The deer mouse, Peromyscus maniculatus, exhibits altitude-associated variation in hemoglobin oxygen affinity. To examine the structural basis of this functional variation, the structure of the hemoglobin has been solved.
Keywords: hemoglobin, deer mouse, oxygen affinity, Peromyscus maniculatus
Abstract
The deer mouse, Peromyscus maniculatus, exhibits altitude-associated variation in hemoglobin oxygen affinity. To examine the structural basis of this functional variation, the structure of the hemoglobin was solved. Recombinant hemoglobin was expressed in Escherichia coli and was purified by ion-exchange chromatography. Recombinant hemoglobin was crystallized by the hanging-drop vapor-diffusion method using polyethylene glycol as a precipitant. The obtained orthorhombic crystal contained two subunits in the asymmetric unit. The refined structure was interpreted as the aquo-met form. Structural comparisons were performed among hemoglobins from deer mouse, house mouse and human. In contrast to human hemoglobin, deer mouse hemoglobin lacks the hydrogen bond between α1Trp14 in the A helix and α1Thr67 in the E helix owing to the Thr67Ala substitution. In addition, deer mouse hemoglobin has a unique hydrogen bond at the α1β1 interface between residues α1Cys34 and β1Ser128.
1. Introduction
The oxygen-carrying protein hemoglobin is a heterotetramer consisting of two α and two β subunits (Perutz et al., 1960 ▶; Perutz, 1968 ▶, 1978 ▶). Two distinct structures have been described: the deoxygenated tense (T) state with a domed heme and the oxygenated relaxed (R) state with a flat heme (Perutz, 1970 ▶). Shifts in quaternary structure induced by changes in pH and temperature result in a change in the oxygen-binding affinity of hemoglobin (Monod et al., 1965 ▶; Perutz, 1972 ▶). Recently, high-resolution structures of human hemoglobin in the deoxy, oxy and carbonmonoxy forms have been reported (Park et al., 2006 ▶).
Various approaches have been used to understand the molecular mechanisms of oxygen binding in hemoglobin, including time-resolved crystallography (Srajer et al., 2001 ▶; Schmidt et al., 2005 ▶; Knapp et al., 2006 ▶). Analysis of photolysis products showed that the α and β subunits are radically different. In particular, the T state of hemoglobin is the result of cooperative movements of the heme plane itself and phenylalanine 98 toward the heme plane in the α subunit (Adachi et al., 2003 ▶). Theoretical studies indicated the presence of two additional structural transition states called Q1 and Q2 during the T-to-R state transition in human hemoglobin (Cammarata et al., 2010 ▶; Fischer et al., 2011 ▶). Pump-probe X-ray solution scattering experiments of photolysis products showed three other intermediate structures between the R and T states of clam hemoglobin (Kim et al., 2012 ▶).
To understand the structural basis of adaptive variation in hemoglobin oxygen affinity, hemoglobin from the deer mouse, Peromyscus maniculatus, was selected as a research model. The partial pressure of oxygen required to saturate 50% of hemoglobin (P 50) was used as a measure of hemoglobin oxygen affinity. Hemoglobin variants that predominate in highland populations of deer mice possess lower P 50 values than those in lowland populations as an adaptation to the highland environment (Storz, Sabatino et al., 2007 ▶; Storz & Moriyama, 2008 ▶; Storz et al., 2009 ▶, 2010 ▶). Here, we report the expression, purification, crystallization and structure analysis of recombinant lowland deer mouse hemoglobin; the stucture has been deposited in the Protein Data Bank (PDB entry 4h2l).
2. Materials and methods
2.1. Protein expression and purification
Recombinant deer mouse hemoglobin was expressed as previously reported, with slight modifications (Natarajan et al., 2011 ▶). To achieve an improved yield, the obtained crude lysate was immediately dialyzed against CO-saturated Tris–EDTA buffer pH 8.3 consisting of 20 mM Tris acetate and 0.5 mM EDTA for at least 12 h and was kept unfrozen. Purification was performed by two-step ion-exchange chromatography using quaternary amine (Q) Sepharose followed by sulfopropyl (SP) Sepharose (GE Healthcare, Pittsburgh, USA). After Q chromatography, fractions with 418 nm (Soret):280 nm (protein) absorbance ratios of above 2.5 were subjected to SP chromatography. After SP chromatography, samples with a Soret:protein peak ratios of higher than 4.5 were considered to be pure. A typical SDS–PAGE image and a representative spectrum are shown in Supplementary Figs. S1 and S21, respectively.
2.2. Crystallization
Purified recombinant deer mouse hemoglobin (Perutz, 1968 ▶) was crystallized by the hanging-drop vapor-diffusion method (Bajaj & Moriyama, 2007 ▶). The initial crystallizations were carried out using 1 ml reservoirs each containing 18–28%(w/v) polyethylene glycol (PEG) 3350 in 50 mM sodium potassium phosphate as a precipitant. Each 6.5 µl protein drop contained an initial concentration of 0.2 mM heme as determined from the oxyhemoglobin absorption peaks at 541 and 577 nm (Antonini & Brunori, 1971 ▶). Crystals were grown in an incubator at 299 K. After we had observed the PEG cloud point of hemoglobin (the point at which precipitation occurs), crystallization conditions were optimized using additives (Additive Screen, Hampton Research). Of the additives tested, calcium chloride dihydrate gave crystals with sharp edges. The crystallization conditions were further optimized by varying the concentration of calcium chloride dihydrate. The crystals obtained had hexagonal shapes with dimensions of 0.2 × 0.2 × 0.04 mm. A picture of a typical drop is shown in Supplementary Fig. S3.
2.3. Data-collection and structure solution
A complete data set was collected from a single crystal using Cu Kα radiation generated by a rotating-anode generator operated at 40 kV and 20 mA (MicroMax-007 with Blue optics, Rigaku, The Woodlands, Texas, USA) under a nitrogen-gas flow at 93 K. The crystal belonged to the orthorhombic space group C2221, with unit-cell parameters a = 76.7, b = 80.1, c = 86.8 Å. The obtained diffraction spots were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The structure of deer mouse hemoglobin was solved by the molecular-replacement method using PHENIX (Adams et al., 2010 ▶; Afonine et al., 2012 ▶). Deoxy human hemoglobin (PDB entry 2dn2; Park et al., 2006 ▶) was used as the search model. The crystal diffracted to beyond 1.8 Å in Bragg spacing. Although the orthorhombic crystal contained two subunits in the asymmetric unit, as is often found in oxyhemoglobin, the deer mouse hemoglobin showed lower temperature factors when using the deoxyhemoglobin conformation in the molecular-replacement process. The initial crystallographic R factor was 23%, with an R free value of 28%. To reduce the R factor, the model was refined by reducing the rotamer outliers. Coot was used for structure modification (Emsley & Cowtan, 2004 ▶) and model refinement was performed using REFMAC5 (Murshudov et al., 2011 ▶) from CCP4 (Winn et al., 2011 ▶). PHENIX was used for comprehensive statistical analysis and the refined structure showed a crystallographic R factor of 16% with an R free value of 20%. Details of data collection and structural refinement are given in Table 1 ▶. Based on the observations of the collected images, the reason for the high mean I/σ(I) value in the highest resolution shell was that the crystal diffracted to higher resolution and the detector was placed at a distance determined by the geometrical configuration of the diffractometer.
Table 1. Statistics of data processing and model refinement.
Values in parentheses are for the highest resolution shell.
| Space group | C2221 |
| Unit-cell parameters () | |
| a | 76.68 |
| b | 80.06 |
| c | 86.85 |
| Wavelength () | 1.542 [CuK] |
| Resolution range () | 501.78 (1.811.78) |
| No. of reflections recorded | 79999 |
| No. of unique reflections recorded | 25578 (1243) |
| Multiplicity | 3.1 (3.0) |
| Completeness (%) | 98.32 (96.33) |
| Mean I/(I) | 37.20 (18.94) |
| Wilson B factor (2) | 18.77 |
| R merge (%) | 5.0 (17.9) |
| R factor (%) | 16.20 (20.93) |
| R free (%) | 20.10 (23.98) |
| No. of atoms | |
| Total | 2692 |
| Protein | 2171 |
| Ligands | 86 |
| Water | 435 |
| Protein residues | 287 |
| R.m.s.d., bonds () | 0.007 |
| R.m.s.d., angles () | 1.06 |
| Ramachandran favored (%) | 99 |
| Ramachandran outliers (%) | 0 |
| Clashscore | 4.90 |
| Average B factor (2) | |
| Overall | 19.70 |
| Protein | 17.90 |
| Solvent | 29.30 |
2.4. Analysis of the solution UV–Vis spectrum during crystallization
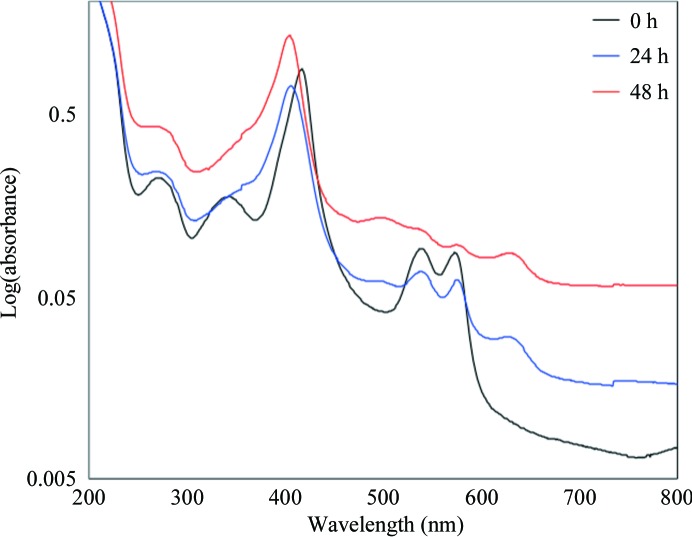
To analyze the oxidation state of hemoglobin (Antonini & Brunori, 1971 ▶) during crystallization, the UV–Vis spectra of the protein drops were analyzed every 24 h. A total of 15 identical crystallization setups were prepared. At each time point, three drops were randomly chosen; 3 µl of the protein drop was taken from each drop and diluted in 110 µl water (Table 2 ▶, Fig. 1 ▶).
Table 2. Peak wavelengths and absorbance of deer mouse hemoglobin during crystallization.
Abs, absorbance; N/D, not detected.
| Soret | Visible | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Time (h) | (nm) | Abs | (nm) | Abs | (nm) | Abs | (nm) | Abs | (nm) | Abs |
| 0 | 417 | 0.9152 | 539 | 0.0980 | 573 | 0.0928 | N/D | N/D | N/D | N/D |
| 24 | 406 | 0.6948 | 539 | 0.0674 | 576 | 0.0606 | 493 | 0.0599 | 630 | 0.2930 |
| 48 | 405 | 1.2084 | 537 | 0.1181 | 576 | 0.1014 | 497 | 0.1234 | 628 | 0.0649 |
Figure 1.
Solution UV–Vis spectrum changes in deer mouse hemoglobin during crystallization. 3 µl samples of the protein drops from the crystallization setups were diluted with water and their spectra were recorded.
3. Results
3.1. Heme-configuration analysis
The 2F o − F c electron-density map indicates the presence of a heme-bound O atom in both the α and β subunits (Supplementary Fig. S6). Since electron density for only one O atom was found around the heme, the obtained deer mouse hemoglobin was determined as being in the aquo-met form. The configuration of the hemes, either dome-shaped as in the T state or flat as in the R state, was determined by calculating the relative distance of each Fe atom from the plane of the porphyrin ring. The calculated distances were compared with those in three forms of human hemoglobin: the deoxy form (PDB entry 2dn2; Park et al., 2006 ▶), the oxy form (PDB entry 2dn1; Park et al., 2006 ▶) and the carbonmonoxy form (PDB entry 2dn3; Park et al., 2006 ▶). Using the coordinates of the four N atoms in the porphyrin ring, the heme plane was defined and the distance of the Fe atom from the plane in each subunit was calculated as shown in Table 3 ▶. According to the calculations, the Fe atoms in deoxy human hemoglobin were located 0.31–0.39 Å below the defined planes. In contrast, the Fe atom in the α subunit of deer mouse hemoglobin was located 0.07 Å below the defined plane, similar to the heme configuration of oxy human hemoglobin. Likewise, the deer mouse heme configuration in the β subunit was similar to that of oxy human hemoglobin. Therefore, the configuration of heme in deer mouse hemoglobin appeared to be flat as in the R state.
Table 3. Projected distance between the Fe atom and the heme plane ().
The coordinates of the four N atoms in the porphyrin ring, NA, NB, NC and ND, were used to define the heme plane and to calculate the distance to the Fe atom. For each subunit, heme planes were defined using combinations of any three N-atom combinations, NANBNC, NBNCND, NCNDNA and NDNANB, and the Fe-atom distances were calculated separately for each case. The distances shown in the table are mean values. A negative sign indicates that the Fe atom was located below the defined plane and a positive sign indicates that the Fe atom was above the plane.
3.2. Analysis of the solution UV–Vis spectrum during the crystallization process
While purified deer mouse hemoglobin in the carbonmonoxy form is stable for more than three weeks, the crystal structure contained the aquo-met form. To examine the changes in the oxidation status of deer mouse hemoglobin in the crystallization setup, the UV–Vis spectrum of the protein drop was analyzed. At the time of crystallization setup, deer mouse hemoglobin had peak wavelengths at 417 (Soret peak), 539 and 573 nm (Table 2 ▶); these indicated that the deer mouse hemoglobin was in the carbonmonoxy form. After 24 h the Soret peak was shifted to 406 nm and two additional peaks were observed at 490 and 630 nm, while peaks at 539 and 576 nm indicated the presence of liganded hemoglobin. In the drops after 48 h, more than 90 crystals with dimensions of 200 µm were present in the aggregated background. Despite this, we still took 3 µl of the solution from the drop. At the 48 h time point, no detectable peaks were present at 540 and 577 nm. These results indicate that the majority of deer mouse hemoglobin in the protein drops was in the aquo-met form. Therefore, changes in the deer mouse hemoglobin form were likely to occur during crystallization. When PEG was excluded from the drop, the same spectral changes were still observed (data not shown).
3.3. Secondary- and quaternary-structure comparisons
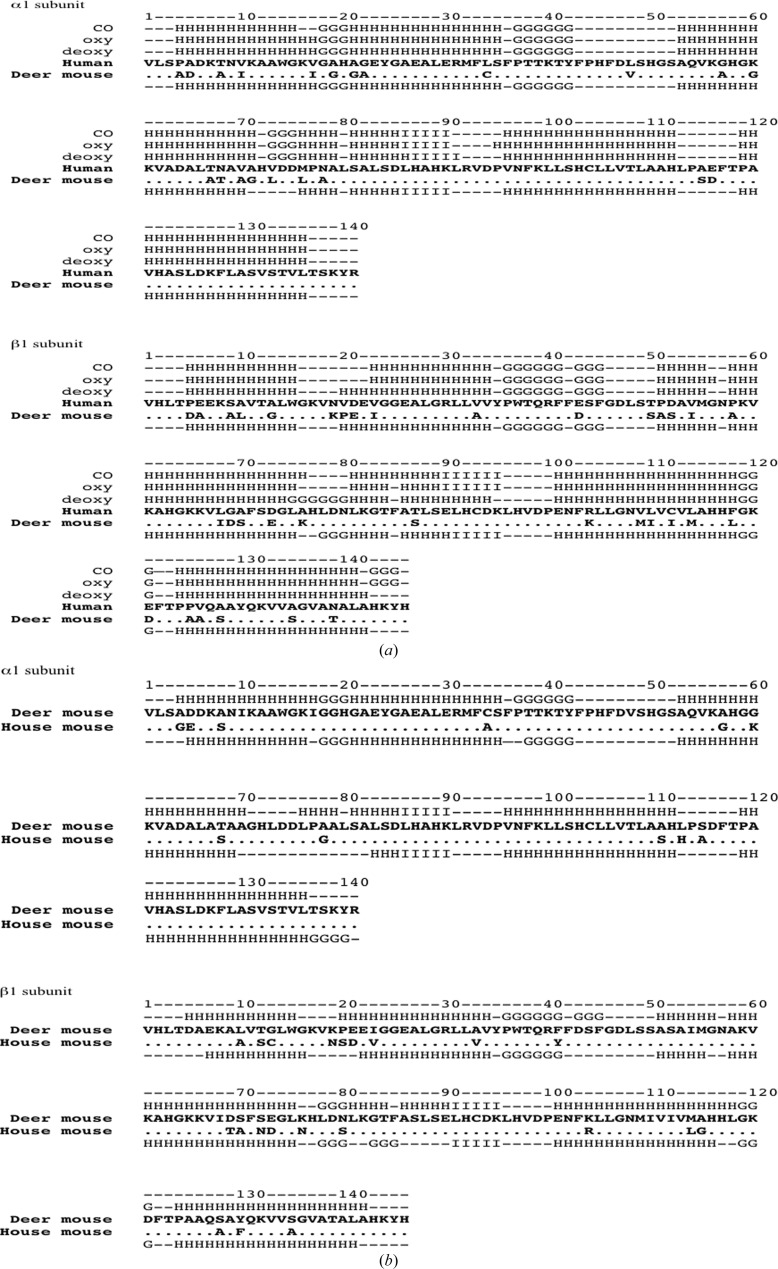
Using DSSP (Kabsch & Sander, 1983 ▶), the secondary structures of deer mouse hemoglobin and the three forms of human hemoglobin were compared (Fig. 2 ▶). The compositions of the secondary structures in the α subunit were similar for deer mouse hemoglobin and all three forms of human hemoglobin, except that residues 73–75 in deer mouse hemoglobin were not assigned as a helical structure as is the case in human hemoglobin. In contrast, the secondary structures of the deer mouse hemoglobin β subunits were similar to those of deoxy human hemoglobin. Deer mouse and deoxy human hemoglobin had two extended helices, one of which begins at residue 20 and the other at residue 78 in deer mouse hemoglobin and at residue 77 in human deoxy hemoglobin. In addition, the three helices at the C-terminus of the β-subunit starting at residue 143 were present in both oxy and carbonmonoxy human hemoglobin but were missing in deer mouse and deoxy human hemoglobin.
Figure 2.
Sequences and secondary-structure comparisons of deer mouse, house mouse and human hemoglobin. (a) Comparisons between deer mouse hemoglobin and three forms of human hemoglobin. Sequences of deer mouse and human hemoglobin α and β subunits were aligned separately; conserved amino-acid residues are denoted by dots. Secondary structures of three forms of human hemoglobin [the deoxy, oxy and carbonmonoxy (CO) forms] are presented above the human globin sequences and that of deer mouse hemoglobin is presented below the human sequence. H, α-helix; G, three-helix motif; I, five-helix motif; –, residues with no assigned secondary structure. (b) Comparisons between deer mouse hemoglobin and house mouse (Mus musculus) hemoglobin. The sequences of the α and β subunits of deer mouse and house mouse hemoglobin (PDB entry 3hrw) were aligned separately; identical amino-acid residues between the two species are represented as dots in the house mouse sequence. The secondary structure of deer mouse hemoglobin is presented above its sequence and that of house mouse hemoglobin is presented below its sequence.
The deer mouse hemoglobin structure was superimposed with those of deoxy and oxy human hemoglobin. The comparisons showed that deer mouse hemoglobin had differences in the α1 subunit at His45 (Supplementary Fig. S6), the carboxyl group of the heme and Lys61; however, these are owing to crystal packing. The interactions of symmetry-related subunits show that the NE2 atom of the His45 imidazole ring forms a hydrogen bond to the O atom of His116 in the β subunit. The O1A atom of the heme porphyrin ring interacts with the NZ atom of Lys19 in the β subunit. Lys61 did not interact with a particular amino-acid residue; however, Lys61 was positioned owing to its interaction with an ordered water molecule in the crystal.
The β1 subunits of deer mouse and human hemoglobin were also superimposed for comparison. One notable difference was found at β1His146, which has different positions in deoxy and oxy human hemoglobin. Deer mouse hemoglobin appears to be similar to human deoxy hemoglobin; however, electron density for β1His146 was very weak because electron density for one of the N atoms in the imidazole ring was observed at 1.2σ in the 2F o − F c electron-density map.
3.4. Subunit-interface comparison
The subunit interfaces of deer mouse and human hemoglobin were compared using the PISA algorithm (Krissinel & Henrick, 2007 ▶). Tetramers of deer mouse hemoglobin as well as liganded human hemoglobin were generated by crystallographic symmetry operations. The analyzed interfaces were α1–β2, α1–α2, α1–β1 and β1–β2, as listed in Supplementary Table S1. According to PISA, the interfaces of deer mouse hemoglobin were similar to those of deoxy human hemoglobin in terms of the numbers of hydrogen bonds and salt bridges. In the comparison of the α1–β2 interface, deer mouse and deoxy human hemoglobin could form salt bridges between Arg92 in the α1 submit (α1Arg92) and residue 43 in the β2 subunit, whereas liganded human hemoglobin lacked the potential for salt-bridge formation. Comparison of the α1–α2 interface also revealed similarities between deer mouse and deoxy human hemoglobin; however, the interfaces in deer mouse hemoglobin were tighter than in deoxy human hemoglobin. Three potential atom pairs were identified for hydrogen-bond formation in deoxy human hemoglobin: the N atom of Arg141 in the α1 subunit and the O atom of Asp126 in the α2 subunit (α1Arg141 NH2–α2Asp126 OD2), Asp126 OD2–Arg141 NH1 and Lys127 NZ–Arg141 OXT. In the case of deer mouse hemoglobin, there are five potential atom pairs: Val1 N–Ser138 O, Val1 N–Arg141 O, Val1 N–Arg141 OXT, Asp6 OD1–Arg141 NH1 and Ala123 O–Arg141 NH2.
4. Discussion
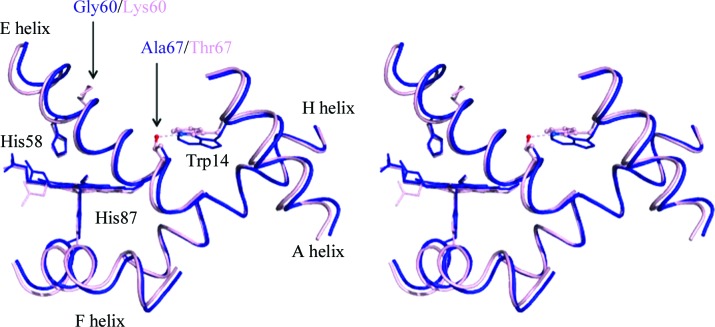
The P 50 values of human and lowland deer mouse hemolysates were 1216 and 1929 Pa, respectively (Clementi et al., 2003 ▶; Storz et al., 2010 ▶) in 100 mM HEPES buffer pH 7.4 with 100 mM Cl− at 310 K. Under these conditions, lowland deer mouse hemoglobin has the higher P 50 value. At the oxygen-binding site, the distal histidine residues αHis58 and βHis63 are located in the E helix and the proximal histidine residues αHis87 and βHis92 are located in the F helix. The mobility of helices E and F is affected by interactions between helices A and E as well as between helices F and H (Balakrishnan, Tsai et al., 2004 ▶). Deer mouse hemoglobin has an alanine substitution at position 67; thus, the hydrogen bond to residue 14 that creates the interaction between helices A and E is not formed (Fig. 3 ▶). Although the lack of a hydrogen bond between residues 14 and 67 did not affect the oxygen-binding affinity (Balakrishnan, Case et al., 2004 ▶), the lack of this hydrogen bond promotes deligation (Balakrishnan, Tsai et al., 2004 ▶). Therefore, lowland deer mouse has a reduced capability to retain oxygen at the heme and this could lead to low oxygen-binding affinity. In addition, helix E of deer mouse hemoglobin has a glycine substitution at position 60, where the human hemoglobin has a positively charged lysine. This substitution potentially increases the mobility of the helix. The P 50 values of hemoglobins from several species of house mouse (genus Mus) under the same conditions were in the range 2133–2400 Pa (Runck et al., 2010 ▶; Storz et al., 2012 ▶). Further structural characterization of house mouse hemoglobin (PDB entry 3hrw; S. S. Sundaresan, P. Ramesh & M. N. Ponnuswamy, unpublished work) was not performed owing to differences in the amino-acid sequence. The sequence of the house mouse hemoglobin used for comparative analyses is similar to that of the hemoglobin of the BALB/c strain of M. musculus (Storz, Baze et al., 2007 ▶; Runck et al., 2009 ▶); however, there are differences at the fifth and sixth residues in the β subunit, which are Asp-Ala in the compared structure (3hrw) and Pro-Glu in the BALB/c strain.
Figure 3.
Superimposed structures of the interactions of the A and E helices in the α subunits of deer mouse hemoglobin and deoxy human hemoglobin. The A, E, F and H helices in the deer mouse hemoglobin (blue) and deoxy human hemoglobin (light pink) α1 subunits are shown. Residues that differ between the two species are shown in color and conserved residues are shown in black. In deoxy human hemoglobin, the A–E helix interaction between α1Thr14 NE1 (marine blue) and α1Thr67 OG1 (red) is represented by a pink dotted line.
In deer mouse hemoglobin, limited freedom in the sliding contacts α1β2 and α2β1 (Tsai et al., 1999 ▶; Shikama & Matsuoka, 2003 ▶) might contribute to its higher P 50 value. Deer mouse hemoglobin has a hydrogen bond between α1Arg92 NH1 and β2Gln39 O which is lacking in human hemoglobin. There are no amino-acid substitutions at these positions (Fig. 2 ▶ and Supplementary Table S1). Furthermore, both deer mouse and deoxy human hemoglobin have a potential hydrogen-bonding atom pair consisting of α1Arg92 and residue 43 in β2. Owing to an aspartic acid substitution at position 43, deer mouse hemoglobin has a tighter interface between α1 and β2. The increased number of hydrogen bonds could reduce the subunit rotation.
The α1β1 (or α2β2) contact surfaces are also important to consider because the nature of the intradimer contact influences the susceptibility to heme autoxidation (Shikama & Matsuoka, 2003 ▶). In the case of deer mouse hemoglobin there is the potential for hydrogen-bond formation between the atom pair α1Cys34 O and β1Ser128 OG. This bond cannot form in human hemoglobin as leucine is present at position α34 and alanine at position β128.
As future structural characterizations of deer mouse hemoglobin with regard to oxygen-binding affinity, deoxy and oxy states of hemoglobin from both lowland and highland populations will be examined and time-resolved protein crystallography will be used to determine the effects of specific amino-acid substitutions on the hemoglobin oxygen-binding affinity.
Supplementary Material
PDB reference: deer mouse hemoglobin, 4h2l
Supporting information file. DOI: 10.1107/S1744309113005708/fw5397sup1.pdf
Acknowledgments
This work was funded by grants from the Nebraska Tobacco Settlement Biomedical Research Development Funds, the School of Biological Science Special Funds, Nebraska INBRE Program, University of Nebraska-Lincoln, the National Institutes of Health/National Heart, Lung and Blood Institute (R01 HL087216 and HL087216-S1), the National Science Foundation (DEB-DEB-0614342 and IOS-0949931) and the Danish Council for Independent Research, Natural Sciences (10-084565) and the Faculty of Science and Technology, Aarhus University.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: FW5397).
References
- Adachi, S., Park, S.-Y., Tame, J. R. H., Shiro, Y. & Shibayama, N. (2003). Proc. Natl Acad. Sci. USA, 100, 7039–7044. [DOI] [PMC free article] [PubMed]
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Antonini, E. & Brunori, M. (1971). Hemoglobin and Myoglobin in their Reactions with Ligands Amsterdam: North-Holland.
- Bajaj, M. & Moriyama, H. (2007). Acta Cryst. F63, 409–411. [DOI] [PMC free article] [PubMed]
- Balakrishnan, G., Case, M. A., Pevsner, A., Zhao, X., Tengroth, C., McLendon, G. L. & Spiro, T. G. (2004). J. Mol. Biol. 340, 843–856. [DOI] [PubMed]
- Balakrishnan, G., Tsai, C.-H., Wu, Q., Case, M. A., Pevsner, A., McLendon, G. L., Ho, C. & Spiro, T. G. (2004). J. Mol. Biol. 340, 857–868. [DOI] [PubMed]
- Cammarata, M., Levantino, M., Wulff, M. & Cupane, A. (2010). J. Mol. Biol. 400, 951–962. [DOI] [PubMed]
- Clementi, M. E., Petruzzelli, R., Filippucci, M. G., Capo, C., Misiti, F. & Giardina, B. (2003). Eur. J. Physiol. 446, 46–51. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fischer, S., Olsen, K. W., Nam, K. & Karplus, M. (2011). Proc. Natl Acad. Sci. USA, 108, 5608–5613. [DOI] [PMC free article] [PubMed]
- Kabsch, W. & Sander, C. (1983). Biopolymers, 22, 2577–2637. [DOI] [PubMed]
- Kim, K. H., Muniyappan, S., Oang, K. Y., Kim, J. G., Nozawa, S., Sato, T., Koshihara, S., Henning, R., Kosheleva, I., Ki, H., Kim, Y., Kim, T. W., Kim, J., Adachi, S. & Ihee, H. (2012). J. Am. Chem. Soc. 134, 7001–7008. [DOI] [PMC free article] [PubMed]
- Knapp, J. E., Pahl, R., Srajer, V. & Royer, W. E. (2006). Proc. Natl Acad. Sci. USA, 103, 7649–7654. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Monod, J., Wyman, J. & Changeux, J.-P. (1965). J. Mol. Biol. 12, 88–118. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Natarajan, C., Jiang, X., Fago, A., Weber, R. E., Moriyama, H. & Storz, J. F. (2011). PLoS One, 6, e20176. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Park, S.-Y., Yokoyama, T., Shibayama, N., Shiro, Y. & Tame, J. R. H. (2006). J. Mol. Biol. 360, 690–701. [DOI] [PubMed]
- Perutz, M. (1968). J. Cryst. Growth, 2, 54–56.
- Perutz, M. F. (1970). Nature (London), 228, 726–739. [DOI] [PubMed]
- Perutz, M. F. (1972). Nature (London), 237, 495–499. [DOI] [PubMed]
- Perutz, M. F. (1978). Sci. Am. 239, 92–125. [DOI] [PubMed]
- Perutz, M. F., Rossmann, M. G., Cullis, A. F., Muirhead, H., Will, G. & North, A. C. (1960). Nature (London), 185, 416–422. [DOI] [PubMed]
- Runck, A. M., Moriyama, H. & Storz, J. F. (2009). Mol. Biol. Evol. 26, 2521–2532. [DOI] [PMC free article] [PubMed]
- Runck, A. M., Weber, R. E., Fago, A. & Storz, J. F. (2010). Genetics, 184, 1121–1131. [DOI] [PMC free article] [PubMed]
- Schmidt, M., Ihee, H., Pahl, R. & Srajer, V. (2005). Methods Mol. Biol. 305, 115–154. [DOI] [PubMed]
- Shikama, K. & Matsuoka, A. (2003). Eur. J. Biochem. 270, 4041–4051. [DOI] [PubMed]
- Srajer, V., Ren, Z., Teng, T.-Y., Schmidt, M., Ursby, T., Bourgeois, D., Pradervand, C., Schildkamp, W., Wulff, M. & Moffat, K. (2001). Biochemistry, 40, 13802–13815. [DOI] [PubMed]
- Storz, J. F., Baze, M., Waite, J. L., Hoffmann, F. G., Opazo, J. C. & Hayes, J. P. (2007). Genetics, 177, 481–500. [DOI] [PMC free article] [PubMed]
- Storz, J. F. & Moriyama, H. (2008). High Alt. Med. Biol. 9, 148–157. [DOI] [PMC free article] [PubMed]
- Storz, J. F., Runck, A. M., Moriyama, H., Weber, R. E. & Fago, A. (2010). J. Exp. Biol. 213, 2565–2574. [DOI] [PMC free article] [PubMed]
- Storz, J. F., Runck, A. M., Sabatino, S. J., Kelly, J. K., Ferrand, N., Moriyama, H., Weber, R. E. & Fago, A. (2009). Proc. Natl Acad. Sci. USA, 106, 14450–14455. [DOI] [PMC free article] [PubMed]
- Storz, J. F., Sabatino, S. J., Hoffmann, F. G., Gering, E. J., Moriyama, H., Ferrand, N., Monteiro, B. & Nachman, M. W. (2007). PLoS Genet. 3, e45. [DOI] [PMC free article] [PubMed]
- Storz, J. F., Weber, R. E. & Fago, A. (2012). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 161, 265–270. [DOI] [PMC free article] [PubMed]
- Tsai, C.-H., Shen, T.-J., Ho, N. T. & Ho, C. (1999). Biochemistry, 38, 8751–8761. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: deer mouse hemoglobin, 4h2l
Supporting information file. DOI: 10.1107/S1744309113005708/fw5397sup1.pdf