Abstract
Interacting sets of actin assembly factors work together in cells, but the underlying mechanisms have remained obscure. We used triple-color single molecule fluorescence microscopy to image the tumor-suppressor Adenomateous polyposis coli (APC) and the formin mDia1 during filament assembly. Complexes consisting of APC, mDia1, and actin monomers intiated actin filament formation, overcoming inhibition by capping protein and profilin. Upon filament polymerization, the complexes separated, with mDia1 moving processively on growing barbed ends while APC remained at the site of nucleation. Thus, the two assembly factors directly interact to initiate filament assembly, and then separate but retain independent associations with either end of the growing filament.
Regulation of actin assembly is a fundamental requirement in all eukaryotic cells, and a growing number of factors have been identified that either inhibit or promote this process. For example, the combined presence of the actin monomer binding protein profilin and the filament end-binding capping protein (CapZ), strongly suppresses both spontaneous filament nucleation and elongation. Thus, filament assembly in vivo requires nucleation and elongation factors to overcome these barriers to assembly (1-3). The formation of most cellular actin structures depends on two or more such factors, which often interact directly. A formin is a component of many actin assembly-promoting factor (APF) pairs that likely function together in vivo: the formin FMN/Capu and Spire (4), the formin Bni1 and Bud6 (5), the formin mDia1 and Adenomatous polyposis coli (APC) (6), and the formin dDia2 and dVASP (7).
The dimeric formin-homology 2 (FH2) domain of formins processively tracks the growing barbed end of the actin filament, protecting it from capping proteins (8-10). Adjacent FH1 domains recruit profilin-actin complexes and can increase the rate of elongation at barbed ends (11). While profilin enhances formin-mediated filament elongation, its presence also strongly suppresses filament nucleation by formins (12). Collaboration of formins with other APFs that bind multiple actin monomers (4-6, 13) may be required to overcome the inhibitory effects of profilin and capping protein. However, direct evidence for this hypothesis has been lacking. To address this, we reconstituted mDia1-APC mediated actin assembly with purified, fluorescently tagged proteins, and used multi-wavelength single-molecule TIRFM (total internal reflection fluorescence microscopy) to directly visualize and define the mechanisms promoting collaborative filament assembly.
For single-molecule imaging, we purified a SNAP-tagged C-terminal fragment of APC (APC-C, residues 2130-2843), encompassing its ‘Basic’ domain that is sufficient to mediate actin nucleation, and the domain that binds EB1 (a microtubule end-binding protein) (6, 14). SNAP-APC-C labeled with SNAP-surface-647 (herein named SNAP-647-APC-C) displayed identical activities to APC-C in pyrene-actin assembly assays (Fig. S1A). Photobleaching data suggested that most SNAP-647-APC-C molecules are dimeric (Figs. S1B and C, movie 1), consistent with hydrodynamic studies on MBP-tagged APC-C (6).
APC, like Spire and Bud6, has been proposed to catalyze actin nucleation by binding actin monomers to form an F-actin seed (4-6). We employed dual-color TIRFM to directly visualize surface-adsorbed SNAP-647-APC-C molecules, appearing as discrete spots, during the assembly of Oregon Green (OG)-actin filaments (Fig. 1A, movie 2). From some of these spots, we observed actin filaments emerge and grow primarily from their barbed ends, and APC did not alter the growth rate in the presence or absence of profilin (Fig. S2A). Further, APC-C did not block polymerization at pointed ends (Fig. S2B), suggesting that it stays bound to the filament at the site of nucleation.
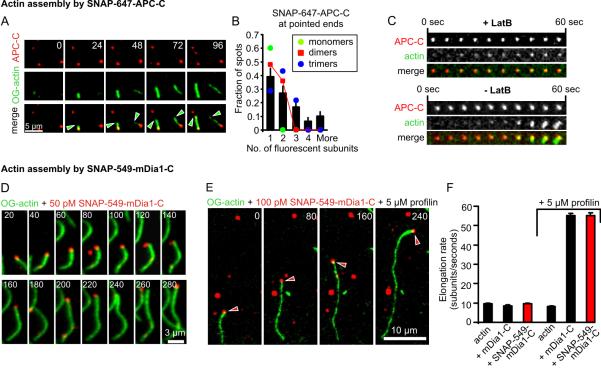
Figure 1. Single molecule analysis of the APC-C- and mDia1-C mediated actin assembly processes.
(A) Dual-color TIRFM of OG-actin filaments (1 μM, 10% labeled, green) originating from SNAP-647-APC-C-spots (10 nM, red). Green arrowheads mark the growing barbed ends. (B) Fluorescence intensity analysis of number of SNAP-647-APC-C subunits at filament ends. (C) Micrographs of OG-actin accumulation at SNAP-647-APC-C spots in absence and presence of latB. (D, E) Dual-color TIRFM of OG-actin filaments (1 μM, 10% labeled, green) being elongated by single SNAP-549-mDia1-C molecules (red) without (D) and with (E) profilin. Time is given in seconds. Red arrowheads in E mark the formin molecule associated with the barbed end. (F) Average elongation rates of mDia1-C and SNAP-549-mDia1-C-assembled filaments without and with profilin. Error bars represent SE, n ≥ 12, N=3.
For the majority of surface-tethered filaments (88/106 observed), APC fluorescence remained visible at the nucleation site 10 min after nucleation, suggesting a thermodynamically stable association. We also observed filaments with APC-C associated at their ends freely diffusing near the surface, suggesting that the stable association is not due to surface tethering. At concentrations ≥ 10 nM SNAP-647-APC-C, we occasionally observed APC-C accumulating on filament sides, which led to bundling (Fig. S3; movie 3), consistent with previous reports (15).
Quantifying the oligomeric state of APC-C at the onset of nucleation suggested that an APC-C dimer was sufficient to nucleate filament assembly, although a minority of the filaments originated from spots consisting of more than two APC-C subunits (Fig. 1B). When we included latrunculin B (latB), which associates with actin monomers and blocks polymerization, OG-actin still accumulated in some of the SNAP-647-APC-C spots, however, no filaments polymerized (Fig. 1C; movie 4). These observations suggest that nucleation by APC involves monomer recruitment, as opposed to capture of spontaneously formed F-actin intermediates.
We next purified SNAP-tagged mDia1-C, which contains FH1, FH2, and diaphanous autoregulatory (DAD) domains, and fluorescently labeled it (SNAP-549-mDia1-C). Single molecule photobleaching experiments suggested that the SNAP-549-mDia1-C preparation is predominantly dimeric but also contains higher order oligomers (Figs. S4A and B). SNAP-549-mDia1-C and unlabeled mDia1-C stimulated indistinguishable rates of pyrene-actin assembly (Fig. S4C). Using dual color TIRFM, we observed single SNAP-549-mDia1-C molecules translocating with the growing barbed ends of filaments both in the presence and absence of profilin (Figs. 1D and E; Fig. S5, movies 5, 6 and 7). Processive association of the formin was verified by additional lines of evidence (Figs. S5 and S6, movie 7). mDia1-C and SNAP-549-mDia1-C each elongated filaments at equivalent rates in presence and absence of profilin (Fig. 1F). Similar results were obtained with SNAP-549-mDia1-C in the presence of 1 nM CapZ. Our observations directly confirm that individual formin molecules processively track filament barbed ends, with or without profilin, and accelerate elongation in a profilin-dependent manner (11).
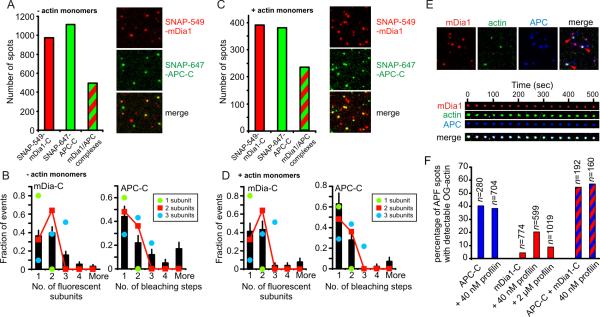
APC binds mDia formins in solution (16). Therefore, we employed dual-color TIRFM to investigate interactions between SNAP-647-APC-C and SNAP-549-mDia1-C molecules. After mixing the two labeled proteins in solution, we examined the composition of complexes that adsorbed to the surface. We found that 51 ± 2.3% (SEM) of SNAP-549-mDia-C spots colocalized with SNAP-647-APC-C spots (Fig. 2A). Coincidental colocalization was minimal (1.7%; see Methods). Taking into account the labeling stoichiometries of both proteins, our analysis is consistent with a model in which most of the fluorescent spots contain two SNAP-647-APC-C and two SNAP-549-mDia1-C molecules (Figs. 2A and 2B). Similar results were obtained in the presence of latB-sequestered actin monomers (60 ± 3.9% (SEM) colocalization; Figs. 2C and 2D). These data suggest that actin monomers neither interfere with APC-formin interactions nor change the oligomeric state of the complex, and may even modestly promote APC-formin interactions. Using triple-color TIRFM, we observed 54.8 ± 6.6 % (SEM) of APC/mDia1 complexes stably occupied by LatB-OG-actin (Fig. 2E and F). These assemblies may represent nucleation intermediates that are normally short-lived during filament assembly but can be trapped by latB. In the absence of mDia1, OG-actin accumulation was observed in 40.5 ± 4.6% (SEM) of SNAP-647-APC-C spots, but in the absence of APC-C, OG-actin accumulation was observed in only 4.0 ± 0.7% (SEM) of SNAP-549-mDia1-C spots (Fig. 2F). These observations suggest that APC-C is the primary actin monomer-recruiting factor in the APC/mDia1 complex. Profilin did not change OG-actin accumulation in SNAP-647-APC-C spots, but resulted in an increase in OG-actin accumulation to 20.2 ± 1.8% (SEM) in SNAP-549-mDia1 spots, likely representing profilin-actin recruitment by FH1 domains. Consistent with this view, competition with a 50-fold excess of profilin over OG-actin reduced the number of formin spots with OG-actin accumulation to 8.5 ± 0.9% (SEM).
Figure 2. Association of APC-C and mDia1-C and formation of tripartite complexes with G-actin.
(A) Colocalization of SNAP-647-APC-C and SNAP-549-mDia1-C in surface adsorbed complexes. The proteins (200 nM each) were preincubated for 10 min, diluted 200-fold, and visualized by TIRFM. Spots emitting fluorescence from SNAP-549, SNAP-647 or both were counted. (B) Oligomeric state of APC-C/mDia1-C complexes (n = 91) determined by fluorescence intensity analysis for mDia1-C and photobleaching step analysis for APC-C (see methods). (C,D) Identical to A and B, except that 20 μM latB-sequestered actin monomers were included during pre-incubation (100 nM in final reaction). In D, n = 43. Error bars represent SE. (E) Tripartite complexes formed by preincubating 200 nM SNAP-647-APC-C, 200 nM SNAP-549-mDia1-C, 8 μM latB-OG-actin and then diluting 200-fold prior to surface adsorption. Field of view (top) and time-lapse record of a single spot containing all three proteins (bottom). (F) Fraction of APC/mDia1 spots that contain detectable OG-actin in presence or absence of profilin (see Supplement).
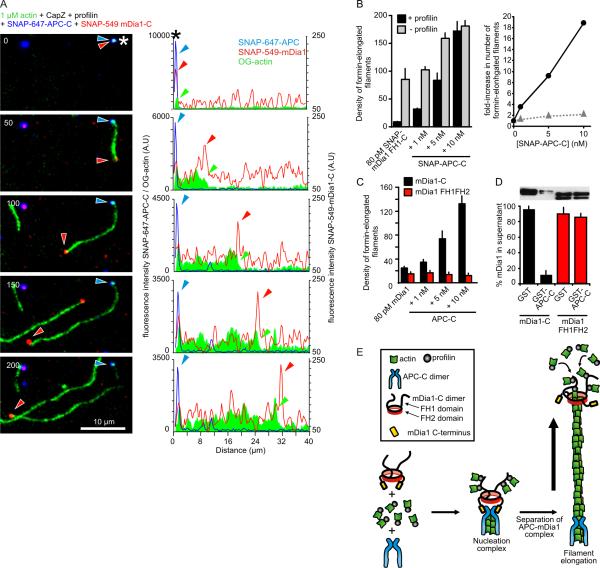
Next, we used triple-color TIRFM to define the sequence of events and spatial locations of SNAP-549-mDia1-C and SNAP-647-APC-C during collaborative filament assembly. We included CapZ and profilin to reconstitute cellular barriers to actin assembly. Tripartite complexes consisting of SNAP-549-mDia1-C, SNAP-647-APC-C and OG-actin were observed (asterisks, Fig. 3A). Filaments grew from some of these spots. Concomitant with filament growth, APC and mDia1 invariably separated (n>100), leaving SNAP-647-APC-C at the pointed end and SNAP-549-mDia1-C translocating on the growing barbed end (Fig. 3A; Fig. S8; movie 8). The same was observed in the absence of profilin (Fig. S9; movie 9). Most nucleation complexes had already separated by the start of imaging, as indicated by the presence of many short filaments carrying SNAP-549-mDia1-C at their barbed ends and SNAP-647-APC-C at their pointed ends (Fig. S10). Interestingly, SNAP-647-APC-C molecules were never seen bound to processively moving formins (n>100), suggesting that mDia1 molecules in the process of catalyzing barbed end polymerization may not be capable of interacting with APC-C. Filaments bearing APC-C on one end and mDia1 on the other end showed rates of elongation indistinguishable from rates observed for mDia1 without APC-C (Fig. S11). Thus, elongation of filaments is likely to be purely mDia1-catalyzed with no contribution from APC.
Figure 3. Single molecule visualization of APC-C and mDia1-C collaborating to assemble actin filaments.
(A) Triple-color TIRFM of the assembly of 1 μM OG-actin (green) in the presence of 50 pM SNAP-549-mDia1-C (red), 5 nM SNAP-647-APC-C (blue), 1 nM CapZ and 5 μM profilin. Asterisks indicates APC/mDia1/actin nucleation complex. Images (left; time in seconds) and corresponding profiles of fluorescence intensity along the length of the filament (right). Arrowheads mark barbed end (green), mDia1 (red), and APC (blue). (B) (Left) Density of formin-elongated filaments per 100 × 100 μm area for reactions containing 80 pM SNAP-549-mDia1-C, 1 nM CapZ, variable concentrations of SNAP-APC-C, ± 5 μM profilin. Error bars represent SD, N=3. (Right) APC-C dependent fold increase in density of formin-elongated filaments in the presence (black) and absence (grey) of profilin. (C) Same as in B, except using non-SNAP-tagged mDia1-C and tail-less mDia1 FH1FH2. (D) Western blot (anti-6His antibody) showing levels of mDia1-C and mDia1 FH1FH2 (both tagged with 6His) in supernatants after depletion by GST- (control) or GST-APC-C beads. Error bars represent SD, N=2. (E) Proposed ‘rocket launcher’ model for mDia1-APC collaboration during actin filament assembly (details in text).
Increasing concentrations of APC-C produced increasing numbers of SNAP-549-mDia1-C-elongated filaments in the presence of profilin and CapZ (Fig. 3B; Fig. S12A). Quantitatively similar results were obtained using unlabeled APC-C and mDia1-C (Fig. 3C). Moreover, longer preincubation of mDia1-C and APC-C increased the number of mDia1-C-elongated filaments by another ~2-fold (Fig. S12B). These effects of APC-C were less pronounced in the absence of profilin (Fig. 3B), suggesting that APC-C plays a key role in overcoming the nucleation barrier imposed by profilin.
Two other APFs (Spire and Bud6) were recently shown to bind to formin C-terminal tail sequences (4, 5, 13, 17). Similarly, we found that APC-C-coated beads depleted soluble mDia1-C but not mDia1-FH1FH2 (which lacks tail sequences) from supernatants (Fig. 3D), suggesting that APC binds mDia1 tail sequences. Moreover, APC-C failed to increase actin assembly activity in combination with mDia1-FH1FH2 (red bars, Fig. 3C and Fig. S13). These observations strongly suggest that direct interactions between APC and mDia1 tail sequences are required for their collaborative effects in actin assembly.
Taken together, our results suggest a mechanism (Fig. 3E) in which APC-C dimers recruit actin monomers and bind the tail region of mDia1-C to form a tripartite nucleation complex. At the onset of actin polymerization, the complex separates, leaving APC-C stably associated near the pointed end, while mDia1-C is propelled away on the rapidly growing barbed end (Fig. 3E). By this mechanism, APC plays the central role in assembling the nucleation seed, and the formin then detaches and serves as the elongation catalyst. It remains to be seen whether the same mechanism applies to other pairs of APFs that are known to interact, e.g., Spire/Capu and Bud6/Bni1 (4, 5). This work may have important implications for APC biological function. Full-length APC is a large protein that binds to numerous other cellular factors and functions in a wide variety of regulatory pathways (18). Thus, the mechanism for APC/mDia1 collaboration described here may result in site-specific actin assembly, and possibly tethering of polymerizing actin filaments at their pointed-ends to APC-binding partners (e.g., IQGAP1, microtubule plus ends) (19, 20). These phenomena may serve to promote directed cell migration or other functions that require coordinated reorganization of the actin cytoskeleton with respect to other cellular structures.
Supplementary Material
Acknowledgments
We thank B. McCartney, H. Higgs, J. Moseley, S. Reck-Petersen, and B. Smith for helpful comments on the manuscript. We are also grateful to K. Okada and A. Deaconescu for purifying CapZ and generating the SNAP-APC-C plasmid, and B. Smith for advice on single molecule imaging. The project was supported by grants from DFG (BR 4116/1-1 to D.B.) and NIH (GM43369 and GM81648 to J.G., GM083137 to B.G.).
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movemente. Science. 2009 Nov 27;326:1208. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009 Feb;21:28. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010 Apr;11:237. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007 Oct 8;179:117. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graziano BR, et al. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011 Aug 31; doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okada K, et al. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010 Jun 28;189:1087. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirenbeck A, et al. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci U S A. 2006 May 16;103:7694. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruyne D, et al. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002 Jul 26;297:612. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno H, et al. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011 Jan 7;331:80. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 10.Zigmond SH, et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003 Oct 14;13:1820. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 11.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006 Jan 27;124:423. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 12.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010 Jan;11:62. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 13.Vizcarra CL, et al. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci U S A. 2011 Jul 19;108:11884. doi: 10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Materials and methods are available as supporting material on Science Online.
- 15.Moseley JB, et al. Regulated binding of adenomatous polyposis coli protein to actin. J Biol Chem. 2007 Apr 27;282:12661. doi: 10.1074/jbc.M610615200. [DOI] [PubMed] [Google Scholar]
- 16.Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004 Sep;6:820. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 17.Zeth K, et al. Molecular basis of actin nucleation factor cooperativity: crystal structure of the Spir-1 kinase non-catalytic C-lobe domain (KIND)*formin-2 formin SPIR interaction motif (FSI) complex. J Biol Chem. 2011 Sep 2;286:30732. doi: 10.1074/jbc.M111.257782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCartney BM, Nathke IS. Cell regulation by the Apc protein. Apc as master regulator of epithelia. Current Opinion in Cell Biology. 2008 Apr;20:186. doi: 10.1016/j.ceb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004 Dec;7:871. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Mimori-Kiyosue Y, Shiina N, Tsukita S. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J Cell Biol. 2000 Feb 7;148:505. doi: 10.1083/jcb.148.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moseley JB, Maiti S, Goode BL. Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 2006;406:215. doi: 10.1016/S0076-6879(06)06016-2. [DOI] [PubMed] [Google Scholar]
- 22.Gould CJ, et al. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011 Mar 8;21:384. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soeno Y, Abe H, Kimura S, Maruyama K, Obinata T. Generation of functional beta-actinin (CapZ) in an E. coli expression system. J Muscle Res Cell Motil. 1998 Aug;19:639. doi: 10.1023/a:1005329114263. [DOI] [PubMed] [Google Scholar]
- 24.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003 Jan;21:86. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 25.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971 Aug 10;246:4866. [PubMed] [Google Scholar]
- 26.Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984 Dec 18;23:6631. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- 27.Moseley JB, et al. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004 Feb;15:896. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005 Feb;88:1387. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitsprecher D, et al. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008 Nov 19;27:2943. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004 Oct 12;101:14725. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramalingam N, et al. Phospholipids regulate localization and activity of mDia1 formin. Eur J Cell Biol. 2010 Oct;89:723. doi: 10.1016/j.ejcb.2010.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.