Abstract
DNA damaging agents typically induce an apoptotic cascade in which p53 plays a central role. However, absence of a p53-mediated response does not necessarily abrogate programmed cell death, due to the existence of p53-independent apoptotic pathways, such as those mediated by the pro-apoptotic molecule ceramide. We compared ceramide levels before and after DNA damage in human osteosarcoma (U2OS) and colon cancer (HCT116) cells that were either expressing or deficient in p53. When treated with mitomycin C, p53-deficient cells, but not p53-expressing cells, showed a marked increase in ceramide levels. Microarray analysis of genes involved in ceramide metabolism identified acid ceramidase (ASAH1, up-regulated), ceramide glucosyltransferase (UGCG, down-regulated), and galactosylceramidase (GALC, up-regulated) as the three genes most affected. Experiments employing pharmacological and siRNA agents revealed that inhibition of UGCG is sufficient to increase ceramide levels and induce cell death. When inhibition of UGCG and treatment with mitomycin C were combined, p53-deficient, but not p53-expressing cells, showed a significant increase in cell death, suggesting that the regulation of sphingolipid metabolism could be used to sensitize cells to chemotherapeutic drugs.
Keywords: Ceramide, Apoptosis, Ceramide glucosyltransferase, p53, HPV16 E6
1. Introduction
Despite major advances in cancer research over the past decades, many questions regarding the disease process still remain that hamper the development of safe and effective treatment options. Currently, most cancers are treated using surgery, radiation, chemotherapy, or a combination of these. The majority of chemotherapeutic agents, including mitomycin C, achieve their results through apoptosis induced by DNA damage [1], and the best-understood pathway linking damage to the cellular DNA with the induction of apoptosis is the pathway mediated by p53. However, this is not the only pathway, as many anti-cancer agents, including cytosine arabinoside, daunorubicin and etoposide, can also be effective in a p53-negative context. Interestingly, each of these agents has been associated with increased levels of the ubiquitous sphingolipid, ceramide [2–8], suggesting that their clinical use may trigger ceramide-mediated pathways as well as pathways mediated by p53 [9,10]. In fact, when certain cancer cells are treated with anti-cancer drugs, apoptosis can occur following ceramide accumulation, and one factor contributing to chemotherapy resistance is defective ceramide signaling [10–13].
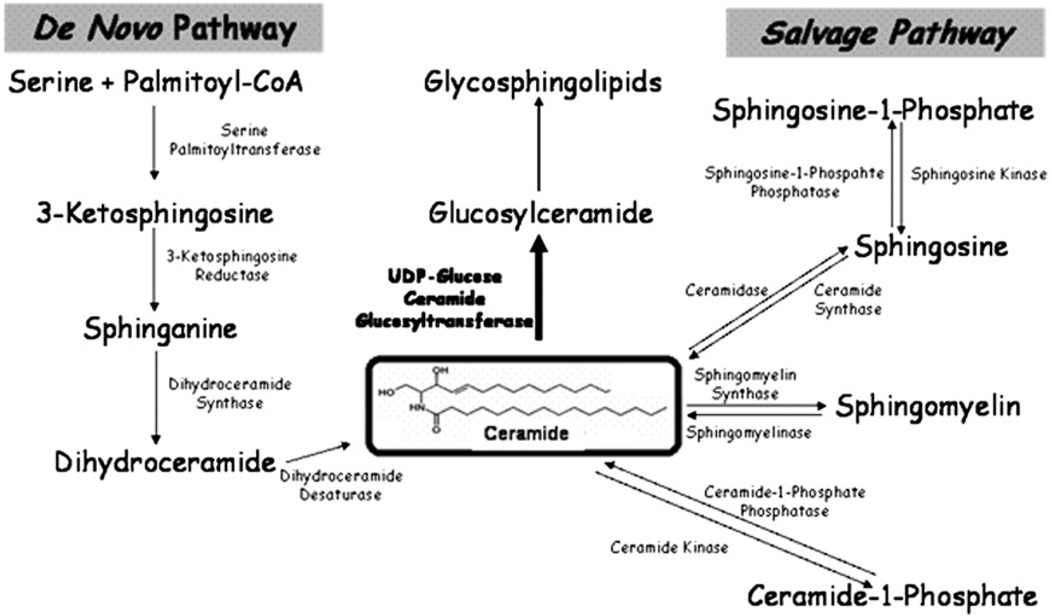
Ceramide, also known as N-acyl sphingosine and consisting of a sphingoid base covalently attached to an acyl chain by an amide bond, forms the central backbone of all sphingolipids (Fig. 1). Sphingolipids are structural components of the biological membranes of all eukaryotic organisms, and mutations in sphingolipid-metabolizing enzymes and disruptions in sphingolipid equilibria cause several severe diseases. For example, hereditary neuropathy is due to a primary mutation in the Long Chain Base 1 (LCB1) subunit of Serine Palmitoyltranferase (SPT) [14,15], and Gaucher's disease, the most common glycosphingolipidosis, is caused by an accumulation of glucosylceramide due to a deficiency in glucocerebrosidase [16]. In addition to their structural roles, several of these sphingolipids also function as signaling molecules involved in the regulation of the cell cycle, survival and apoptosis.
Fig. 1.
Sphingolipid metabolism. Ceramide can be generated by either de novo synthesis, which begins with the condensation of serine and palmitoyl coA, or through the action of enzymes such as ceramide synthase. Ceramide can also be metabolized by enzymes such as UDP-glucose ceramide glucosytransferase (UGCG).
Ceramide itself plays an important role in cellular processes such as signal transduction (by acting as a second messenger) [7,17], cell–cell adhesion [18], caspase-dependent apoptosis [19], and senescence [20]. Ceramide mediates apoptosis triggered by numerous mechanisms, including treatment with TNF-α and UV irradiation [21–24], though less is known regarding its connection to chemically-mediated DNA damage. The mechanism of ceramide-mediated cell death is thought to involve the mitochondria, the loss of mitochondrial membrane integrity, the bcl-2 family members that regulate the release of substances such as cytochrome c from the mitochondria, and the caspases that intersect with the mitochondrial pathway. For example, it has been shown that ceramide can form complexes in the mitochondrial membranes that function as channels [25]. Ceramide-mediated apoptosis may also involve activation of the JNK pathway [26–28] as well as interactions with receptor-mediated apoptosis [10].
In addition to its structural and signaling properties, ceramide also serves as the precursor for the synthesis of many sphingolipids, including sphingomyelin, ceramide phosphate and glucosylceramide, the product of UDP-glucose ceramide glucosyltransferase (UGCG) (Fig. 1). The absolute and relative levels of the various sphingolipids, including ceramide, are regulated by changes in the activity of enzymes controlling the synthesis and breakdown of ceramide. The current paradigm focuses primarily on regulation at the level of either the first enzyme involved in de novo synthesis, SPT, or of the enzymes involved in the breakdown of more complex sphingolipids, sphingomyelinases [4,23,29–31]. However, these may not be the only critical control points [32,33], and it has been suggested that the balance between SPT and UGCG can function as a key regulatory ‘rheostat’ for sphingolipid metabolism [34]. In fact, UGCG has the potential to serve as a crucial control point within the sphingolipid metabolism pathway (Fig. 1) for decisions involving cell growth and death in at least two ways. First, UGCG catalyzes the first committed step in the formation of glycosphingolipids by facilitating the synthesis of glucosylceramides, which are required for the synthesis of new cell membranes. Second, in order to create these new glucosylceramides, UGCG must metabolize ceramide, resulting in a decrease in the concentration of that biologically-active, pro-apoptotic mediator. The connection between higher levels of UGCG and lower levels of apoptosis is consistent with the observation that expression of UGCG is elevated in multidrug resistant cell lines [10], and the idea that cell growth requires a sufficient level of compounds catalyzed by UGCG is consistent with studies showing that knockout UGCG-deficient mice die as embryos [35].
Ceramide, like p53, can perform an integrative function by accepting input from numerous stimuli and pathways [17], and the importance of this integration point could become particularly significant in cells lacking a functional p53 pathway. A general consensus regarding the detailed molecular mechanisms connecting the administration of chemotherapeutic drugs with the resulting increases in ceramide and apoptosis has not yet been achieved, and may well differ between drugs and between cell types. Some cross-talk between p53 and ceramide is likely to exist, but it has also been proposed that ceramide-mediated cell cycle arrest and cell death are p53-independent [36]. Clearly, an enhanced understanding of how and when each of these pathways is activated will be necessary in order to determine which molecules should be targeted during drug design. In the experiments described in this study, we utilized a cell line derived from a human osteosarcoma (U2OS) to show that in the absence of p53, mitomycin C treatment elicits a down-regulation of UGCG, an increase in ceramide, and apoptosis. We also demonstrated that inhibiting UGCG increases the susceptibility of p53-deficient cells, but not p53-expressing cells, to mitomycin C, suggesting that agents which modulate the ceramide metabolic pathway may synergize with genotoxic agents in a chemotherapeutic context.
2. Materials and methods
2.1. Materials
Monoclonal antibodies directed against UGCG (Abnova, Taipei City, 114 Taiwan) and β-actin (Sigma, St Louis, MO) were dissolved in PBS, and solutions were stored at −20 °C prior to use. 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) (Sigma) was prepared as previously reported [37]. Mitomycin C (Sigma) was prepared at a stock concentration of 5 mg/ml in dimethyl sulfoxide (DMSO) and stored at 4 °C until ready for use. Thre-1-Phenyl-2-decanoyl-amino-3-morpholino-1-propanol hydrochloride (PDMP) (Sigma), thymidine (Fisher), RNase A (Invitrogen, Carlsbad, CA), propidium iodide (Sigma), TriZol (Invitrogen, Carlsbad, CA), and ceramides (C2, C6, C16, C17, C18, C20, and C24) (Avanti Polar Lipids, Alabaster, AL) were prepared according to manufacturer's instructions. All organic solvents were HPLC gradient, ACS certified, and were of the highest purity and quality commercially available.
2.2. Cell culture
U2OS cells, derived from a human osteosarcoma, were obtained from the ATCC (Manassas, VA). HCT116 human colorectal cancer cells and HCT116 p53-null mutant cells were a kind gift of Dr. B. Vogelstein (Johns Hopkins University) [38]. All cell lines were cultured in McCoy's 5A medium (Invitrogen) supplemented to contain 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 µg/ml) (Sigma). Construction of the pHA-E6S and pHA-E6AS plasmids, which respectively contain either the sense or the antisense versions of epitope-tagged E6 (HA-E6) under the control of the CMV promoter, and establishment of the U2OSE64b and U2OSE6AS cell lines by stable transfection with these plasmids has been described previously [37].
2.3. Cell treatment
To determine cell survival following mitomycin C treatment, cells were seeded into 96-well plates (2 × 104 cell/well, 100 µl total volume) and allowed to adhere overnight. Mitomycin C was then added to the indicated final concentration, and the cells were incubated for the specified time periods prior to measuring viability by the MTT assay. To measure ceramide levels following mitomycin C treatment, cells were plated in 6 cm plates and allowed to adhere overnight. The indicated concentrations of mitomycin C were added and cells were harvested at the indicated times. To inhibit UGCG, cells were plated and allowed to adhere overnight. 25 mM PDMP was then added to a final concentration of 25 µM and the cells were incubated for the specified time periods.
2.4. p53 ELISA
The p53 ELISA assay was performed according to the protocol of Filippova et al. [39].
2.5. Cell viability assays
Following treatment of cells as described earlier, the incubation medium was removed and exchanged with 80 µl of fresh medium. 20 µl of MTT was then added (5 mg/ml stock), and cells were incubated at 37 °C for 2 h. The medium was removed, and 200 µl of DMSO was added and incubated with shaking for 10 min. Afterward, the solution was mixed by pipetting and the absorbance of each well was measured at 490 nm. Three independent experiments, each in triplicate, were conducted.
2.6. Targeted gene silencing using siRNA
Cells were transfected with pre-designed small interfering RNAs (siRNA) for the human UGCG gene from Ambion (Ambion, Austin, TX) or with control siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) using the siRNA transfection kit as recommended by the manufacturer (Santa Cruz Biotechnology, Santa Cruz, CA). Briefly, cells were seeded in media lacking antibiotics and incubated at 37 °C overnight to 40% confluency. The transfection mixtures were then prepared by adding siRNA to siTransfection Medium (Santa Cruz Biotechnology) in one tube and siTransfection Reagent (Santa Cruz Biotechnology) to siTransfection Medium (Santa Cruz Biotechnology) in another tube and incubating the contents of the tubes at room temperature for 5 min. The amounts of the reagents were calculated based on the values indicated in the protocol and were scaled up or down in proportion to the surface area of the plate; the final concentration was 25–30 nM as recommended. After 5 min, the mixtures were combined and incubated at room temperature for 20 min to facilitate the development of a siRNA–siRNA transfection reagent complex. During this time, the media covering the cells was replaced with fresh media without antibiotics and at the end of the 20 min incubation, the complex was added to the cells. The cells were then incubated at 37 °C for the indicated times and the indicated procedures carried out. Levels of silencing of the UGCG gene were evaluated by RT-PCR and immunoblot analyses as described. Three independent experiments, each in quadruplicate were conducted.
2.7. Microarray
Total RNA was extracted using Trizol Reagent (Invitrogen) and subsequently purified using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA samples of three independent replicates for each time point were further processed and hybridized to Affymetrix Human Genome U133A and U133B expression arrays according to standard recommended protocols (Affymetrix, Santa Clara, CA) at the DNA MicroArray Facility, University of California, Irvine, CA. To extract and analyze microarray data, we used the Microarray Suite 5.0, GCOS 1.2 and Data Mining Tools 3.0 programs (Affymetrix, Santa Clara, CA). To compare data from different arrays, the signal intensity of arrays was globally scaled to 500 and normalization was done using a probe set of 100 constitutively expressed transcripts provided by Affymetrix. The Signal Log Ratio was calculated by comparing transcripts between mitomycin C-treated and untreated cells. Generation of detected (present or absent) and changed (increased or decreased) calls was done using the Wilcoxon's test and default parameters of p-value cut-offs. During the first step to identify genes with altered expression, we discarded genes that had changed calls in less than 6 out of 9 comparative files (Rank test) as well as genes that had “absent” calls in all microarrays. Remaining genes were further analyzed using both a Student's t test and the Mann–Whitney test (p<0.05 cut-offs). Genes were considered up-regulated if they passed the Mann–Whitney and Rank tests, and down-regulated if they passed the Rank and t tests. Normalized expression data were further analyzed using online-based Gene Sifter microarray analysis tool.
2.8. Reverse transcriptase–polymerase chain reaction
Reverse transcriptase–polymerase chain reaction (RT–PCR) analysis was used to estimate transcript levels in total RNA samples isolated from cell cultures. 10 µg of total RNA was used as a template for cDNA synthesis with SuperScript III reverse transcriptase and an oligo(dT) primer according to the manufacturer's instructions (Invitrogen). The cDNA obtained was normalized by PCR with primers specific to cofilin1 (CFL1), a gene that showed no change in expression in microarray experiments (forward 5′-CCTTCCCAAACTGCTTTTGAT-3′ and reverse 5′-CTGGTCCTGCTTCCATGAGTA-3′). Serial dilutions of the normalized cDNA samples were used to find the linear range of amplification for gene-specific DNA fragments. PCR was performed at 94 °C for 30 s, at 57 °C for 30 s, and at 72 °C for 1 min for 35 cycles using Taq DNA polymerase and the recommended protocol (NEB, Beverly, MA). For the UGCG gene, primers were forward 5′-TTCAATCCAGAATGATCAGGT-3′ and reverse 5′-TATAGTTGGGTCCCATAATGC-3′. To perform real-time quantative PCR, cDNA was used with the QuantiTect SYBR PCR kit (Invitrogen) on a CFX96 PCR System (Bio-Rad) according to the manufacturer's protocols.
2.9. Immunoblotting
Cells (1 × 106) were lysed in 50 to 100 µl lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, with one tablet of protease inhibitor mixture [Roche Molecular Biochemicals] per 10 ml of buffer added just prior to use) for 10 min on ice. Lysates (25 µg total protein/lane) were then subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon P membranes (Millipore). After blocking the membranes with 5% nonfat milk in TBST (50 mM Tris–Cl, pH 7.5, 150 mM NaCl, 0.1% Tween 20), primary antibodies (1:10,000 dilution) were applied and membranes were incubated for 1 h, with rocking, at room temperature. After washing with TBST, peroxidase-coupled secondary antibodies were added for detection. The membranes were then washed again with TBST following 1 h of incubation in the secondary antibodies (1:5000 dilution). Detection of the proteins was performed by using the chemiluminescent Super-Signal West Femto or Pico Maximum Sensitivity substrate (Pierce).
2.10. Lipid extraction and ceramide quantification
Lipids were extracted and ceramides were measured according to previously developed protocols [6]. Briefly, cells were harvested at the indicated times, washed in PBS, concentrated by centrifugation, transferred in a glass tube in 0.2 ml of PBS, and resuspended in 2 ml of methanol with 10 ng C17 added as an internal standard. Lipids were extracted twice using chloroform and methanol (2:1). High performance liquid chromatography (HPLC) was performed using Agilent Technologies Model 1200 (Agilent Technologies, Santa Clara, CA). Lipids were separated on a Pursuit 3 Diphenyl reversed-phase column, 50 × 2.0 mm i.d. 3 µm (Varian, Walnut Creek, CA) using the mobile phases — A: 0.1% formic acid, and 25 mM ammonium acetate in water, B: 100% acetonitrile. The ion source employed was the electrospray ionization interface (ESI) of an Agilent triple quadrupole 6410 mass spectrometer (Agilent Technologies, Santa Clara, CA) and ionization was performed in the positive ion mode. The multiple reaction monitoring (MRM) mode was employed for monitoring precursor ions, m/z 324.3 for C2, 380.4 for C6, 538.4 for C16, 566.5 for C18, 594.6 for C20, and 650.7 for C24 ceramide and product ion m/z 264.2 for all species. The MRM transition for C17, which served as the internal standard (IS), was m/z 552.5 (precursor ion)–m/z 264.2 (product ion). The instrumental MassHunter software with qualitative analysis and quantitative features was used to process the data. Concentration of ceramide species was calculated based on standard curves of known ceramide concentrations (from 0.1 to 100 ng/ml) and normalized to the concentration of phospholipid phosphate, as measured using the EnzyChrom Phospholipid Assay EPLP-100 Kit according to the manufacturer's protocol (BioAssay Systems, Hayward, CA).
2.11. Cell synchronization
Cells were synchronized according to the method of Yoshizawa-Sugata [40]. Briefly, cells were seeded overnight for attachment, then treated with 2.5 mM thymidine for 16 h. Cells were released into the cell cycle for 9 h in media containing no thymidine after which cells were treated once more with 2.5 mM thymidine for another 16 h. Cells for growth control were then re-released into the cell cycle for 48 h before harvest.
2.12. Flow cytometry
Flow cytometric analysis was performed according to the method of Ormerod [41] with modifications. Cells (1 × 106) were detached using Cellstripper (Mediatech, Inc., Herdon, VA), centrifuged, then resuspended in 200 µl ice cold PBS. Cells were vortexed lightly for 5 s, and then 2 ml ice cold 70% ethanol was added with vigorous shaking. Cells were then fixed on ice for 30 min, after which they were centrifuged and resuspended in 400 µl ice cold PBS followed by the addition of 50 µl of RNase solution (1 mg/ml) and 50 µl of propidium iodide (400 µg/ml working solution). Cells were incubated for 30 min in the dark at room temperature. 10,000 cells per sample were analyzed using a FACSCalibur (BD Biosciences).
2.13. Statistical analysis
Each experiment was performed at least three times, and each assay point was measured in triplicate. Values are expressed as means+/−standard deviation. Results shown are from representative experiments. Student's one- or two-tailed t tests, as appropriate, were used for statistical analysis, with values of <0.05 considered to be statistically significant.
3. Results
3.1. Mitomycin C induces apoptosis in both p53-expressing and p53-deficient cells
p53 is the most commonly mutated gene in human cancers [42]. Therefore, understanding p53-independent mechanisms of cell death is critical for the enhanced design of target therapeutics [43]. In general, cells with defective p53 signaling are thought to be more resistant to apoptosis than are their p53wt counterparts [44,45], though there are exceptions and many cancer cell lines with defective p53 are more sensitive to DNA damage in vitro than are cells expressing the wt protein [46–48]. Previously, we have shown that similar viability profiles are obtained when mitomycin C is used to treat either p53-deficient or p53-expressing U2OS cells [49]. This data is consistent with clinical data showing that cervical tumor cells, in which p53 levels are very low due to E6-mediated degradation, are not necessarily resistant to either chemotherapy or radiation [45]. These studies suggest that while cells expressing wt p53 respond to DNA damage in a p53-dependent manner, cells lacking p53 can utilize other mechanisms to achieve the same result.
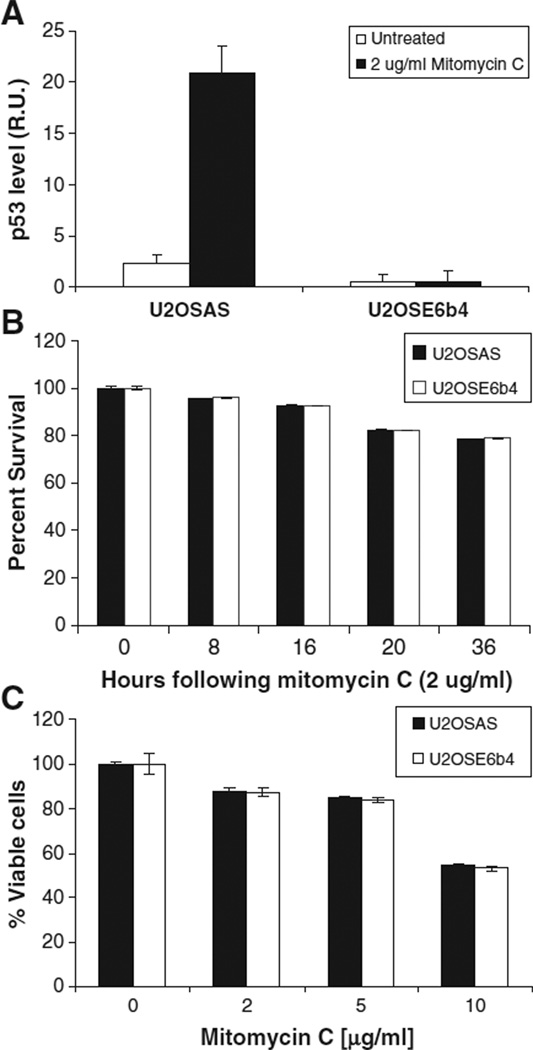
The E6 oncoprotein of Human Papillomavirus 16 (HPV16) mediates the accelerated degradation of p53. For this reason, cells stably transfected with the sense version of E6 display low or undetectable levels of p53 (U2OSE6b4), while those transfected with the anti-sense version express intact and functional p53 (U2OSAS) [49]. We monitored the expression of p53 in these two cell lines following treatment with low doses of the DNA damaging agent mitomycin C (Fig. 2A) and observed no p53 increases in U2OSE6b4 cells. We then re-examined the cell viability of both p53-expressing and p53-deficient U2OSAS and U2OSE6b4 cells by treating with 2 µg/ml mitomycin C, then assaying with MTT at the indicated times (Fig. 2B), or by treating cells with different drug concentrations for 24 h (Fig. 2C). The results obtained indicate that, consistent with our previous results [49], the cell viability is similar in both the presence and absence of p53.
Fig. 2.
Mitomycin C induces apoptosis in both p53-expressing and p53-deficient cells. A) U2OSAS (p53-expressing) and U2OSE6b4 (p53-deficient) cells were treated with 2 µg/ml mitomycin C, and a p53 ELISA was performed after 16 h. The normalized level of p53 is presented in Relative Units (RU). B) U2OSAS and U2OSE6b4 cells were treated with 2 µg/ml mitomycin C, and cell viability was estimated as a percent of untreated cells using the MTT assay at the indicated times. Data points represent the mean of triplicate measurements, error bars represent the standard deviation, and results are presented as percentages of untreated cells. C) U2OSAS and U2OSE6b4 cells were treated with mitomycin C for 24 h. Cell viability was estimated as a percent of untreated cells using the MTT assay at the indicated times.
3.2. Ceramide species composition is cell-type specific and does not change significantly following DNA damage
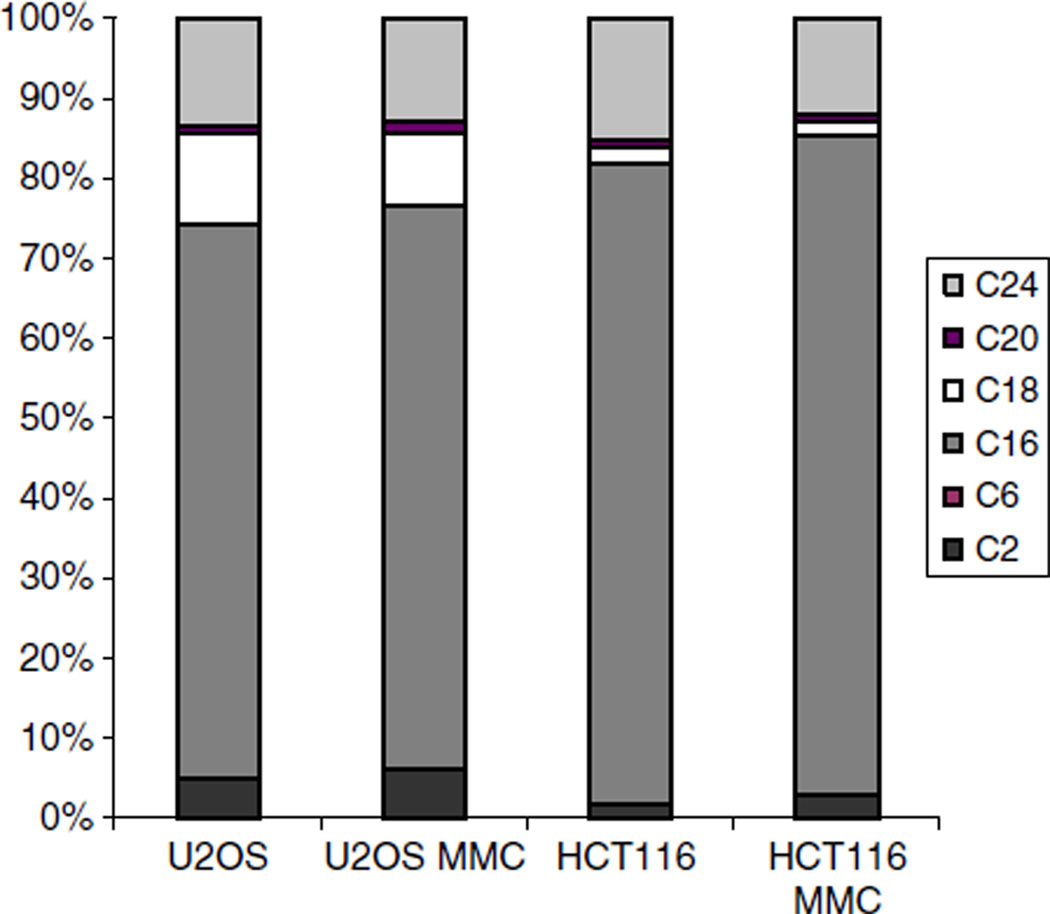
Ceramides found in mammalian cells are represented by even-numbered fatty acid chains (C2–C24) linked to a sphingosine backbone [50], and the levels and composition of these various ceramide species are believed to be tissue-specific and to vary during differentiation. We therefore assessed how the relative levels of the various ceramide species compared and varied before and after the DNA damage response in both the U2OS and the HCT116 cell lines (Fig. 3). Comparison of species abundance in U2OS and HCT116 cells showed that in both cell lines, C16 accounted for the majority of ceramide molecules (approximately 70% for U2OS and 80% for HCT116 cells). However, there are some differences between the two cell lines. While both cell lines had the same proportion of C24 (10–12%), they differed in their composition of other species in that U2OS cells displayed significantly higher levels of C2 and C18 ceramide molecules. We could not detect the presence of C6 in HCT116 cells, and found only a barely-detectable level in U2OS cells. Monitoring ceramide species composition during mitomycin C treatment showed that the relative proportions did not change significantly following treatment. Three major ceramide species, C16, C18, and C24, represent almost 95% of the total ceramide content in both HCT and U2OS cell lines. In the following experiments, unless indicated otherwise, we used the sum of these three species to represent the ceramide concentration.
Fig. 3.
The profile of ceramide molecules is cell-specific and does not change significantly after mitomycin C treatment. U2OS and HCT116 cells were treated with 2 µg/ml mitomycin C for 36 h. Ceramide levels of C2, C6, C16, C18, C20, and C24 were quantified for both untreated and treated (MMC) cells. 100% corresponds to the sum of all measured ceramide species.
3.3. Mitomycin C induces ceramide accumulation in p53-deficient cells
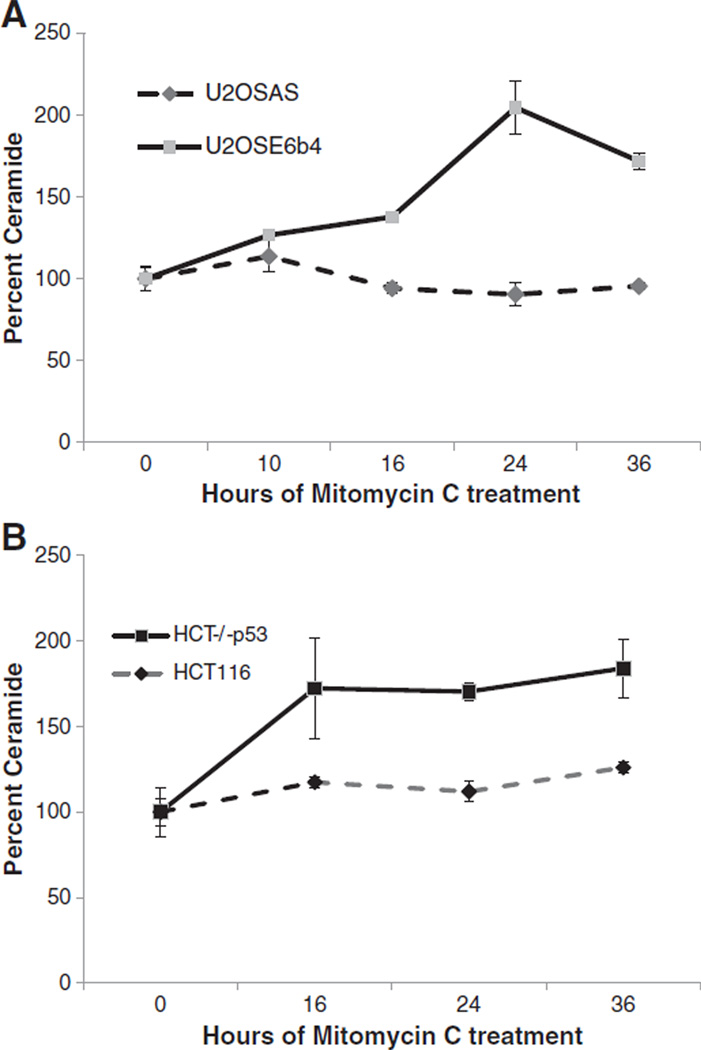
To examine how ceramide levels were affected by genotoxic stress in the presence and absence of the p53 response, we plated each of the four cell lines for 24 h to allow growth to 50% confluency, treated cells with mitomycin C for up to 36 h, then extracted lipids, quantified ceramide by HPLC-MS/MS and normalized against phospholipids. The results (Fig. 4A and B) revealed that ceramide levels increased sharply in both cells lines lacking p53 (U2OSE6b4 and HCT−/−p53). In contrast, the U2OSAS and HCT116 cells, which expressed p53, did not display this dramatic increase. These results indicate that DNA damage can indeed trigger an accumulation of ceramide, with the extent of this accumulation dependent upon the cellular p53 status.
Fig. 4.
Mitomycin C induces ceramide accumulation in p53-deficient cells. A) U2OSAS and U2OSE6b4 cells were treated with 2 µg/ml mitomycin C and harvested. Lipids were extracted and ceramide was quantified using HPLC-MS/MS as described in Materials and methods. B) HCT116 and HCT116 p53 null cells were treated with 2 µg/ml mitomycin C and harvested. Lipids were extracted and ceramide was quantified using HPLC-MS/MS as described in Materials and methods. Experiments were done in triplicate, and the error bars represent the standard deviation.
3.4. UDP-glucose ceramide glucosyltransferase is down- regulated following mitomycin C treatment
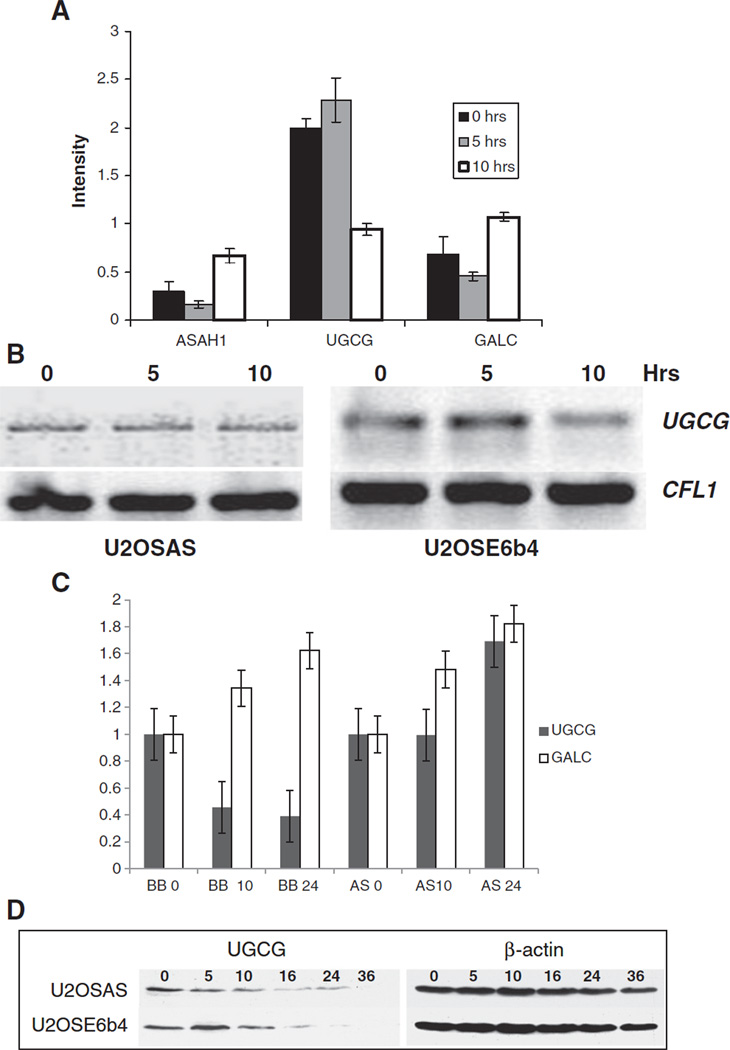
Ceramide biosynthesis consists of three major pathways and includes dozens of enzymes that may regulate its concentration in cell. De novo synthesis creates ceramide by condensation of palmitate and serine (Fig. 1). It also can be generated by the action of dihydroceramide desaturase, which converts dihydroceramide to ceramide; sphingomyelinase, which converts sphingomyelin to ceramide; and galactosylceramidase, which converts galactosylceramide to ceramide. In turn, ceramide can be metabolized by the action of ceramidase, which converts ceramide to sphingosine; sphingomyelin synthase, which converts ceramide to sphingomyelin; and UGCG, which converts ceramide to glucosylceramide (Fig. 1) [51]. To identify which ceramide-related genes are most affected in our system, we performed a microarray analysis to find genes with altered expression patterns following DNA damage (Fig. 5A). This analysis revealed three genes involved in the ceramide metabolic pathway that significantly changed their expression within 10 h of mitomycin C treatment. Acid ceramidase (ASAH1) and galactosylceramidase (GALC) were up-regulated, while UGCG was down-regulated by two-fold following mitomycin C-induced DNA damage. UGCG is well-known for its role in synthesizing glycosphingolipids, which are important in cellular processes such as cell-cell recognition, cell cycle regulation, differentiation, survival, and cell death [52], and has been proposed to serve as a ‘rheostat’ enzyme in these processes. Semi-quantitative PCR confirmed the down-regulation of UGCG (Fig. 5B). Microarray results were also confirmed by quantitative real-time RT-PCR, which showed a decrease of over 50% in UGCG transcripts in U2OSE6b4 cells treated with mitomycin C. This decrease was accompanied by an increase in GALC transcription, while no UGCG down-regulation was detected in U2OSAS cells (Fig. 5C). At the protein level, there was a slow decrease of UGCG in the p53-expressing U2OSAS cells, and a more rapid decrease in the p53-deficient U2OSE6b4 cells (Fig. 5D). As analyzed by densitometry, the UGCG protein level in U2OSE6b4 cells dropped to 5–15% after 24 h treatment with mitomycin C, while in U2OSAS cells, approximately 40% remained as compared to the untreated cells.
Fig. 5.
UGCG is down-regulated following DNA damage. A) U2OSE6b4 (p53-deficient) cells were treated with 2 µg/ml mitomycin C for 0, 5, or 10 h. Microarray analysis of three independent replicates was performed to identify changes in gene expression. Normalized data were analyzed using the Gene Sifter online analysis tool. The expression profiles of three genes: acid ceramidase (ASAH1), ceramide glucosyltransferase (UGCG), and galactosylceramidase (GALC) at 0, 5 and 10 h of treatment are shown. Experiments were performed in triplicate, and the error bars represent the standard deviation. B) U2OSAS and U2OSE6b4 cells were treated with 2 µg/ml mitomycin C for 0, 5, or 10 h. RNA was extracted using the TRizol reagent, and RT-PCR was performed using the iScript system and the forward and reverse primers for either UGCG or the house-keeping gene Cofilin 1 (CFL1). C) UGCG and GALC transcripts were quantified by real time-PCR of RNA isolated from U2OSAS (AS columns) and U2OSE6b4 (BB columns) cells treated with 2 µg/ml mitomycin C for 10 and 24 h. Black columns represent levels of the UGCG transcript, white columns the levels of the GALC transcript. D) U2OSAS and U2OSE6b4 cells were treated with 2 µg/ml mitomycin C for up to 36 h. Cells were lysed with lysis buffer and the lysates were analyzed by immunoblotting using either anti-UGCG or anti-β-actin antibodies, which served as loading controls.
3.5. Inhibition of UGCG is sufficient to cause ceramide accumulation and induce apoptosis associated with caspase activation
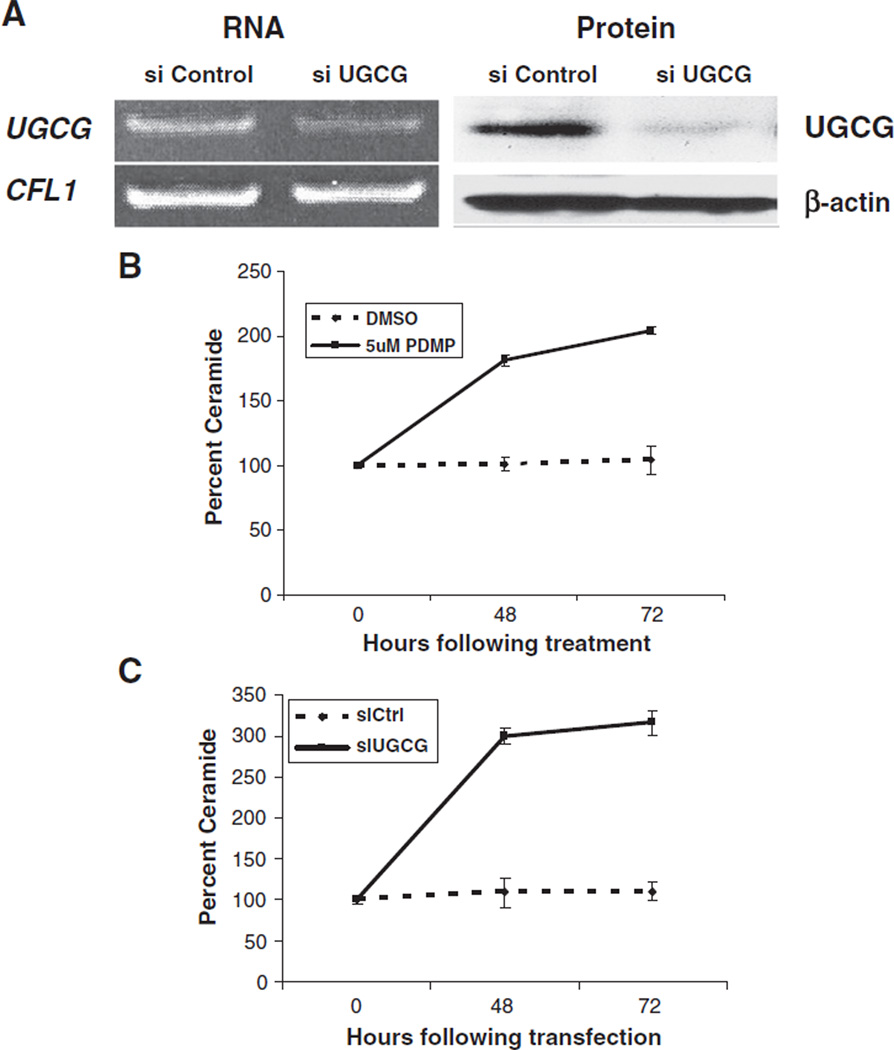
The studies described earlier suggest that UGCG down-regulation contributes to the accumulation of ceramide in cells lacking p53. This enzyme catalyzes the first committed glycosylation step of glycosphingolipid (GSL) biosynthesis — the formation of glucosylceramide from ceramide and UDP-glucose [52,53], which then goes on to serve as the base for over 300 glycosphingolipids [17,54]. Since this occurs in the forward direction, utilizing ceramide, it should follow that blocking the action of this enzyme would cause an accumulation of its substrate (ceramide). In order to test this prediction, we decreased the level of active UGCG in two ways. First, we used the pharmacological agent PDMP, thre-1-Phenyl-2-decanoyl-amino-3-morpholino-1-propanol hydrochloride, to decrease UGCG activity [55], and second, we utilized small interfering RNA (siRNA) to inhibit UGCG by lowering its expression. RT-PCR and immunoblotting were used to monitor the efficiency of siRNA-mediated UGCG inhibition at both the transcriptional and translational levels (Fig. 6A). Densitometric analysis of RT-PCR gels and immunoblots indicated that siRNA inhibition decreased the UGCG level by approximately 80%. Inhibition of UGCG with either 5 µM PDMP (Fig. 6B) or 30 nM siRNA directed against UGCG (Fig. 6C) for up to 72 h led to a significant increase in ceramide levels as compared to the controls, DMSO (used as a vehicle for PDMP) and control siRNA, respectively.
Fig. 6.
Ceramide levels increase following inhibition of UGCG. A) Control siRNA (siControl) and siRNA directed against UGCG (siUGCG) were transfected into the parental cell line, U2OS, for 30 h. Left panel — RNA was extracted using the TRizol reagent, and then RT-PCR was performed using the iScript kit system (Bio-Rad) and the forward and reverse primers for either UGCG or the house-keeping gene Cofilin 1 (CFL1). Right panel — Cells were lysed with mammalian buffer and the lysates were analyzed by immunoblotting with either anti-UGCG or anti-β-actin antibodies, which served as loading controls. B) and C) U2OS cells were either treated with DMSO or 5 µM PDMP (B) or transfected with control siRNA (siCtrl) or siRNA directed against UGCG (siUGCG) (C) for 0, 48, or 72 h. Cells were harvested at the indicated times, lipids were extracted as described in Materials and methods, and C18 ceramide was quantified using HPLC-MS/MS. Experiments represent the mean of the triplicate measurements, error bars represent the standard deviation, and results are presented as percentages.
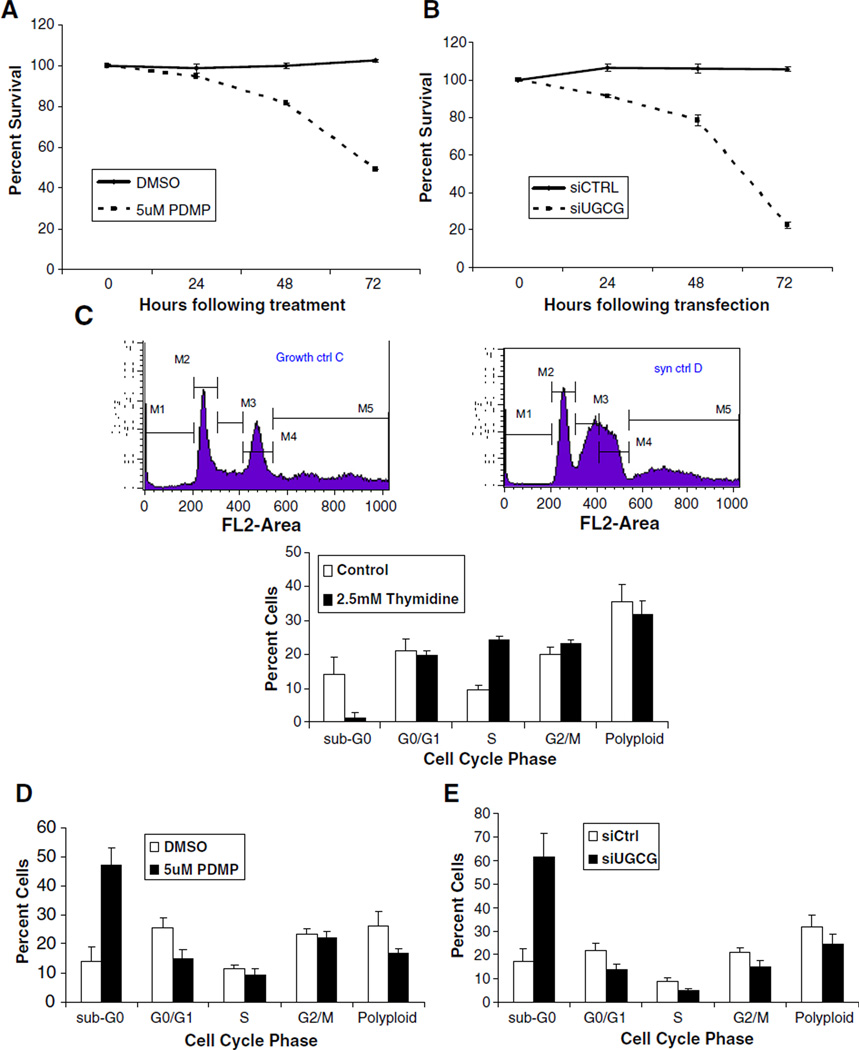
Ceramide is a pro-apoptotic mediator, and thus cells are expected to undergo apoptosis when inhibition of UGCG causes an accumulation of ceramide. We therefore determined cell viability following UGCG inhibition with 5 µM PDMP (Fig. 7A) or siRNA directed against UGCG (Fig. 7B). When UGCG was inhibited, viable cell numbers decreased due to either cell cycle arrest or cell death. For both treatments, the greatest differences in viability were noted at 48 and 72 h post-treatment. It may be that at the doses administered, the siRNA treatment is more effective in inhibiting UGCG activity.
Fig. 7.
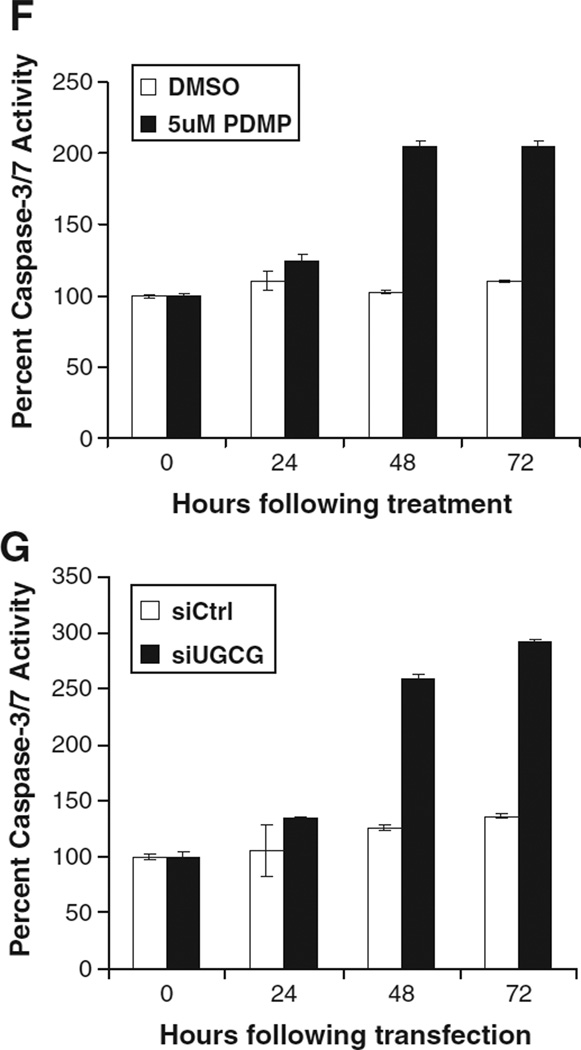
Inhibition of UGCG induces cell death and caspase 3/7 activation. U2OS cells were treated with either DMSO or 5 µM PDMP (A) or transfected with control siRNA (siCtrl) or siRNA directed against UGCG (siUGCG) (B) for 0, 24, 48, or 72 h. A cell viability assay was performed using MTT. Data represents the mean of the triplicates, error bars represent the standard deviation, and results are presented as percentages. C) U2OS cells were synchronized using 2.5 mM thymidine as described in Materials and methods. Cells were harvested, stained with propidium iodide (PI), and then flow cytometry was performed. Control (white bars) represent cells synchronized with thymidine, followed by removal of media containing thymidine, and the cells allowed to return to their normal cycling for another 48 h. Flow cytometry graphs represent a typical sample. Stages of the cell cycle are represented as: M1: Sub-G0, M2: G1, M3: S, M4: G2/M, and M5: Polyploid. Bar graphs represent the mean of triplicate experiments. D) and E) Synchronized U2OS cells were either: treated with DMSO or 5 µM PDMP (D) or transfected with control siRNA (siCtrl) or siRNA directed against UGCG (siUGCG) (E) for 48 h. Cells were then harvested, stained with propidium iodide (PI) and subjected to flow cytometry. F) and G) U2OS cells were either: treated with DMSO or 5 µM PDMP (F) or transfected with control siRNA (siCtrl) or siRNA directed against UGCG (siUGCG) (G) for 0, 24, 48, or 72 h. Caspase 3/7 activity was measured using the Caspase-Glo 3/7 assay as described in the Materials and methods. Data represents the mean of the triplicate measurements, error bars represent standard deviation, and results are presented as percentages. Caspase activity from the untreated cells (0) served as the reference.
A ceramide-induced increase in cell death is not the only explanation for these results, however, as it has been reported that the accumulation of ceramide induces a G0/G1 cell cycle arrest in human KB cells [34]. To determine whether inhibition of UGCG leads to G0/G1 cell cycle arrest in U2OS cells, we utilized the fluorescent DNA dye propidium iodide and flow cytometry to investigate possible changes in the cell cycle. Prior to inhibition with either PDMP or siRNA, we synchronized cells into the S phase using 2.5 mM thymidine [40]. The left upper panel of Fig. 7C shows a typical, un-synchronized U2OS cell population, while the right panel shows the synchronized cells enriched in the S phase. After synchronization and UGCG inhibition, we analyzed the distribution of cells over the cell cycle using flow cytometry. The results demonstrated that after inhibiting UGCG activity by treatment with either PDMP or siUGCG, the G0/G1 population did not exceed that of the control (Fig. 7D and E). However, there was a substantial increase in the sub-G0 population (approximately 3-fold) when cells were treated with 5 µM PDMP for 48 h as compared to the control vehicle treatment (Fig. 7D), and also when cells were transfected with siUGCG as compared to transfection with siCtrl (Fig. 7E). These results suggest that the inhibition of UGCT initiated apoptosis rather than G0/G1 cell cycle arrest.
To determine whether the inhibition of UGCG resulted in caspase activation, we examined caspase 3/7 activity following treatment with 5 µM PDMP (Fig. 7F) and siUGCG (Fig. 7G). Caspase 3/7 activity increased over time when UGCG was inhibited by either method, confirming that the cell death observed was indeed accompanied by activation of caspase-3/7. Overall, we can conclude that inhibition of UGCG leads to accumulation of ceramide, activation of caspase 3/7, and increases in the sub-G0 population of cells.
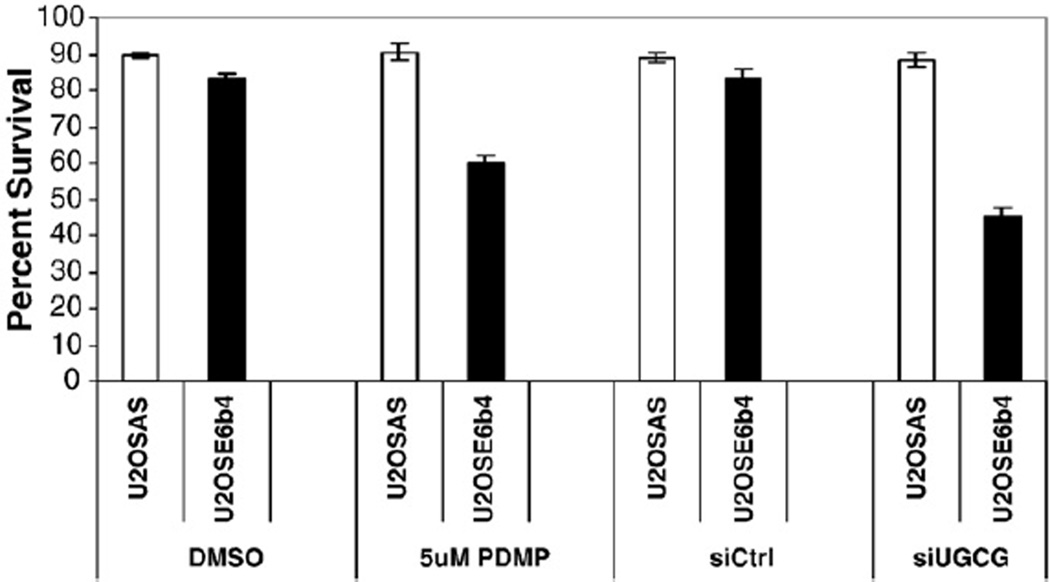
3.6. Inhibition of UGCG increases the sensitivity of p53-deficient cells to mitomycin C
Studies have shown that the cytotoxic effect of chemotherapy can be decreased when ceramide generation is impaired, but increased when the metabolism of ceramide is blocked (see review) [10]. This suggests that by targeting ceramide metabolism, it may be possible to reverse drug resistance or to increase the effect of drugs in chemotherapy. Furthermore, the conversion of ceramide to glucosylceramide by UGCG has been noted as one mechanism of drug resistance in some cancer cells [56,57]. We therefore predicted that inhibition of UGCG could lead to increased cytotoxicity of mitomycin C in cells where UGCG plays a role in the post-DNA damage response, such as in the p53-deficient cells used in this study. We therefore compared the sensitivity of U2OSAS and U2OSE6b4 cells to mitomycin C treatment with and without UGCG inhibition. As shown in Fig. 8, inhibition of UGCG had no effect on the susceptibility of p53-expressing cells to mitomycin C-induced apoptosis. On the other hand, inhibition of UGCG in p53-deficient cells led to a marked increase in their susceptibility to mitomycin C, confirming our hypothesis.
Fig. 8.
Down-regulation of UGCG can enhance p53-independent cell death induced by DNA damage. U2OSAS and U2OSE6b4 cells were either treated with DMSO or 5 µM PDMP (first two sets of bars) or transfected with control siRNA (siCtrl) or siRNA directed against UGCG (siUGCG) (last two sets of bars) for 30 h, then treated with 2 µg/ml mitomycin C for 24 h. A cell viability assay was performed using MTT. Experiments represent the mean of the triplicate measurements, error bars represent the standard deviation, and results are presented as percentages.
4. Discussion
Previously published reports have indicated that the presence or absence of wild-type p53 does not accurately predict responses to endocrine, cytotoxic, biological or radio-therapies for cancer [45,58–60]. Furthermore, advanced cancers often acquire resistance to chemotherapy without accompanying changes in their p53 status [61]. Such data suggests that factors other than p53 can regulate and mediate apoptosis induced by DNA damage. In this study, we investigated the p53-independent response to DNA damage induced by mitomycin C using two isogenic pairs of cell lines. U2OSAS and U2OSE6b4 cells differ only by the presence or absence of the HPV 16 E6 gene and hence in their p53 levels, while the HCT116 p53−/− cell line is a p53-null version of the HCT116 original cell line, which expresses wild-type p53 [38]. Our original observation that both p53-expressing and p53-deficient U2OS cells demonstrate similar viability when treated with 2 µg/ml mitomycin C sparked an interest in these alternate pathways, and the results reported here demonstrate that DNA damage-induced apoptosis in the absence of a normal p53 response is accompanied by an increase in ceramide levels (Fig. 4), which coincides with a down-regulation of UGCG (Fig. 5). This represents an important finding in the field of sphingolipid signaling and apoptosis, especially as it relates to p53-independent signaling.
The detection and quantification of ceramide has advanced rapidly in recent years [6], Application of high performance liquid chromatography coupled to mass spectrometry with multiple-reaction-monitoring mode allowed us to effectively identify and quantify six types of ceramide molecules from the same small starting sample [62]. A comparison of the proportions of ceramide species between cells derived from colon cancer and from osteosarcoma indicated that the composition is very cell specific. Furthermore, this composition did not change significantly during the response to DNA damage (Fig. 3). The later observation allowed us to monitor changes in total ceramide levels based on changes in the concentration of the major ceramide species found in both cell lines: C16, C18, and C24.
Though the mechanism of ceramide-mediated cell death is not completely understood, it is generally believed to involve the mitochondria. Consistent with this model, we found that caspases were activated following inhibition of UGCG (Fig. 7F and G). Other studies have also shown that the executioner caspase 3 is activated in ceramide-induced apoptosis [63,64].
Ceramide levels in cells are regulated by a complex system involving dozens of enzymes. Data from numerous laboratories, as well as our own microarray data (Fig. 5A), confirm a complex regulatory structure that involves both increased and decreased expression of genes involved in all three pathways of sphingolipid metabolism (Fig. 1). Analysis of gene expression in the p53-deficient U2OSE6b4 cells revealed that in this particular system, the ASAH1, UGCG, and GALC genes showed the highest differences in expression levels following DNA damage. Of these, UGCG was the most responsive to genotoxic treatment (Fig. 5A), prompting us to further investigate its role in ceramide regulation. Of course, changes in the expression levels of other enzymes may also contribute.
UGCG is known to be involved in several important cellular processes such as cell differentiation, cell cycle regulation, cell survival and cell death. For example, the absence of this gene in UGCG-null mutant mice causes embryonic lethality due to elevation of apoptosis [35], demonstrating its essential role in development. Another study demonstrated the importance of the balance between ceramide and its glucosyl derivatives in carcinogenesis by showing that increased tumor growth resulted when glucosylceramide was injected into mice bearing Ehrilich ascites tumor cells, while injection of a UGCG inhibitor resulted in a marked anti-tumor effect [65]. Several studies have demonstrated that ceramide glucosylation plays a significant role in the regulation of cell resistance to chemotherapeutic drugs, including multidrug resistance (MDR) [61,66]. For example, it has been shown that the abnormal resistance of certain cancer cells to chemotherapeutic agents is accompanied by an increased accumulation of glucosylceramides, and that transfection of UGCG into MCF-7 cells leads to adriamycin resistance [67]. However, inhibition of the same gene in two other cell lines, U937 and HL-60, with the UGCG-specific inhibitors PDMP and PPMP, led to a decrease in apoptosis caused by the DNA damaging agent daunorubicin [66], showing that the influence exerted by the glucosylation of ceramide on apoptosis is cell specific and may depend on other factors, such as the presence or absence of a functional p53 response.
Mitomycin C is used in combination chemotherapy regimens to treat a variety of tumors. However, its mechanism of action is a topic of debate. Our unexpected observation that the viability of both p53-expressing and p53-deficient U2OS cells was similar following mitomycin C treatment [49] suggested to us that an examination of the molecular events following DNA damage could both address the question of mechanism and suggest novel and effective approaches for dealing effectively with p53-negative cancer cells. In this study, we show that mitomycin C is able to down-regulate UGCG, increase ceramide levels, and induce apoptosis in cells lacking p53. We then found that this DNA-damaging drug, when administered along with agents that inhibit UGCG, becomes even more potent in killing p53-deficient tumor cells (Fig. 8). Though this response may be cell type and/or drug specific, this type of combination may provide a novel approach for effectively targeting p53-deficient cells during chemotherapy, especially in cases of acquired drug resistance. Variations on this approach could involve the combination of ceramide-elevating drugs with an inhibitor of UGCG [13,34,68], with or without the addition of DNA-damaging agents. The feasibility of using ceramide-elevating drugs to enhance cancer chemotherapy was recently demonstrated by the sensitization of multiple cancer cell lines to doxorubicin when treated with exogenous cell-permeable C6 ceramide [69]. Clearly, an enhanced understanding of ceramide apoptotic pathways, how they are activated, how they function and how they are controlled, can lead to enhancements in cancer treatment. Such pathways are expected to offer a number of therapeutic targets for novel drug development, and an understanding of which pathways are operative in a given tumor will enable a better and more individualized targeting of treatments.
Acknowledgements
This work was supported by the National Institutes of Health (N.I.H.) Grant R01 CA095461 (to P.D.H.) from the National Cancer Institute. We are grateful to Dr. B. Vogelstein (Johns Hopkins University) for providing us with the HCT 116 cell lines. We would also like to thank Dr. Nathan Wall for his aid in the flow cytometry experiments.
References
- 1.Iyer VN, Szybalski W. Mitomycins and porfiromycin: chemical mechanism of activation and cross-linking of DNA. Science. 1964;145:55–58. doi: 10.1126/science.145.3627.55. [DOI] [PubMed] [Google Scholar]
- 2.Grant S, Freemerman AJ, Birrer MJ, Martin HA, Turner AJ, Szabo E, Chelliah J, Jarvis WD. Effect of 1-beta-D-arabinofuranosylcytosine on apoptosis and differentiation in human monocytic leukemia cells (U937) expressing a c-Jun dominant-negative mutant protein (TAM67) Cell Growth Differ. 1996;7:603–613. [PubMed] [Google Scholar]
- 3.Strum JC, Small GW, Pauig SB, Daniel LW. 1-beta-d-arabinofuranosylcytosine stimulates ceramide and diglyceride formation in HL-60 cells. J. Biol. Chem. 1994;269:15493–15497. [PubMed] [Google Scholar]
- 4.Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Kolesnick RN. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell Death Differ. 1995;82:405–414. doi: 10.1016/0092-8674(95)90429-8. [DOI] [PubMed] [Google Scholar]
- 5.Jaffrezou J-P, Levade T, Bettaieb A, Andrieu N, Bezombes C, Maestre N. Daunorubicin-induced apoptosis: triggering ceramide generation through sphingomyelin hydrolysis. EMBO J. 1996;15:2417–2424. [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes TA, Duerksen-Hughes PJ, Filippova M, Filippov V, Zhang K. C18 ceramide analysis in mammalian cells employing reversed-phase high-performance liquid chromatography tandem mass spectrometry. Anal. Biochem. 2008;378:80–86. doi: 10.1016/j.ab.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry DK, Carton J, Shah AK, Meredith F, Uhlinger DJ, Hannun YA. Serine palmitoyltransferase regulates de novo ceramide generation during etoposide-induced apoptosis. J. Biol. Chem. 2000;275:9078–9084. doi: 10.1074/jbc.275.12.9078. [DOI] [PubMed] [Google Scholar]
- 8.Tepper AD, deVries E, van Blitterswijk WJ, Borst J. Ordering of ceramide formation, caspase actgivation, and mitochondrial changes during KCKDK95- and DNA damage-induced apoptosis. J. Clin. Invest. 1999;103:971–978. doi: 10.1172/JCI5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds CP, Maurer BJ, Kolesnick RN. Ceramide synthesis and metabolism as a target for cancer therapy. Cancer Lett. 2004;206:169–180. doi: 10.1016/j.canlet.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 10.Senchenkov A, Litvak DA, Cabot MC. Targeting ceramide metabolism — a strategy for overcoming drug resistance. J. Natl. Cancer Inst. 2001;93:347–357. doi: 10.1093/jnci/93.5.347. [DOI] [PubMed] [Google Scholar]
- 11.Lin CF, Chen CL, Lin YS. Ceramide in apoptotic signaling and anticancer therapy. Curr. Med. Chem. 2006;13:1609–1616. doi: 10.2174/092986706777441986. [DOI] [PubMed] [Google Scholar]
- 12.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 13.Sietsma H, Veldman RJ, Kok JW. The involvement of sphingolipids in multidrug resistance. J. Membr. Biol. 2001;181:153–162. doi: 10.1007/s00232-001-0033-1. [DOI] [PubMed] [Google Scholar]
- 14.Dawkins JL, Hulme DJ, Brahmbhatt SB, Auer-Grumbach M, Nicholson GA. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause heriditary sensory neuropathy type I. Nat. Genet. 2001;27:309–312. doi: 10.1038/85879. [DOI] [PubMed] [Google Scholar]
- 15.Bejaoui K, Wu C, Scheffler MD, Haan G, Ashby P, Wu L, deJong P, Brown RHJ. SPTLC1 is mutated in hereditary sensory neuropathy, type I. Nat. Genet. 2001;27:261–262. doi: 10.1038/85817. [DOI] [PubMed] [Google Scholar]
- 16.Diaz-Font A, Chabas A, Grinberg D, Vilageliu L. RNAi-mediated inhibition of the glucosylceramide synthase (GCS) gene: a preliminary study towards a therapeutic strategy for Gaucher disease and other glycosphingolipid storage diseases. Blood Cells Mol. Dis. 2006;37:197–203. doi: 10.1016/j.bcmd.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Hannun YA. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 18.Pettus BJ, Chalfant CE, Hannun YA. Ceramide in apoptosis: an overview and current perspectives. Biochim. Biophys. Acta. 2002;1585:114–125. doi: 10.1016/s1388-1981(02)00331-1. [DOI] [PubMed] [Google Scholar]
- 19.Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 20.Venable ME, Lee JY, Smyth MJ, Bielawska A, Obeid LM. Role of ceramide in cellular senescence. J. Biol. Chem. 1995;270:30701–30708. doi: 10.1074/jbc.270.51.30701. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Yu Y, Shuyu S, Duerksen-Hughes PJ. Ceramide and other sphingolipids in cellular responses. Cell Biochem. Biophys. 2004;40:323–350. doi: 10.1385/CBB:40:3:323. [DOI] [PubMed] [Google Scholar]
- 22.Dbaibo GS. Regulation of the stress response by ceramide. Biochem. Soc. Trans. 1997;25:557–561. doi: 10.1042/bst0250557. [DOI] [PubMed] [Google Scholar]
- 23.Merrill AJ, Jr, Schmeltz E-M, Dillehay DL, Spiegel S, Shayman JA, Schroeder JJ. Sphingolipids - the enigmatic lipid class: biochemistry, physiology, and pathophysiology. Toxicol. Appl. Pharmacol. 1997;142:208–225. doi: 10.1006/taap.1996.8029. [DOI] [PubMed] [Google Scholar]
- 24.Ohanian J, Ohanian V. Sphingolipids in mammalian cell signalling. Cell. Mol. Life Sci. 2001;58:2053–2068. doi: 10.1007/PL00000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannun YA, Obeid LM. Ceramide: an intracellular signal for apoptosis. Trends Biochem. Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 26.Brenner B, Koppenhoef C, Linderkamp O, Lang F, Gulbins E. Fas- or ceramide-induced apoptosis is mediated by a Rac1-regulated activation of Jn N-terminal kinase/p38 kinases and GADD153. J. Biol. Chem. 1997;272:22173–22181. doi: 10.1074/jbc.272.35.22173. [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Ma W, Ding M, Bowden GT, Dong Z. Direct evidence for an important role of sphingomyelinase in ultraviolet-induced activation of c-Jun N-terminal kinase. J. Biol. Chem. 1997;272:27753–27757. doi: 10.1074/jbc.272.44.27753. [DOI] [PubMed] [Google Scholar]
- 28.Verjeij M, Bose R, Lin XH, Yao B, Jarvis WD, Grant S. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 29.Merrill AHJ. De novo sphingolipid biosynthesis: a necessary, but dangerous, pathway. J. Biol. f. 2002;277:25843–25846. doi: 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- 30.Hannun YA, Obeid LM. The ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 31.Linn SC, Kim HS, Keane EM, Andras LM, Wang E, Merrill AHJ. Regulation of de novo sphingolipid biosynthesis and the toxic consequences of its disruption. Biochem. Soc. Trans. 2001;29:831–835. doi: 10.1042/0300-5127:0290831. [DOI] [PubMed] [Google Scholar]
- 32.Devod VN, Dedova IV, Merrill AHJ, Nicholson GA. Activity of a partially inhibited serine palmitoyltransferase is sufficient for normal sphingolipid metabolism and viability of HSN1 patient cells. Biochim. Biophys. Acta. 2004;1688:168–175. doi: 10.1016/j.bbadis.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Osuchowski MF, Johnson VJ, He Q, Sharma RP. Myriocin, a serine palmitoyltransferase inhibitor, alters regional brain neurotransmitter levels without concurrent inhibition of the brain sphingolipid biosynthesis in mice. Toxicol. Lett. 2004;147:87–94. doi: 10.1016/j.toxlet.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 34.Bieberich E. Integration of glycosphingolipid metabolism and cell-fate decisions in cancer and stem cells: review and hypothesis. Glycoconj. J. 2004;21:315–327. doi: 10.1023/B:GLYC.0000046274.35732.47. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita T, Wada R, Sasaki T, Deng C, Bierfreund U, Sandhoff K, Proia RL. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9142–9147. doi: 10.1073/pnas.96.16.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokoyama K, Suzuki M, Kawashima I, Karasawa K, Nojima S, Enomoto T, Tai T, Suzuki A, Setaka M. Changes in composition of newly synthesized sphingolipids of HeLa cells during the cell cycle — suppression of sphingomyelin and higher-glycosphingolipid synthesis and accumulation of ceramide and glucosylceramide in mitotic cells. Eur. J. Biochem. 1997;249:450–455. doi: 10.1111/j.1432-1033.1997.00450.x. [DOI] [PubMed] [Google Scholar]
- 37.Filippova M, Song H, Connolly JL, Dermody TS, Duerksen-Hughes PJ. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 2002;277:21730–21739. doi: 10.1074/jbc.M200113200. [DOI] [PubMed] [Google Scholar]
- 38.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 39.Filippova M, Duerksen-Hughes PJ. Inorganic and dimethylated arsenic species induce cellular p53. Chem. Res. Toxicol. 2003;16:423–431. doi: 10.1021/tx025606a. [DOI] [PubMed] [Google Scholar]
- 40.Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J. Biol. Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 41.Ormerod MG. Cell-cycle analysis of asynchronous populations. Methods Mol. Biol. 2004;263:345–354. doi: 10.1385/1-59259-773-4:345. [DOI] [PubMed] [Google Scholar]
- 42.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 43.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 44.Corazzari M, Lovat PE, Oliverio S, Di Sano F, Donnorso RP, Redfern CP, Piacentini M. Fenretinide: a p53-independent way to kill cancer cells. Biochem. Biophys. Res. Commun. 2005;331:810–815. doi: 10.1016/j.bbrc.2005.03.184. [DOI] [PubMed] [Google Scholar]
- 45.Velculescu VE, El-Deiry WS. Biological and clinical importance of the p53 tumor suppressor gene. Clin. Chem. 1996;42:858–868. [PubMed] [Google Scholar]
- 46.Fan S, Smith ML, Rivet DJ, II, Duba D, Zhan Q, Kohn KW, Fornace AJ, Jr, O'Connor PM. Disruption of p53 function sensitizes breast cancer MCF-7 cells to cisplatin and pentoxifylline. Cancer Res. 1995;55:1649–1654. [PubMed] [Google Scholar]
- 47.Hawkins DS, Demers GW, Galloway DA. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 1996;56:892–898. [PubMed] [Google Scholar]
- 48.Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ., Jr p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol. Cell. Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filippov V, Filippova M, Duerksen-Hughes PJ. The early response to DNA damage can lead to activation of alternative splicing activity resulting in CD44 splice pattern changes. Cancer Res. 2007;67:7621–7630. doi: 10.1158/0008-5472.CAN-07-0145. [DOI] [PubMed] [Google Scholar]
- 50.Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv. Exp. Med. Biol. 2010;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okazaki T, Kondo T, Kitano T, Tashima M. Diversity and complexity of ceramide signalling in apoptosis. Cell. Signal. 1998;10:685–692. doi: 10.1016/s0898-6568(98)00035-7. [DOI] [PubMed] [Google Scholar]
- 52.Bleicher RJ, Cabot MC. Glucosylceramide synthase and apoptosis. Biochim. Biophys. Acta. 2002;1585:172–178. doi: 10.1016/s1388-1981(02)00338-4. [DOI] [PubMed] [Google Scholar]
- 53.Komori H, Ichikawa S, Hirabayashi Y, Ito M. Regulation of UDP-glucose:ceramide glucosyltransferase-1 by ceramide. FEBS Lett. 2000;475:247–250. doi: 10.1016/s0014-5793(00)01696-3. [DOI] [PubMed] [Google Scholar]
- 54.Ichikawa S, Sakiyama H, Suzuki G, Hidari KI, Hirabayashi Y. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4638–4643. doi: 10.1073/pnas.93.10.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radin NS, Shayman JA, Inokuchi J. Metabolic effects of inhibiting glucosylceramide synthesis with PDMP and other substances. Adv. Lipid Res. 1993;26:183–213. [PubMed] [Google Scholar]
- 56.Lucci A, Cho WI, Han TY, Giuliano AE, Morton DL, Cabot MC. Glucosylceramide: a marker for multiple-drug resistant cancers. Anticancer Res. 1998;18:475–480. [PubMed] [Google Scholar]
- 57.Lavie Y, Cao H, Bursten SL, Giuliano AE, Cabot MC. Accumulation of glucosylceramides in multidrug-resistant cancer cells. J. Biol. Chem. 1996;271:19530–19536. doi: 10.1074/jbc.271.32.19530. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton A, Piccart M. The contribution of molecular markers to the prediction of response in the treatment of breast cancer: a review of the literature on HER-2, p53 and BCL-2. Ann. Oncol. 2000;11:647–663. doi: 10.1023/a:1008390429428. [DOI] [PubMed] [Google Scholar]
- 59.Ring AE, Ellis PA. Predictors of resonse to systemic therapy in breast cancer. Forum (Genova) 2002;12:19–32. [PubMed] [Google Scholar]
- 60.Rosen EM, Fan S, Rockwell S, Goldberg ID. The molecular and cellular basis of radiosensitivity: implications for understanding how normal tissues and tumors respond to therapeutic radiation. Cancer Invest. 1999;17:56–72. [PubMed] [Google Scholar]
- 61.Liu YY, Yu JY, Yin D, Patwardhan GA, Gupta V, Hirabayashi Y, Holleran WM, Giuliano AE, Jazwinski SM, Gouaze-Andersson V, Consoli DP, Cabot MC. A role for ceramide in driving cancer cell resistance to doxorubicin. FASEB J. 2008;22:2541–2551. doi: 10.1096/fj.07-092981. [DOI] [PubMed] [Google Scholar]
- 62.Zhang K, Haynes T-AS, Filippova M, Filippov V, Duerksen-Hughes PJ. Quantification of ceramide levels in mammalian cells by high performance liquid chromatography coupled to tandem mass spectrometry with multiple-reaction-monitoring mode (HPLC-MS/MS-MRM) Anal. Methods. 2011;3:1193–1197. [Google Scholar]
- 63.Mizushima N, Koike R, Kohsaka H, Kushi Y, Handa S, Yagita H, Miyasaka N. Ceramide induces apoptosis via CPP32 activation. FEBS Lett. 1996;395:267–271. doi: 10.1016/0014-5793(96)01050-2. [DOI] [PubMed] [Google Scholar]
- 64.Smyth MJ, Perry DK, Zhang J, Poirier GG, Hannun YA, Obeid LM. prICE: a downstream target for ceramide-induced apoptosis and for the inhibitory action of Bcl-2. Biochem. J. 1996;316(Pt 1):25–28. doi: 10.1042/bj3160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Irwin MS. Family feud in chemosensitvity: p73 and mutant p53. Cell Cycle. 2004;3:319–323. [PubMed] [Google Scholar]
- 66.Grazide S, Terrisse AD, Lerouge S, Laurent G, Jaffrezou JP. Cytoprotective effect of glucosylceramide synthase inhibition against daunorubicin-induced apoptosis in human leukemic cell lines. J. Biol. Chem. 2004;279:18256–18261. doi: 10.1074/jbc.M314105200. [DOI] [PubMed] [Google Scholar]
- 67.Liu YY, Han TY, Giuliano AE, Cabot MC. Expression of glucosylceramide synthase, converting ceramide to glucosylceramide, confers adriamycin resistance in human breast cancer cells. J. Biol. Chem. 1999;274:1140–1146. doi: 10.1074/jbc.274.2.1140. [DOI] [PubMed] [Google Scholar]
- 68.Inokuchi J, Mason I, Radin NS. Antitumor activity via inhibition of glycosphingolipid biosynthesis. Cancer Lett. 1987;38:23–30. doi: 10.1016/0304-3835(87)90196-0. [DOI] [PubMed] [Google Scholar]
- 69.Ji C, Yang B, Yang YL, He SH, Miao DS, He L, Bi ZG. Exogenous cell-permeable C6 ceramide sensitizes multiple cancer cell lines to Doxorubicin-induced apoptosis by promoting AMPK activation and mTORC1 inhibition. Oncogene. 2010;29:6557–6568. doi: 10.1038/onc.2010.379. [DOI] [PubMed] [Google Scholar]