The CARD domain of human CARMA1 was crystallized. The crystals belonged to space group P212121, with unit-cell parameters a = 45.73, b = 53.37, c = 91.89 Å. The crystals were obtained at 293 K and diffracted to a resolution of 3.2 Å.
Keywords: NF-κB, CARMA1 signalosome, CARD domains
Abstract
The CARMA1 signalosome, which is composed of CARMA1 [caspase recruitment domain (CARD) containing MAGUK protein 1], BCL10 (B-cell lymphoma 10) and MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1), is a molecular-signalling complex that performs pivotal functions in T-cell receptor (TCR) and B-cell receptor (BCR) mediated NF-κB activation. In this study, the CARD domain of human CARMA1 (CARMA1 CARD), corresponding to amino acids 14–109, was overexpressed in Escherichia coli using an engineered C-terminal His tag. CARMA1 CARD was then purified to homogeneity and crystallized at 293 K. Finally, X-ray diffraction data were collected to a resolution of 3.2 Å from a crystal belonging to space group P212121 with unit-cell parameters a = 45.73, b = 53.37, c = 91.89 Å.
1. Introduction
CARMA1 (also called CARD11) belongs to the MAGUK (membrane-associated guanylate kinase) family of scaffold proteins that assist in the recruitment and assembly of signalling molecules in the cytoplasmic membrane. During immune-cell activation, CARMA1 forms an intracellular multimolecular complex with BCL10 and MALT1 that is known as the CARMA1 signalosome, creating a main signalling complex for activation of NF-κB followed by production of IL-2. CARMA1 contains an N-terminal CARD (caspase recruitment domain) domain followed by a coiled-coil domain and a C-terminal MAGUK domain (Bertin et al., 2001 ▶; Gaide et al., 2001 ▶). The CARD domain is one subfamily of the death-domain (DD) superfamily, which comprises the death-domain (DD) subfamily, the death-effector domain (DED) subfamily, the caspase recruitment domain (CARD) subfamily and the pyrin-domain (PYD) subfamily (Park, 2011 ▶; Bae & Park, 2011 ▶; Kwon et al., 2012 ▶). The death-domain superfamily is one of the largest classes of protein-interaction modules and plays a pivotal role in apoptosis, inflammation and innate immune signalling pathways (Park et al., 2007 ▶; Weber & Vincenz, 2001 ▶; Reed et al., 2004 ▶). MALT1 contains an N-terminal DD followed by two immunoglobulin-like (Ig) domains and a C-terminal caspase-like domain. MALT1 functions as a ubiquitin E3 ligase inducing K63-linked ubiquitination and as a protease that can cleave the deubiquitinating enzyme A20 and BCL10 (Zhou et al., 2004 ▶; Oeckinghaus et al., 2007 ▶; Rebeaud et al., 2008 ▶; Coornaert et al., 2008 ▶). BCL10 is composed of an N-terminal CARD and a C-terminal Ser/Thr-rich region and has been shown to induce apoptosis and to activate NF-κB. For assembly of the CARMA1 signalosome for immune-cell activation, the interaction of the CARMA1 CARD and the BCL10 CARD is critical. BCL10 also associates with MALT1 via interaction between its C-terminal Ser/Thr-rich region and the first Ig domain of MALT1 (Uren et al., 2000 ▶; Lucas et al., 2001 ▶).
Despite the fundamental importance of the CARMA1 signalosome in innate and adaptive immunity, limited structural information is available. In particular, structural studies of the CARD domain and the CARD–CARD interaction in the CARMA1 signalosome have been difficult because CARD domains are subject to self-aggregation under physiological conditions (Jang & Park, 2011 ▶). A CARD domain comprised of a six-helical bundle fold is a well known structure in the death-domain superfamily. The most similar CARD to CARMA1 CARD is Apaf-1 CARD, which shares 22% sequence identity. The stoichiometry of the CARD domain is important to understand the function of each CARD-containing protein. Although most CARD domains exist as monomers in solution, several self-dimerized forms of CARD have been reported, including RIPK2 and NOD2 (Wagner et al., 2009 ▶).
As a first step towards elucidating the molecular basis of assembly of the CARMA1 signalosome and further understanding the homotypic interaction of CARD in the immune-cell signalling pathway, we overexpressed, purified and crystallized the CARD of human CARMA1 (CARMA1 CARD). Crystals were obtained at 293 K and diffracted to a resolution of 3.2 Å. Details of the structure of CARMA1 CARD should enable us to understand the assembly mechanism of the CARMA1 signalosome via the CARD–CARD interaction.
2. Materials and methods
2.1. Expression and purification
To express C-terminally His-tagged protein, the human CARMA1 (accession No. NP_115791) CARD corresponding to amino acids 14–109 was amplified by PCR using forward (5′-GGGCATATGACGCTGAAGGATGAAGAGGA-3′) and reverse (5′-GGGCTCGAGCTTTCCAGTCACCAGTTTGTAAAGT-3′) primers. The PCR product was then digested with the restriction enzymes NdeI and XhoI (Enzynomics, Republic of Korea), after which it was inserted into the pOKD vector, which had been cut with the same restriction enzymes. The pOKD vector is a home-made vector which is a modification of the commercially available pET26b vector (Novagen, USA; Dzivenu et al., 2004 ▶).
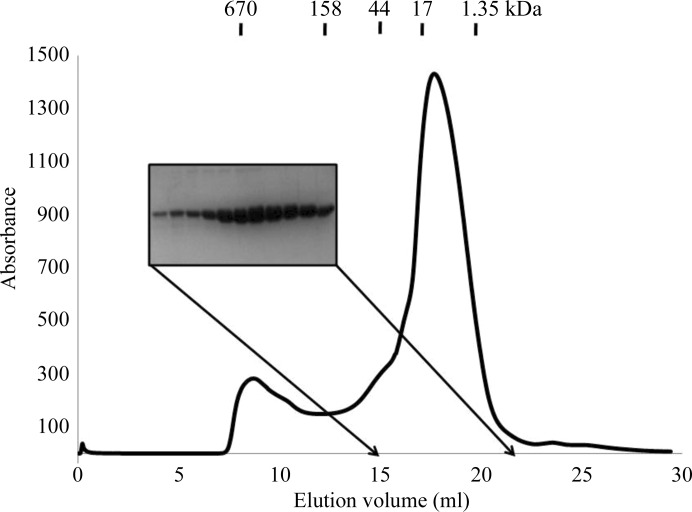
The plasmid was transformed into Escherichia coli BL21 (DE3) competent cells and its expression was induced by treating the bacteria with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 293 K. Cells expressing CARMA1 CARD were pelleted by centrifugation, resuspended and lysed by sonication in 50 ml lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl, 25 mM imidazole). The lysate was then centrifuged at 16 000 rev min−1 for 30 min at 277 K, after which the supernatant fractions were applied onto a gravity-flow column (Bio-Rad) packed with Ni–NTA affinity resin (Qiagen). The unbound bacterial proteins were subsequently removed from the column using wash buffer (20 mM Tris pH 8.0, 500 mM NaCl, 60 mM imidazole). The C-terminally His-tagged CARMA1 CARD was eluted from the column using elution buffer (20 mM Tris pH 8.0, 500 mM NaCl, 250 mM imidazole). Eluted fractions that contained greater than 95% homogeneous CARMA1 CARD, as shown by SDS–PAGE, were collected and applied onto a Superdex 200 gel-filtration column (GE Healthcare) that had been pre-equilibrated with a solution consisting of 20 mM Tris pH 8.0, 150 mM NaCl. A gel-filtration standard (Bio-Rad) containing a mixture of molecular-mass markers (thyroglobulin, 670 000 Da; globulin, 158 000 Da; ovalbumin, 44 000 Da; myoglobulin, 17 000 Da; vitamin B12, 1350 Da) was used for size calibration. The extinction coefficient of CARMA1 CARD was calculated as 14 565 M −1 cm−1 at 280 nm.
CARMA1 CARD (molecular mass 11 286 Da, including the C-terminal His tag) eluted at around 18 ml and was collected and concentrated to 5–6 mg ml−1. The protein concentration was measured using a protein-assay kit (Bio-Rad) and was determined using the Bradford method (Bradford, 1976 ▶). The peak was then confirmed to contain CARMA1 CARD by SDS–PAGE (Fig. 1 ▶). Purified CARMA1 CARD contained the extra residues LEHHHHHH at the C-terminus, which were not removed.
Figure 1.
Purification of human CARMA1 CARD. Gel-filtration chromatography and SDS–PAGE of human CARMA1 CARD.
2.2. Crystallization
The crystallization conditions were initially screened by the hanging-drop vapour-diffusion method at 293 K using screening kits from Hampton Research (Crystal Screen, Crystal Screen 2, Natrix, MembFac, Index, Crystal Screen Cryo and Crystal Screen Lite) and Emerald BioStructures (Wizard I, II, III and IV). Initial crystals were grown by equilibrating a mixture consisting of 1 µl protein solution (5–6 mg ml−1 protein in 20 mM Tris pH 8.0, 150 mM NaCl) and 1 µl reservoir solution [condition No. 47 of Wizard III; 0.2 M ammonium sulfate, 0.1 M MES pH 6.5, 30% polyethylene glycol (PEG) monomethyl ether (MME) 5000] against 0.4 ml reservoir solution. Crystallization was further optimized by searching over a range of protein, PEG MME 5000 and ammonium sulfate concentrations. Crystals appeared within 10 d and grew to maximum dimensions of 0.05 × 0.05 × 0.5 mm in the presence of 30% PEG 5000 MME, 0.2 M ammonium sulfate, 0.1 M sodium acetate pH 6.5 (Fig. 2 ▶).
Figure 2.
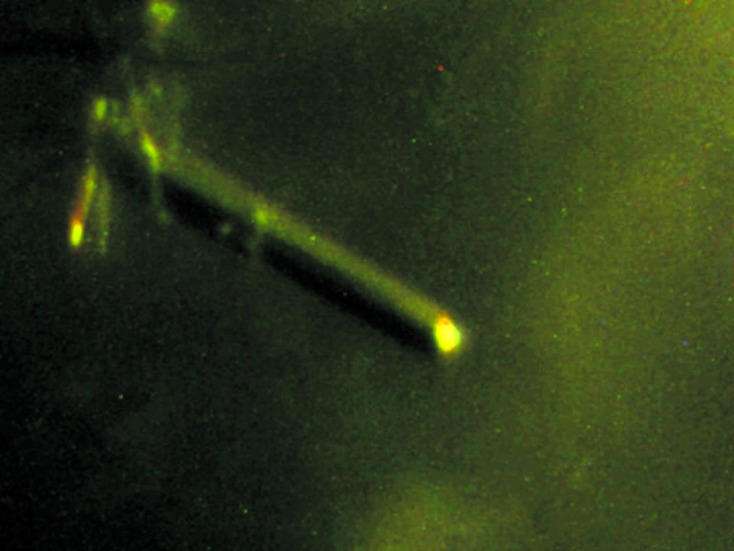
Crystals of human CARMA1 CARD. Crystals grew in 10 d in the presence of 30% PEG 5000 MME, 0.2 M ammonium sulfate, 0.1 M sodium acetate pH 6.5. The approximate dimensions of the crystals were 0.05 × 0.05 × 0.5 mm.
2.3. Crystallographic data collection
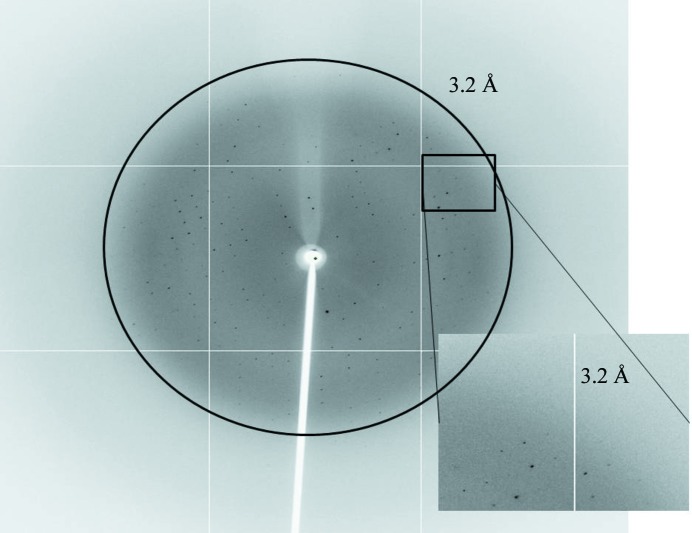
For data collection, the crystals were transiently soaked in a solution corresponding to the reservoir solution supplemented with 30%(v/v) glycerol. The soaked crystals were then flash-cooled in liquid nitrogen and a 3.2 Å resolution native diffraction data set was collected at 110 K on beamline 5C (SB II) at Pohang Accelerator Laboratory (PAL), Republic of Korea (Fig. 3 ▶). Images were recorded at a wavelength of 1.000 Å with a rotation angle of 1° and an exposure time of 1 s per image. The data sets were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The data-collection statistics are summarized in Table 1 ▶.
Figure 3.
Diffraction image (1° oscillation) of a CARMA1 CARD crystal with a 3.2 Å resolution limit.
Table 1. Diffraction data statistics of the CARMA1 CARD crystal.
Values in parentheses are for the highest resolution shell.
| X-ray source | SB II (5C), PAL |
| Wavelength (Å) | 1.0000 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 45.73, b = 53.37, c = 91.89 |
| Resolution limits (Å) | 50–3.2 (3.31–3.20) |
| No. of observations | 28354 |
| No. of unique reflections | 3460 |
| Multiplicity | 8.9 (8.8) |
| Mean I/σ(I) | 21.0 (5.0) |
| Completeness (%) | 99.9 (100) |
| R merge † (%) | 15.9 (55.0) |
R
merge = 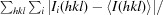
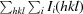 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
3. Results and discussion
The molecular basis of the mechanism of assembly of the CARMA1 signalosome is unknown. To obtain a better understanding of this process, we overexpressed, purified and crystallized CARMA1 CARD, which is a well known protein-interaction module and is critical for interacting with BCL10 during CARMA1 signalosome assembly.
His-tag affinity chromatography followed by gel-filtration chromatography produced 95% pure CARMA1 CARD and no contaminating bands were observed upon SDS–PAGE analysis (Fig. 1 ▶). The calculated monomeric molecular weight of CARMA1 CARD including the C-terminal His tag was 11 286 Da and it eluted at approximately 12 kDa, suggesting that it may exist as a monomer in solution (Fig. 1 ▶).
A tiny initial crystal that diffracted poorly was obtained using solution No. 47 of the Wizard III screen (Emerald BioStructures; 30% PEG 5000 MME, 0.2 M ammonium sulfate, 0.1 M MES pH 6.5). Optimization of the crystallization conditions using a range of protein, PEG 5000 MME and salt concentrations and pH values led to improved crystals for diffraction (Fig. 2 ▶). The optimized crystals grew to dimensions of 0.05 × 0.05 × 0.5 mm in 10 d and diffracted to 3.2 Å resolution (Fig. 3 ▶). The crystals were obtained at 293 K and belonged to space group P212121, with unit-cell parameters a = 45.73, b = 53.37, c = 91.89 Å. Diffraction data statistics are shown in Table 1 ▶. Assuming the presence of two molecules in the crystallographic asymmetric unit, the Matthews coefficient (V M) was calculated to be 2.34 Å3 Da−1, which corresponds to a solvent content of 47.35% (Matthews, 1968 ▶). Although self-rotation analysis indicated that the crystal packing allows twofold noncrystallographic symmetry (NCS), the lack of a clear twofold NCS peak suggested that the possibility of the presence of a single molecule in the asymmetric unit cannot be excluded. The molecular-replacement (MR) phasing method was conducting using Phaser (McCoy et al., 2007 ▶). Apaf-1 CARD, which share 22% sequence identity, was the most similar CARD to CARMA1 CARD and was used as a search model for MR phasing. No promising solutions were found in the initial MR attempts. We have also produced a selenomethionine-substituted crystal for SAD phasing. Efforts to determine the structure of CARMA1 CARD are presently under way.
Acknowledgments
We are grateful to Dr Yeon Gil Kim of SB II (5C) at Pohang Accelerator Laboratory. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-010870).
References
- Bae, J. Y. & Park, H. H. (2011). J. Biol. Chem. 286, 39528–39536. [DOI] [PMC free article] [PubMed]
- Bertin, J., Wang, L., Guo, Y., Jacobson, M. D., Poyet, J.-L., Srinivasula, S. M., Merriam, S., DiStefano, P. S. & Alnemri, E. S. (2001). J. Biol. Chem. 276, 11877–11882. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Coornaert, B., Baens, M., Heyninck, K., Bekaert, T., Haegman, M., Staal, J., Sun, L., Chen, Z.-J., Marynen, P. & Beyaert, R. (2008). Nature Immunol. 9, 263–271. [DOI] [PubMed]
- Dzivenu, O. K., Park, H. H. & Wu, H. (2004). Protein Expr. Purif. 38, 1–8. [DOI] [PubMed]
- Gaide, O., Martinon, F., Micheau, O., Bonnet, D., Thome, M. & Tschopp, J. (2001). FEBS Lett. 496, 121–127. [DOI] [PubMed]
- Jang, T.-H. & Park, H. H. (2011). J. Biotechnol. 151, 335–342. [DOI] [PubMed]
- Kwon, D., Yoon, J. H., Shin, S.-Y., Jang, T.-H., Kim, H.-G., So, I., Jeon, J.-H. & Park, H. H. (2012). Nucleic Acids Res. 40, D331–D336. [DOI] [PMC free article] [PubMed]
- Lucas, P. C., Yonezumi, M., Inohara, N., McAllister-Lucas, L. M., Abazeed, M. E., Chen, F. F., Yamaoka, S., Seto, M. & Nunez, G. (2001). J. Biol. Chem. 276, 19012–19019. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Oeckinghaus, A., Wegener, E., Welteke, V., Ferch, U., Arslan, S. C., Ruland, J., Scheidereit, C. & Krappmann, D. (2007). EMBO J. 26, 4634–4645. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Park, H. H. (2011). Apoptosis, 16, 209–220. [DOI] [PubMed]
- Park, H. H., Lo, Y.-C., Lin, S.-C., Wang, L., Yang, J. K. & Wu, H. (2007). Annu. Rev. Immunol. 25, 561–586. [DOI] [PMC free article] [PubMed]
- Rebeaud, F., Hailfinger, S., Posevitz-Fejfar, A., Tapernoux, M., Moser, R., Rueda, D., Gaide, O., Guzzardi, M., Iancu, E. M., Rufer, N., Fasel, N. & Thome, M. (2008). Nature Immunol. 9, 272–281. [DOI] [PubMed]
- Reed, J. C., Doctor, K. S. & Godzik, A. (2004). Sci. STKE, 2004, re9. [DOI] [PubMed]
- Uren, A. G., O’Rourke, K., Aravind, L. A., Pisabarro, M. T., Seshagiri, S., Koonin, E. V. & Dixit, V. M. (2000). Mol. Cell, 6, 961–967. [DOI] [PubMed]
- Wagner, R. N., Proell, M., Kufer, T. A. & Schwarzenbacher, R. (2009). PLoS One, 4, e4931. [DOI] [PMC free article] [PubMed]
- Weber, C. H. & Vincenz, C. (2001). Trends Biochem. Sci. 26, 475–481. [DOI] [PubMed]
- Zhou, H., Wertz, I., O’Rourke, K., Ultsch, M., Seshagiri, S., Eby, M., Xiao, W. & Dixit, V. M. (2004). Nature (London), 427, 167–171. [DOI] [PubMed]