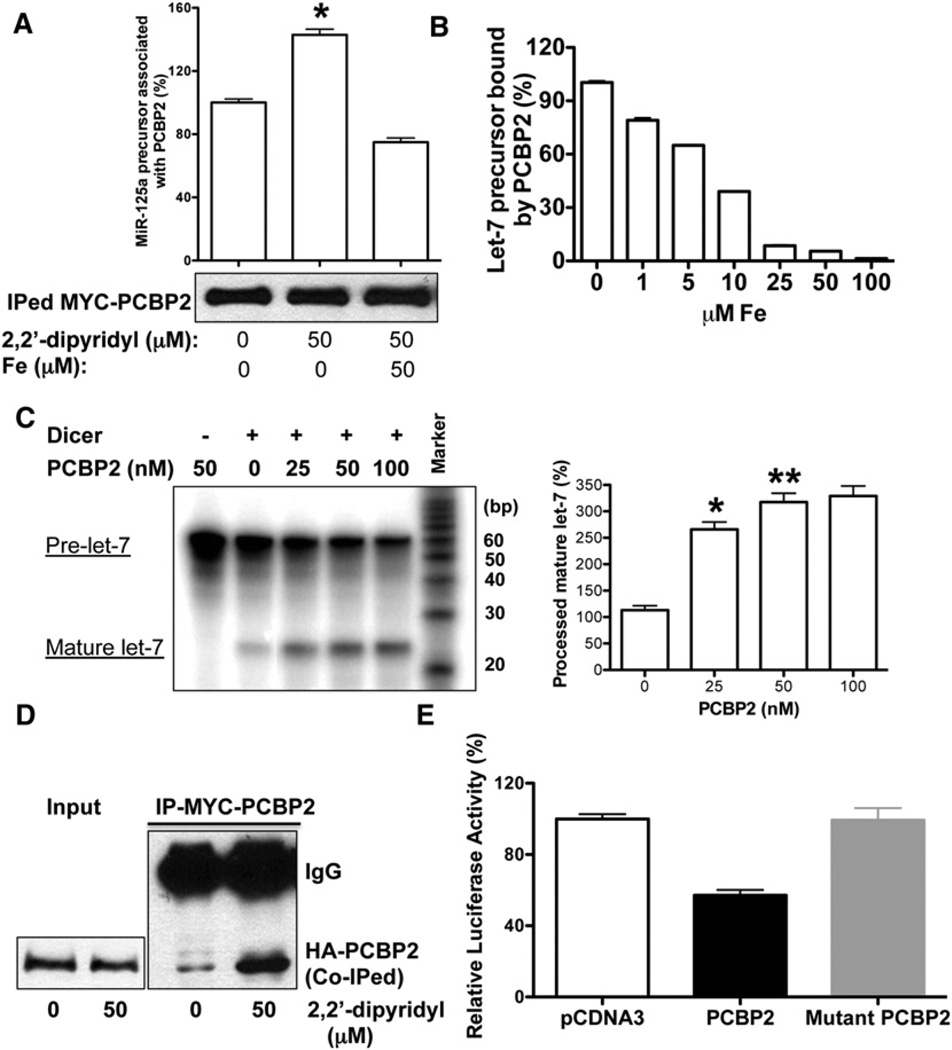
Since PCBP2 could bind to RNA and regulate the processing of miRNA precursors, we next examined whether PCBP2 could bind to miRNA precursors. Using HEK293 cells stably overexpressing pri-miR-125a transfected with MYC-PCBP2 expression vector, we isolated PCBP2-containing protein-RNA complexes through immunoprecipitation and found that PCBP2 is indeed associated with miR-125a precursor, but not pri-miR-125a (Figure 6A). Furthermore, the amount of miR-125a precursor associated with PCBP2 increased significantly with iron chelator treatment, despite the fact that a similar amount of PCBP2 protein was immunoprecipitated (Figure 6A). This enhanced association could be disrupted by the addition of iron to the immunoprecipitation buffer. We also saw similar enhanced associations between PCBP2 and other miRNA precursors as a result of iron chelator (Figure S6A). To further delineate the role of iron in the interaction between PCBP2 and miRNA precursor, we performed a series of in vitro RNA-binding assays using recombinant PCBP2 and let-7 precursor. We found that iron could indeed directly disrupt the interaction between PCBP2 and miRNA precursor, which could be rescued by 2,2′-dipyridyl, but not its inactive isomer, 4,4′-bipyridyl (Figures 6B and S6B). Furthermore, the removal of loop region from miRNA precursor could significantly abolish the binding of PCBP2 to miRNA precursor (Figure S6C). Since PCBP2 was found to prefer binding to C-rich RNAs, we further examined the sequences of the loop regions of these pre-miRNAs, and did not observe the specific C enrichment within these regions (Makeyev and Liebhaber, 2002).
Figure 6. Cytosolic Iron Modulates the Multimerization of PCBP2 and Its Association with miRNA Precursor.
(A) Iron chelator increases the amount of miRNA precursor associated with PCBP2 in vivo. The bottom panel shows the western blot to detect the MYC-PCBP2 in the immunoprecipitated complex using anti-MYC antibody under different conditions. The top panel shows the amount of miR-125a precursor associated with PCBP2 determined by quantitative RT-PCR. Values are mean ± SD for triplicate samples. *p < 0.001.
(B) Iron blocks the binding of PCBP2 to miRNA precursor in vitro. RNA filter-binding assay with recombinant PCBP2 protein (100 nM) and 5′-32P-labeled let-7 precursor in the presence or absence of different dosages of iron is shown. Values are mean ± SD for triplicate samples.
(C) The left panel shows in vitro processing of radioactively labeled let-7 precursor by either Dicer alone or Dicer with PCBP2. Both the precursor and processed miRNAs are indicated. The right panel shows the averages of relative cleavage activity from three independent experiments. *p < 0.001, **p < 0.01.
(D) Iron chelator enhances the multimerization of PCBP2. 293FT cells were transfected with MYC-PCBP2 and HA-PCBP2 expression vectors, which were then treated with iron chelators for 36 hr before immunoprecipitation (IP). Shown is the western blot to detect the coimmunoprecipitated HA-PCBP2 in the immunoprecipitated complex with anti-MYC antibody.
(E) PCBP2 multimerization is required to promote miRNA biogenesis. The cells stably overexpressing shLuciferase were transfected with Luciferase reporter construct along with pCDNA vector alone, pCDNA-PCBP2, or pCDNA Mutant PCBP2. The cells were analyzed at 48 hr after transfection. Values are mean ± SD for triplicate samples. *p < 0.001.
See also Figure S6.
To determine the functional relevance of the PCBP2-Dicer interaction, we performed an in vitro miRNA precursor processing assay. The addition of recombinant PCBP2 could significantly enhance the processing of let-7 miRNA precursor by recombinant Dicer (Figures 6C and S6F). However, PCBP1, the other cytosolic iron chaperone and an ortholog of PCBP2, could not enhance Dicer-mediated miRNA processing in vitro while iron has no significant impact on Dicer activity along, either (Figures S4D and S4E). Similar enhancement could be observed with other miRNA precursor as well (Figure S6G). These findings together suggest that PCBP2 can bind to miRNA precursors and present them to Dicer for more efficient miRNA processing. Cytosolic iron could modulate this process by disrupting the interactions between PCBP2 and miRNA precursors, as well as Dicer.
Finally, previous studies have shown that, as an RNA-binding protein, the multimerization of PCBP2 is required for its interaction with RNA and gene regulation (Bedard et al., 2004; Du et al., 2008). Particularly, the second KH domain of PCBP2 is critical for its multimerization. To determine whether cytosolic iron could modulate the multimerization of PCBP2, we performed coimmunoprecipitation experiments followed by western blot analyses using HEK293 cells cotransfected with PCBP2 expression vectors carrying different tags at the N-terminals, MYC and HA, respectively. As shown in Figure 6D, the multimerization of PCBP2 could be enhanced significantly by the addition of iron chelator, suggesting that iron can indeed modulate the multimerization of PCBP2. To further explore the functional relevance of PCBP2 multimerization in the RNAi pathway, we utilized an identical mutant PCBP2 with the second KH domain mutated, which was published previously, to examine its association with Dicer and effect on the activity of the RNAi pathway (Bedard an et al., 2004). We did not detect the association between mutant PCBP2 and Dicer, and found that the mutant PCBP2 could not enhance the activity of the RNAi pathway (Figures 6E and S6H). Our results indicate that the mutant PCBP2 lacking multimerization activity could not regulate miRNA processing.
DISCUSSION
miRNAs constitute a large family of small regulatory RNAs that have emerged as key posttranscriptional regulators of gene expression in metazoans and plants, revolutionizing our comprehension of the posttranscriptional regulation of gene expression (Krol et al., 2010). miRNAs and their associated proteins are subject to diverse modifications that can impinge on their abundance and function (Kim et al., 2010). Iron is essential for fundamental metabolic processes in cells and organisms. Cellular iron uptake, distribution, and export must be tightly regulated, as insufficient iron concentrations impair the function of numerous iron proteins, whereas excess free iron can oxidize and damage the protein, nucleic acid, and lipid contents of cells (Hentze et al., 2010). Here, through a chemical screen, we identified iron chelators as a class of shRNA-mediated RNAi enhancer and uncovered a role for iron homeostasis in directly modulating the activity of the miRNA/RNAi pathway. Our results suggest potential use of iron chelators to modulate the activity of the miRNA pathway.
Iron is a critical nutrient required for DNA synthesis, energy production, and normal cellular growth. Once iron enters the cells, the majority of iron is routed to the mitochondria, where substantial amounts are needed for heme biosynthesis and maturation of the Fe-S cluster, while the portion not needed for immediate use is stored by ferritin, a ubiquitous and highly conserved multimeric protein. Tight regulation of iron uptake, distribution, and export is mediated through transport, storage, and regulatory proteins (Hentze et al., 2004). The concentration of cytosolic iron is carefully controlled in normal tissue, because cytosolic iron is toxic when present in excess. In the presence of molecular oxygen, iron salts are able to redox cycle between iron (II) and iron (III) and thereby generate highly reactive free radicals, such as hydroxyl radicals, which lead to oxidative tissue damage. Cellular iron levels are predominantly balanced by the IRE/IRP regulatory system (Rouault, 2006). Ferritins are the main iron storage proteins found in animals, plants, and bacteria. The capacity to store iron in ferritin is essential for life in mammals. Both PCBP1 and PCBP2 have been implicated to contribute to ferritin iron loading (Shi et al., 2008). Our results suggest that cytosolic iron, but not heme, could modulate the activity of the miRNA pathway. Whether the administration of iron chelator into an intact organism could affect the in vivo miRNA processing would need to be further tested.
We show that PCBP2, but not PCBP1, mediates this process by associating with Dicer. This specificity is particularly intriguing given the high similarity between these two proteins (Makeyev and Liebhaber, 2002). Indeed, an earlier proteomic analysis of Argonaute-containing mRNA-protein complexes in human cells suggested that PCBP2 (hnRNP E2), but not PCBP1, is associated with Argonaute 1 protein-containing complex with Dicer activity (Höck et al., 2007). These data together suggest that specific domain(s) mediating the association with Dicer activity might only be present in PCBP2 protein, which would need further investigation. Furthermore, our in vitro studies suggest that cytosolic iron could regulate the multimerization of PCBP2, which is required for its interaction with miRNA precursors. These findings reveal a dynamic regulation mediated by cytosolic iron, which is itself being tightly regulated. When cytosolic iron is low, PCBP2 could multimerize, bind to miRNA precursors, and present them to Dicer for more efficient miRNA processing. Excess cytosolic iron will interfere with the binding of PCBP2 to miRNA precursors as well as its association with Dicer, and lead to the reduced production of mature miRNAs.
Disruptions in iron homeostasis from iron deficiency and overload account for some of the most common human diseases. Iron deficiency is the leading cause of anemia in the world, and it interferes significantly with normal cognitive development in children. Conversely, the iron overload observed in common diseases like hemochromatosis and thalassemia results in liver and heart failure (Muckenthaler et al., 2008). Our findings here suggest that disruptions in iron homeostasis could alter the processing of miRNA precursor and the production of selective mature miRNAs, whose precursors are present abundantly at steady state in cells. Thus, the altered expression of cell type-specific miRNAs might contribute to the molecular pathogenesis of human diseases associated with disrupted iron homeostasis.
Iron chelators are being developed to treat different human diseases, including both cancer and neurodegenerative disorders. Many in vitro and in vivo studies have shown that, compared with normal cells, cancer cells are more sensitive to Fe deprivation, because of their marked Fe requirements (Richardson, 2002). To facilitate rapid replication, neoplastic cells have significantly higher levels of ribonucleotide reductase and the transferrin receptor 1. The higher Fe utilization by cancer cells versus their normal counterparts provides a rationale for the selective antitumor activity of iron chelators, so depriving cancer cells of their essential iron is a approach for cancer treatment (Whitnall and Richardson, 2006). The etiology of neurodegenerative diseases, such as Parkinson’s disease, Alzheimer’s disease, amyotrophic lateral sclerosis, and other neurodegenerative diseases, is not yet well understood. Over the last two decades, accumulating evidence has pointed to irondependent oxidative stress, elevated levels of iron, and depletion of antioxidants in the brain as major pathogenic factors (Whitnall and Richardson, 2006). Indeed, iron chelators have shown efficacy in a variety of cellular and animal models of central nervous system injury. More recently, RNAi technology has been under rapid development to treat human diseases, and several pilot RNAi clinical studies are underway to test the therapeutic utility of RNAi-based approaches (Vaishnaw et al., 2010). The identification of iron chelator as an enhancer of shRNA-mediated RNAi could give a push to this development. Our results hold out the hope that combining these two approaches might have a synergistic effect and make for better therapeutic interventions.
In summary, via chemical screen, we identified iron chelators as a class of activator of the miRNA pathway that could promote the processing of both shRNAs and miRNA precursors. We show that cytosolic iron regulates the activity of the miRNA pathway through PCBP2. PCBP2 is associated with Dicer and promotes the processing of miRNA precursors. Cytosolic iron could modulate the association between PCBP2 and Dicer, as well as the multimerization of PCBP2 and its ability to bind to miRNA precursors. Our findings not only reveal the role of iron homeostasis in the regulation of miRNA biogenesis, but also suggest potential use of iron chelators to modulate the activity of the miRNA pathway.
EXPERIMENTAL PROCEDURES
Cells and Small-Molecule Screen
293FT cells, NIH 3T3 cells, shLuciferase cells, shVector control cells, shGAPDH cells, shHtt cells, and RNAi-293-EGFP cells coexpressing EGFP and shEGFP were grown and maintained as described previously (Shan et al., 2008).
RNAi-293-EGFP cells were infected with Lenti-RFP (Lentigen) and then subcloned to generate stable RNAi-293-EGFP/RFP reporter cells. Small molecule libraries, including both The Spectrum Collection and The Structural Diversity Set from the National Cancer Institute (NCI), were used for the chemical screen with RNAi-293-EGFP/RFP cells as described previously (Shan et al., 2008). In brief, cells were seeded into 96-well plates. Two thousand individual drugs from The Spectrum Collection (MicroSource Discovery Systems) and The Structural Diversity Set (NCI) were added into individual wells at a final concentration of 10 µM in 48 hr. GFP and RFP fluorescence intensity was measured in 48 hr on an Analyst HT plate reader (Molecular Devices).
Metal Substitution Assay
RNAi-293-EGFP/RFP cells grown on regular DMEM supplemented with 10% heat-inactivated FBS were seeded into 96-well plates with FBS-free media overnight before metals were added. Metals, including iron, copper, zinc, nickel, maganese, and cobalt, were tested.
DNA Constructs
FLAG-Dicer expression vector was described previously (Paroo et al., 2009). Full-length PCBP1 and PCBP2 complementary DNAs were cloned into pcDNA3 vectors containing MYC or HA tag. Full-length PCBP1 and PCBP2 cDNAs were also cloned into pET-28a (+) to produce recombinant proteins. All the transfection experiments were repeated at least three times.
Immunoprecipitation and Western Blot
The cell lysates were prepared with lysis buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 30 mM EDTA) containing 0.5% Triton X-100 and protease inhibitor cocktail (Roche). Lysis was carried out in ice-cold lysis buffer for 10 min on ice followed by sonication. Protein A agarose beads were used for immunoprecipitation as previously described (Shan et al., 2008). For western blot, protein samples were separated on SDS-PAGE gels and then transferred to PVDF membranes (Millipore). Membranes were processed following the HyGLO Quick Spray western blotting protocol (Denville).
MicroRNA Profiling
Profiling of mature miRNA expression was performed with Applied Biosystems’ TaqMan microRNA assays with 48-plex reverse transcription and individual TaqMan microRNA real-time PCR assays according to protocols provided by the vendor as described previously (Szulwach et al., 2010). Relative quantities of miRNA were determined via the ΔΔCt method (Livak and Schmittgen, 2001). All relative quantity calculations were calibrated to control samples. Three biological replicates were assayed.
Statistical Methods
We used single factor analysis of variance to show significant differences between control and metal chelator treatment. We performed post-hoc t tests (two samples assuming equal variances) to determine significance and indicated the p value. For all the presented data except noted otherwise, we repeated at least three times, and triplicate samples were analyzed each time. A representative experiment with triplicate samples is shown. Values shown are mean ± SD for triplicate samples.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the members of the Jin lab for their assistance, and S. Warren and C. Strauss for their helpful discussions and critical reading of the manuscript. This study was supported by National Institutes of Health (NS051630/MH076090/P50AG025688 to P.J., GM071440 to C.H., and GM084010 and GM091286 to Q.L.). P.J. is the recipient of a Beckman Young Investigator Award, Basil O’Connor Scholar Research Award, and Alfred P. Sloan Research Fellow in Neuroscience. Q.L. is a W.A. “Tex” Moncrief Jr. Scholar in Biomedical Research and is supported by the Welch Foundation (I-1608).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at doi:10.1016/j.cmet.2012.04.021.
REFERENCES
- Bedard KM, Walter BL, Semler BL. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA. 2004;10:1266–1276. doi: 10.1261/rna.7070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fenn S, Tjhen R, James TL. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J. Biol. Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Lieberman J. Running interference: prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat. Struct. Mol. Biol. 2007;14:23–29. doi: 10.1038/nsmb1182. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Hergenrother PJ. Obtaining and screening compound collections: a user’s guide and a call to chemists. Curr. Opin. Chem. Biol. 2006;10:213–218. doi: 10.1016/j.cbpa.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J, O’Day E, Korzh V, Yang H, Lal A, et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002242. e1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. Mill Valley, CA: University Science Books; 1994. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Richardson DR. Iron chelators as therapeutic agents for the treatment of cancer. Crit. Rev. Oncol. Hematol. 2002;42:267–281. doi: 10.1016/s1040-8428(01)00218-9. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Ponka P. Development of iron chelators to treat iron overload disease and their use as experimental tools to probe intracellular iron metabolism. Am. J. Hematol. 1998;58:299–305. doi: 10.1002/(sici)1096-8652(199808)58:4<299::aid-ajh9>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 2002;184:1407–1416. doi: 10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- Rouault TA. Cell biology. An ancient gauge for iron. Science. 2009;326:676–677. doi: 10.1126/science.1181938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL. Small molecules: the missing link in the central dogma. Nat. Chem. Biol. 2005;1:64–66. doi: 10.1038/nchembio0705-64. [DOI] [PubMed] [Google Scholar]
- Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008;26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Bencze KZ, Stemmler TL, Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Theil EC, Goss DJ. Living with iron (and oxygen): questions and answers about iron homeostasis. Chem. Rev. 2009;109:4568–4579. doi: 10.1021/cr900052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, de Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitnall M, Richardson DR. Iron: a new target for pharmacological intervention in neurodegenerative diseases. Semin. Pediatr. Neurol. 2006;13:186–197. doi: 10.1016/j.spen.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Whitnall M, Howard J, Ponka P, Richardson DR. A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. USA. 2006;103:14901–14906. doi: 10.1073/pnas.0604979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, Lazar MA. Reverbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao JS, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, Ramratnam B. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583:3725–3730. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.