The crystal structure of GNA1162 from N. meningitidis has been determined at 1.89 Å resolution and its structural analysis provides important information on its function.
Keywords: GNA1162, outer membrane proteins, lipoproteins, transport systems
Abstract
GNA1162, a predicted lipoprotein from Neisseria meningitidis, is a potential candidate for a universal vaccine against meningococcal disease caused by N. meningitidis serogroup B. Here, the crystal structure of GNA1162 at 1.89 Å resolution determined by single-wavelength anomalous dispersion (SAD) is reported. The structure of GNA1162 appears to be a dimer in the crystallographic asymmetric unit as well as in solution. The overall structure of the dimer indicates that each monomer inserts its C-terminal α5 helix into the hydrophobic groove of the other molecule. Moreover, the β4 strands of each monomer lie antiparallel to each other and interact through multiple main-chain hydrogen bonds. Through structural comparisons and operon predictions, it is hypothesized that GNA1162 is part of a transport system and assists in transport and reassembly. The crystal structure of GNA1162 sheds light on its possible function and provides potentially valuable information for the design of a vaccine against meningococcal disease.
1. Introduction
Meningococcal disease (MD), which is caused by Neisseria meningitidis, is a life-threatening disease with a worldwide distribution. Its incidence rate varies from less than 0.001% in some industrialized countries to 1% in epidemic regions (Al-Tawfiq et al., 2010 ▶). It has been estimated that approximately 500 000 cases occur yearly worldwide, with approximately 10% of cases resulting in death (Wilder-Smith, 2007 ▶). Even with proper and immediate treatment the fatality rate only decreases to 5%, and up to 20% of survivors suffer from various neurologic sequelae (Rosenstein et al., 2001 ▶). N. meningitidis is a Gram-negative oxidase-positive aerobic diplococcus; it is a commensal bacterium in the human nasopharynx and only occasionally causes MD. The World Health Organization reports 12 meningococcal strain serogroups, which are classified according to the various chemical components that make up their polysaccharide capsules. The majority of MD cases result from five major pathogenic serogroups: A, B, C, Y and W135 (Gotschlich et al., 1969 ▶).
Vaccines derived from the polysaccharide (PS) coat of N. meningitidis and from PS conjugated to other proteins are effective ways of preventing MD (de Filippis, 2009 ▶). However, there are still no broad-spectrum vaccines against serogroup B of N. meningitidis (MenB) because the capsular polysaccharide of MenB contains the same components as polysialic acid, which is expressed in many human tissues, and targeting this substance may cause auto-immune diseases or weak immunoreactions (Häyrinen et al., 1995 ▶). Thus, alternative antigens that are conserved among strains and that can effectively stimulate the immune system need to be taken into consideration.
The cell wall of N. meningitidis consists of the cytoplasmic membrane, the periplasm, the outer membrane and the capsule (Hart & Rogers, 1993 ▶). Many different types of proteins are embedded in the outer membrane and are called outer membrane (OM) proteins (OMPs). OM lipoproteins play important physiological roles in the bacterium from cell growth to virulence. For example, GNA1946 is a component of the transport system (Yang et al., 2009 ▶), fHbp is responsible for antigenicity (Cantini et al., 2009 ▶) and NMB0315 is a lysostaphin-type peptidase that is involved in cell-wall degradation during cell growth and separation (Wang et al., 2011 ▶). In addition, other lipoproteins may be involved in colonization, protein folding, immunomodulation etc. (Kovacs-Simon et al., 2011 ▶). In addition, lipoproteins are potential antigens for MD vaccines.
From the whole-genome sequencing of MenB strain MC58, seven representative proteins (GNA33, GNA992, GNA1162, GNA1220, GNA1946, GNA2001 and GNA2132) were identified and found to be highly conserved among different strains, making them likely targets for universal vaccines against MenB (Pizza et al., 2000 ▶). Some of these proteins have been examined in clinical trials. GNA1162 is predicted to be a lipoprotein of 215 amino acids. Sequence-homology results using BLAST indicate that GNA1162 also exists in N. gonorrhoeae, N. lactamica, N. polysaccharea and N. cinerea (UniProt Consortium, 2012 ▶). Thus, GNA1162 is not only a potential antigen for MenB but also for these other pathogens. GNA1162 belongs to the domain of unknown function 799 (DUF799) superfamily of clan TolB_N (Punta et al., 2012 ▶), which contains many putative bacterial lipoproteins. The function of GNA1162 is currently unknown.
Here, we report the crystal structure of GNA1162 from N. meningitidis at 1.89 Å resolution. Structural analysis suggests that GNA1162 may be involved in substance transport and reassembly.
2. Materials and methods
2.1. Cloning and expression
The gene encoding GNA1162 (amino acids 26–180) was cloned into the pET-28a vector (Novagen), from which the N-terminal His tag, thrombin site and T7 tag had been removed. The recombinant plasmid was transformed into Escherichia coli BL21 (DE3) Codon Plus cells. The cells were incubated at 310 K in LB medium supplemented with 30 µg ml−1 kanamycin, induced with a final concentration of 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 298 K overnight on reaching an OD600 of 0.6 and harvested by centrifugation at 5000 rev min−1 for 15 min. For the expression of SeMet-derivatized GNA1162 protein, three mutations (V56M, L86M and A166M) were created using a standard PCR-based mutagenesis method and confirmed by DNA sequencing. The recombinant plasmids were transformed into E. coli B834 cells. The cells were cultured and harvested using the same protocol as described above.
2.2. Protein purification
The cell pellets were resuspended in lysis buffer (20 mM Tris–HCl, 300 mM NaCl, 0.2 mM PMSF pH 8.0) with 0.1 mg ml−1 lysozyme and lysed on ice by sonication. After centrifugation at 18 000 rev min−1 for 45 min, the supernatant was collected and loaded onto an Ni–NTA column pre-treated with equilibration buffer (20 mM Tris–HCl, 300 mM NaCl, 20 mM imidazole pH 8.0). After the column had been washed with three column volumes of the same buffer, GNA1162 was eluted with elution buffer (20 mM Tris–HCl, 300 mM NaCl, 250 mM imidazole pH 8.0). The eluate was then diluted in buffer A (20 mM Tris–HCl, 1 mM EDTA pH 8.0) to a final NaCl concentration of less than 50 mM and loaded onto a Q column (GE Healthcare). The target protein was eluted with buffer B (20 mM Tris–HCl, 1 mM EDTA, 1 M NaCl) using a gradient from 5 to 50%, concentrated and further purified using a Superdex 200 size-exclusion column (GE Healthcare) in buffer C (20 mM Tris–HCl, 1 mM EDTA, 150 mM NaCl pH 8.0) at a flow rate of 0.3 ml min−1. The purity of the protein was assessed by SDS–PAGE and the purified protein was concentrated to 20 mg ml−1 using a Centricon concentrator (Millipore) for crystallization. The SeMet-derivatized GNA1162 protein was expressed by E. coli B834 cells in LeMaster medium (LeMaster & Richards, 1985 ▶) and was purified using the same protocol as was used for the wild-type protein.
2.3. Crystallization and data collection
Several constructs of GNA1162 from N. meningitidis were designed and assessed for protein expression and purity. The proteins were screened with eight crystallization screening kits from Hampton Research (Crystal Screen, Crystal Screen 2, Index, PEGRx, PEG/Ion, Natrix, SaltRx and Grid Screen PEG/LiCl) using the sitting-drop vapour-diffusion method, mixing 1 µl protein solution (20 mg ml−1) with 1 µl reservoir solution. Only the construct containing GNA1162 residues 26–180 yielded crystals. All crystals of wild-type GNA1162 were twinned. Fortunately, SeMet-substituted GNA1162 protein yielded untwinned crystals, which allowed the GNA1162 structure to be determined using the single-wavelength anomalous dispersion (SAD) method. Crystals were grown using 25% ethylene glycol as the reservoir solution. After one week of growth at 277 K, the crystals were harvested and cooled in 40% ethylene glycol. The dimensions of the crystals used for data collection were approximately 0.3 × 0.2 × 0.03 mm. Data sets were collected from SeMet-substituted crystals at 100 K on station BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF) at the experimental peak wavelength for Se. 360 images were recorded using an ADSC Quantum 315r CCD detector. The oscillation angle was 1°, with a crystal-to-detector distance of 250 mm and an exposure time of 1 s. The SeMet-substituted crystals belonged to space group P21, with unit-cell parameters a = 43.9, b = 96.1, c = 45.0 Å, β = 112.8°, and diffracted to 1.89 Å resolution. The data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). Two molecules were present in each asymmetric unit. The Matthews coefficient was 2.44 Å3 Da−1 and the solvent content was 49.6%.
2.4. Structure determination and refinement
The HKL2MAP program (Pape & Schneider, 2004 ▶) yielded six Se sites in one asymmetric unit using SAD data, and the initial phases (31.39–2.30 Å resolution) were calculated and improved using the AutoSol program from the PHENIX package (Zwart et al., 2008 ▶), with an overall figure of merit (FOM) of 0.608. The residues were first built automatically by AutoBuild from PHENIX and then manually built using Coot (Emsley & Cowtan, 2004 ▶) based on 2F obs − F calc and F obs − F calc difference Fourier maps. The structural model was refined using phenix.refine from PHENIX. The final structure had an R cryst value of 23.16% and an R free value of 25.02% in the resolution range 31.39–1.89 Å from the SeMet-derivative data. The data-collection and refinement statistics are summarized in Table 1 ▶. Structural figures were generated with PyMOL (Schrödinger).
Table 1. Data-collection and refinement statistics for SeMet GNA1162 (PDB entry 4hrv).
Values in parentheses are for the highest resolution shell.
| Detector | ADSC Quantum 315r |
| Space group | P21 |
| Unit-cell parameters (, ) | a = 43.9, b = 96.1, c = 45.0, = 112.8 |
| Wavelength () | 0.9792 |
| Resolution range () | 31.41.89 (1.961.89) |
| Total reflections | 27281 |
| No. of unique reflections | 26425 |
| Molecules in asymmetric unit | 2 |
| Multiplicity | 7.3 (6.6) |
| R merge † (%) | 7.8 (76.2) |
| I/(I) | 27.68 (2.28) |
| Completeness (%) | 95.9 (83.0) |
| FOM | 0.608 |
| Refinement | |
| Resolution range () | 31.41.89 |
| R cryst ‡ (%) | 23.16 |
| R free § (%) | 25.02 |
| R.m.s.d., bond lengths () | 0.006 |
| R.m.s.d., bond angles () | 0.969 |
| No. of reflections | |
| Test set | 2007 |
| Working | 24418 |
| No. of protein atoms | 2089 |
| No. of solvent atoms | 36 |
| Ramachandran plot, residues in (%) | |
| Most favoured regions | 93.8 |
| Additional allowed region | 6.2 |
| Generously allowed region | 0 |
| Disallowed region | 0 |
| Average B factor (2) | |
| Protein | 46.1 |
| Solvent | 51.3 |
| DPI () | 0.1347 |
R
merge = 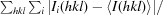
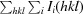 .
.
R
cryst = 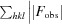
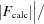
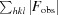 .
.
R free is calculated in the same way as R cryst but from a test set containing 7.6% of the data, which were excluded from refinement.
2.5. Analytical ultracentrifugation
Sedimentation velocity (SV) was performed in a Beckman Coulter XL-I analytical ultracentrifuge using double-sector centrepieces and sapphirine windows. An additional protein-purification step was performed on a Superdex 200 size-exclusion column (GE Healthcare) in 20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA pH 8.0 before the experiments. The experiment was conducted at 40 000 rev min−1 and 285 K using interference detection and double-sector cells loaded with approximately 20 mg ml−1 protein. The buffer composition (density and viscosity) and the protein partial specific volume (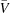 ) were obtained using the SEDNTERP program (available through the Boston Biomedical Research Institute). The data were analysed using the SEDFIT and SEDPHAT programs (Schuck, 2000 ▶, 2003 ▶).
) were obtained using the SEDNTERP program (available through the Boston Biomedical Research Institute). The data were analysed using the SEDFIT and SEDPHAT programs (Schuck, 2000 ▶, 2003 ▶).
2.6. PDB deposition
The coordinates and structure factors for GNA1162 have been deposited in the PDB with accession code 4hrv.
3. Results
3.1. The overall structure of GNA1162
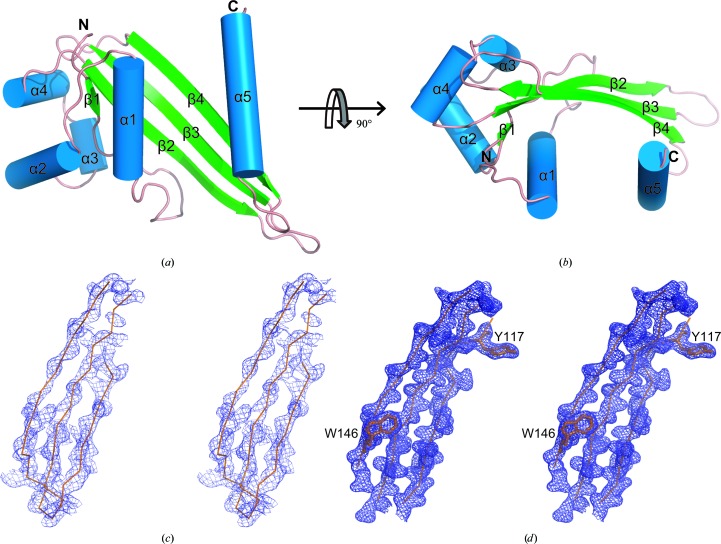
In this study, we solved the structure of GNA1162 (residues 26–180) at a resolution of 1.89 Å using the single-wavelength anomalous dispersion (SAD) method. In the final model, two molecules of GNA1162 are present in each asymmetric unit. Molecule A consists of residues 35–119 and 127–180, while molecule B consists of residues 35–178. The two molecules in the asymmetric unit are very similar, with a root-mean-square deviation (r.m.s.d.) value of 0.58 Å. We will only refer to molecule B in the following discussion. The overall structure of GNA1162 consists of five α-helices (α1–α5) and four β-strands (β1–β4) (Fig. 1 ▶). In detail, the structure begins with a small β-strand (β1) at the N-terminus followed by four α-helices (α1–α4) and three long antiparallel β-strands (β2–β4) that form a β-sheet. Finally, the last α-helix (α5) extends away from the remainder of the structure.
Figure 1.
Crystal structure of GNA1162. (a) A cartoon representation of the structure of GNA1162. The β-strands are shown in green, the α-helices are shown in blue and the connecting loops are shown in salmon. (b) Top view of GNA1162. The electron-density maps derived from SAD phases (c) and the final refinement (d) are shown. The maps are coloured blue and are contoured at 1.5σ.
3.2. Dimer packing in the GNA1162 structure
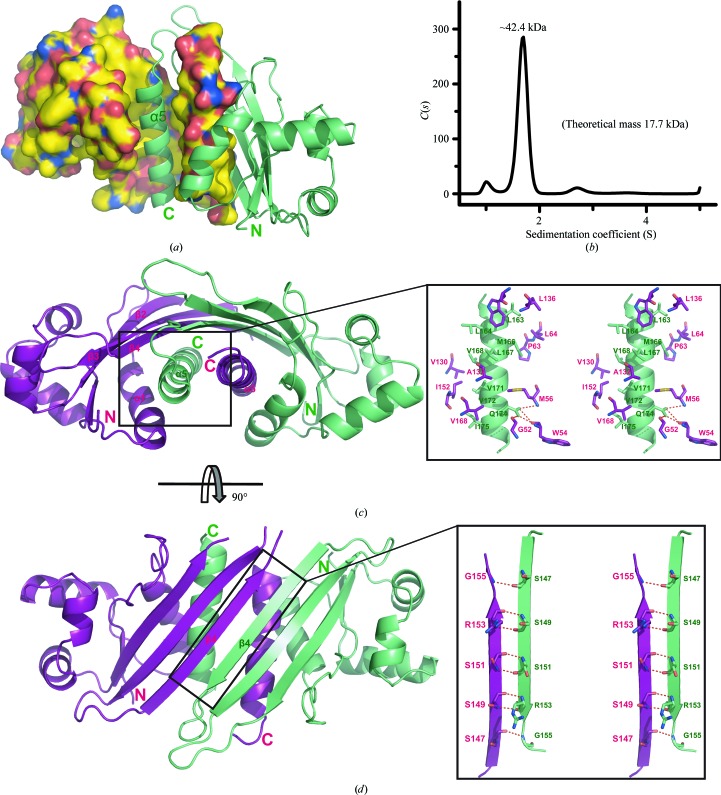
The structure of GNA1162 reveals a dimer in the asymmetric unit with a buried surface area of 2145 Å2 as calculated by AREAIMOL (Winn et al., 2011 ▶). The SV analysis also confirmed that GNA1162 can form a stable homodimer in solution with a molecular mass of approximately 42.4 kDa (Fig. 2 ▶ b). The overall structure of the dimer shows that each molecule inserts its C-terminal α5 helix into the groove of the other molecule (Fig. 2 ▶ a). The interactions within the dimer interface involve the side chains of several hydrophobic residues in the α5 helix of molecule B (Leu163, Leu164, M1et66, Leu167, Val168, Val171, Val172 and Ile175) interacting with the hydrophobic groove (Pro63, Leu64, Val130, Ala132, Leu136, Trp146, Ile152 and Val168), which is formed by helices α1 and α5 and the β-sheet (β2–β4) of molecule A (Fig. 2 ▶ c). In addition, Gln174 of molecule B forms hydrogen bonds to the main-chain atoms of Gly52, Trp54 and Met56 of molecule A. Moreover, the β4 strand of molecule B lies antiparallel to the β4 strand of molecule A to form multiple main-chain hydrogen bonds (Fig. 2 ▶ d). All of the interactions described are reciprocal between the two monomers of the asymmetric unit.
Figure 2.
Dimeric packing of GNA1162. (a) The structure of the GNA1162 dimer. Molecule A is shown as a surface representation. C, N, O and S atoms are coloured yellow, blue, red and orange, respectively. Molecule B is shown as a ribbon representation and is coloured green. (b) SV analysis of the GNA1162 protein (residues 26–180). The difference between the theoretical molecular mass (35.4 kDa) and the SV fit may arise from the molecular shape and sample homogeneity. (c) Interface of α5 (green) and the hydrophobic groove (magenta) formed by α1, α5 and β2–β4. (d) Interface of the β4 strands of both molecules. Molecule A is coloured magenta and molecule B is coloured green. Hydrogen bonds are shown as dotted red lines.
3.3. Structural comparisons of GNA1162
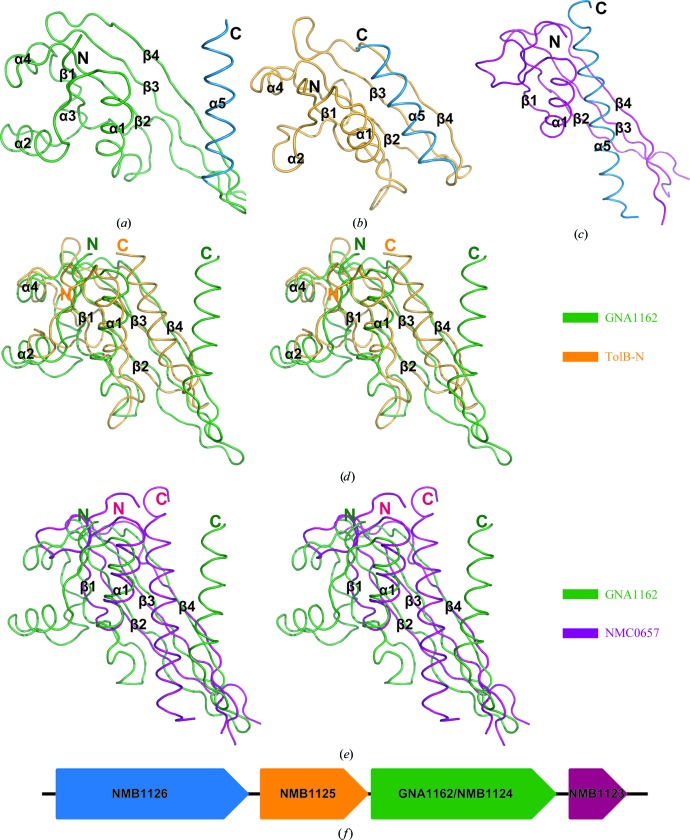
A search for structural homologues of GNA1162 in the PDB using the DALI server (Holm & Sander, 1995 ▶) identified several possible structures. The best match was the N-terminal domain of TolB from Escherichia coli (TolB-N; PDB entry 2hqs; Z-score 8.3; Fig. 3 ▶ b; Bonsor et al., 2007 ▶). Sequence alignment shows only 11.4% amino-acid identity between GNA1162 and TolB. However, their structural comparison yields an r.m.s.d. value of 2.3 Å for 82 equivalent Cα atoms (Fig. 3 ▶ d). Further analysis shows that the β1–β4 strands and α1–α4 helices of GNA1162 resemble those of the TolB-N structure. The largest difference is observed in the α5 helix. In GNA1162 helix α5 is situated away from the core folds, while helix α5 of TolB-N participates in a compact globular structure.
Figure 3.
Structural comparison of GNA1162 with other proteins. Comparison of the overall structures of GNA1162 (a), TolB-N (b) and NMC0657 (c). GNA1162, TolB-N and NMC0657 are coloured green, orange and magenta, respectively. The C-terminal α5 helices are all coloured blue. Stereoviews of the superposition of GNA1162 onto TolB-N and NBC0657 are shown in (d) nd (e), respectively. GNA1162, TolB-N and NMC0657 are coloured green, orange and magenta, respectively. (f) The GNA1162 gene is predicted to be present in the same operon as the NMB1123, NMB1125 and NMB1126 genes. The NMB1123gene is coloured magenta, GNA1162/NMB1124 is coloured green, NMB1125 is coloured orange and NMB1126 is coloured blue. The arrows indicate the direction of transcription.
Another promising structure in the DALI search results was NMC0657 from N. meningitidis serogroup C (PDB entry 3bf2; Z-score 6.0; r.m.s.d. 3.0 Å; Fig. 3 ▶ c). The structural alignment encompasses 75 equivalent Cα atoms of GNA1162. The proteins only share 14.5% amino-acid identity. GNA1162 and NMC0657 have similar β-sheets and helix α1. However, NMC0657 lacks helices α2 and α3 found in GNA1162. Moreover, helix α5 of NMC0657 displays a compact conformation (Fig. 3 ▶ e).
4. Discussion
GNA1162 is conserved among pathogenic Neisseria serogroups and is considered to be a potential candidate for the clinical development of vaccines against MD (Pizza et al., 2000 ▶). Here, we have solved the crystal structure of GNA1162 using the SAD method. Analysis revealed that the structure of GNA1162 is similar to those of the TolB and NMC0657 proteins.
TolB is a member of the Tol–Pal translocation system, which is involved in maintaining the proper structure and function of the cell envelope. In a large system, TolB can bind to TolA through its N-terminus (Godlewska et al., 2009 ▶; Dubuisson et al., 2002 ▶), which is crucial for the function of Tol–Pal. As for NMC0657, sequence comparisons by BLAST indicate that it is a member of the lipopolysaccharide-transport protein E (LptE) superfamily, which was formerly known as rare lipoprotein B (RlpB; Takase et al., 1987 ▶). LptE is a member of a large system responsible for the transport of lipopolysaccharides to the OM and subsequent assembly (Sperandeo et al., 2007 ▶, 2008 ▶; Ruiz et al., 2008 ▶), which is essential in most Gram-negative bacteria. Considering the structural similarity of GNA1162 to proteins such as TolB and LptE which are involved in the transport system, we speculate that GNA1162 acts as an accessory to unidentified transport machinery. GNA1162 may form a large complex with other proteins to assist in transporting materials. GNA1162 is most likely to play an important role in the assembly of these substances.
In addition, the Prokaryotic Operon DataBase (ProOpDB; Taboada et al., 2010 ▶) predicts that the GNA1162 gene (also called NMB1124) is found in the same operon as the genes NMB1123, NMB1125 and NMB1126, as shown in Fig. 3 ▶(f). These gene products form a cluster of proteins that may have similar or complementary functions. Among these proteins, NMB1126 is very similar to CsgG (Finney et al., 2008 ▶), which is a lipoprotein located in the outer membrane that functions by forming a channel and translocating the curli subunits across the outer membrane (Loferer et al., 1997 ▶; Robinson et al., 2006 ▶). Therefore, GNA1162 may also participate in this process as an assistant protein. In the future, further experiments to determine the function of GNA1162 are expected to provide a more complete understanding of its transport and/or assembly mechanisms.
Supplementary Material
PDB reference: GNA1162, 4hrv
Acknowledgments
We are grateful to Mr Zheng Wang for help with the analytical ultracentrifugation experiment and to the staff at beamline BL17U1 of the Shanghai Synchrotron Radiation Facility for excellent technical assistance during data collection. This work was funded by the 973 Program (grants 2013CB910400 and 2012CB917201), NSFC (grant 31170684) and the Fundamental Research Funds for the Central Universities (grant 65020241).
References
- Al-Tawfiq, J. A., Clark, T. A. & Memish, Z. A. (2010). J. Travel Med. 17, Suppl., 3–8. [DOI] [PubMed]
- Bonsor, D. A., Grishkovskaya, I., Dodson, E. J. & Kleanthous, C. (2007). J. Am. Chem. Soc. 129, 4800–4807. [DOI] [PubMed]
- Cantini, F., Veggi, D., Dragonetti, S., Savino, S., Scarselli, M., Romagnoli, G., Pizza, M., Banci, L. & Rappuoli, R. (2009). J. Biol. Chem. 284, 9022–9026. [DOI] [PMC free article] [PubMed]
- Dubuisson, J. F., Vianney, A. & Lazzaroni, J. C. (2002). J. Bacteriol. 184, 4620–4625. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Filippis, I. de (2009). J. Med. Microbiol. 58, 1127–1132.
- Finney, M., Vaughan, T., Taylor, S., Hudson, M. J., Pratt, C., Wheeler, J. X., Vipond, C., Feavers, I., Jones, C., Findlow, J., Borrow, R. & Gorringe, A. (2008). Hum. Vaccin. 4, 23–30. [DOI] [PubMed]
- Godlewska, R., Wiśniewska, K., Pietras, Z. & Jagusztyn-Krynicka, E. K. (2009). FEMS Microbiol. Lett. 298, 1–11. [DOI] [PubMed]
- Gotschlich, E. C., Goldschneider, I. & Artenstein, M. S. (1969). J. Exp. Med. 129, 1385–1395. [DOI] [PMC free article] [PubMed]
- Hart, C. A. & Rogers, T. R. (1993). J. Med. Microbiol. 39, 3–25. [DOI] [PubMed]
- Häyrinen, J., Jennings, H., Raff, H. V., Rougon, G., Hanai, N., Gerardy-Schahn, R. & Finne, J. (1995). J. Infect. Dis. 171, 1481–1490. [DOI] [PubMed]
- Holm, L. & Sander, C. (1995). Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed]
- Kovacs-Simon, A., Titball, R. W. & Michell, S. L. (2011). Infect. Immun. 79, 548–561. [DOI] [PMC free article] [PubMed]
- LeMaster, D. M. & Richards, F. M. (1985). Biochemistry, 24, 7263–7268. [DOI] [PubMed]
- Loferer, H., Hammar, M. & Normark, S. (1997). Mol. Microbiol. 26, 11–23. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pape, T. & Schneider, T. R. (2004). J. Appl. Cryst. 37, 843–844.
- Pizza, M. et al. (2000). Science, 287, 1816–1820. [DOI] [PubMed]
- Punta, M. et al. (2012). Nucleic Acids Res. 40, D290–D301. [DOI] [PMC free article] [PubMed]
- Robinson, L. S., Ashman, E. M., Hultgren, S. J. & Chapman, M. R. (2006). Mol. Microbiol. 59, 870–881. [DOI] [PMC free article] [PubMed]
- Rosenstein, N. E., Perkins, B. A., Stephens, D. S., Popovic, T. & Hughes, J. M. (2001). N. Engl. J. Med. 344, 1378–1388. [DOI] [PubMed]
- Ruiz, N., Gronenberg, L. S., Kahne, D. & Silhavy, T. J. (2008). Proc. Natl Acad. Sci. USA, 105, 5537–5542. [DOI] [PMC free article] [PubMed]
- Schuck, P. (2000). Biophys. J. 78, 1606–1619. [DOI] [PMC free article] [PubMed]
- Schuck, P. (2003). Anal. Biochem. 320, 104–124. [DOI] [PubMed]
- Sperandeo, P., Cescutti, R., Villa, R., Di Benedetto, C., Candia, D., Dehò, G. & Polissi, A. (2007). J. Bacteriol. 189, 244–253. [DOI] [PMC free article] [PubMed]
- Sperandeo, P., Lau, F. K., Carpentieri, A., De Castro, C., Molinaro, A., Dehò, G., Silhavy, T. J. & Polissi, A. (2008). J. Bacteriol. 190, 4460–4469. [DOI] [PMC free article] [PubMed]
- Taboada, B., Verde, C. & Merino, E. (2010). Nucleic Acids Res. 38, e130. [DOI] [PMC free article] [PubMed]
- Takase, I., Ishino, F., Wachi, M., Kamata, H., Doi, M., Asoh, S., Matsuzawa, H., Ohta, T. & Matsuhashi, M. (1987). J. Bacteriol. 169, 5692–5699. [DOI] [PMC free article] [PubMed]
- UniProt Consortium (2012). Nucleic Acids Res. 40, D71–D75. [DOI] [PMC free article] [PubMed]
- Wang, X., Yang, X., Yang, C., Wu, Z., Xu, H. & Shen, Y. (2011). PLoS One, 6, e26845. [DOI] [PMC free article] [PubMed]
- Wilder-Smith, A. (2007). Curr. Opin. Infect. Dis. 20, 454–460. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, X., Wu, Z., Wang, X., Yang, C., Xu, H. & Shen, Y. (2009). J. Struct. Biol. 168, 437–443. [DOI] [PubMed]
- Zwart, P. H., Afonine, P. V., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., McKee, E., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., Storoni, L. C., Terwilliger, T. C. & Adams, P. D. (2008). Methods Mol. Biol. 426, 419–435. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: GNA1162, 4hrv