Abstract
Objectives
Prior candidate gene studies have associated CYP2B6 516G→T [rs3745274] and 983T→C [rs28399499] with increased plasma efavirenz exposure. We sought to identify novel variants associated with efavirenz pharmacokinetics.
Materials and methods
Antiretroviral therapy-naive AIDS Clinical Trials Group studies A5202, A5095, and ACTG 384 included plasma sampling for efavirenz pharmacokinetics. Log-transformed trough efavirenz concentrations (Cmin) were previously estimated by population pharmacokinetic modeling. Stored DNA was genotyped with Illumina HumanHap 650Y or 1MDuo platforms, complemented by additional targeted genotyping of CYP2B6 and CYP2A6 with MassARRAY iPLEX Gold. Associations were identified by linear regression, which included principal component vectors to adjust for genetic ancestry.
Results
Among 856 individuals, CYP2B6 516G→T was associated with efavirenz estimated Cmin (P = 8.5 × 10−41). After adjusting for CYP2B6 516G→T, CYP2B6 983T→C was associated (P = 9.9 × 10−11). After adjusting for both CYP2B6 516G→T and 983T→C, a CYP2B6 variant (rs4803419) in intron 3 was associated (P = 4.4 × 10−15). After adjusting for all the three variants, non-CYP2B6 polymorphisms were associated at P-value less than 5× 10−8. In a separate cohort of 240 individuals, only the three CYP2B6 polymorphisms replicated. These three polymorphisms explained 34% of interindividual variability in efavirenz estimated Cmin. The extensive metabolizer phenotype was best defined by the absence of all three polymorphisms.
Conclusion
Three CYP2B6 polymorphisms were independently associated with efavirenz estimated Cmin at genome-wide significance, and explained one-third of interindividual variability. These data will inform continued efforts to translate pharmacogenomic knowledge into optimal efavirenz utilization.
Keywords: CYP2B6, efavirenz, HIV, pharmacogenomics, pharmacokinetics
Introduction
Efavirenz is one of the most extensively prescribed medications for HIV-1 infection worldwide. Although generally safe and effective, some efavirenz recipients experience virologic failure [1] and/or central nervous system symptoms [2]. Efavirenz is metabolized primarily by cytochrome P450 (CYP) 2B6 [3]. Analyses involving AIDS Clinical Trials Group (ACTG) protocol A5097s first associated the nonsynonymous variant CYP2B6 516G→T (rs3745274) with increased plasma efavirenz exposure [4]. Subsequent studies have consistently replicated this pharmacokinetic association [5–8]. The CYP2B6 516T allele is considerably more frequent in individuals of African ancestry than those of European ancestry [9], which largely explains the somewhat greater mean plasma efavirenz concentrations among populations of African descent [10,11]. A less frequent CYP2B6 polymorphism, 983T→C (rs28399499), also predicts increased plasma efavirenz exposure [12–14]. The CYP2B6 983C allele is also more frequent with African ancestry, although far less frequent overall than 516G→T, and is virtually absent from populations of European ancestry [9]. Functional studies have associated the CYP2B6 516G→T variant with reduced hepatic CYP2B6 mRNA levels in vivo, and with aberrant splicing in vitro [15]. The CYP2B6 983TC variant has also been associated with reduced CYP2B6 mRNA levels in vitro [12,16].
Data are scant regarding efavirenz pharmacokinetic associations beyond CYP2B6 516G→T and 983T→C. Additional CYP2B6 polymorphisms suggested to affect CYP2B6 activity have been extremely infrequent [12,17,18] or have not predicted plasma efavirenz exposure [18,19]. Polymorphisms in genes beyond CYP2B6 reported to affect interindividual variability in efavirenz pharmacokinetics include CYP2A6 [20,21], UGT2B7 [21], CYP3A4 [22], CYP3A5 [4], and CAR [23], although such associations have yet to be replicated.
All previous analyses to identify genetic associations with efavirenz pharmacokinetics have relied upon candidate gene strategies. To determine whether genetic variants beyond CYP2B6 516G→T and 983T→C are associated with interindividual variability in plasma efavirenz exposure, we apply a retrospective genome-wide analysis approach to data from multiple prospective clinical trials of the ACTG.
Materials and methods
Study participants
Treatment-naive individuals were randomized to efavirenz-containing regimens in ACTG studies 384 [24,25], A5095 [1] (including its neurologic substudy A5097s [2]), and A5202 [26,27], with DNA obtained under protocol A5128 [28]. The present analyses involved individuals from these clinical trials who also had available plasma efavirenz assay data. Some participants from 384 and A5097s in these analyses were also included in previous candidate gene studies describing associations between CYP2B6 516G→T, 983T→C, and plasma efavirenz exposure [4,6,14]. Self-identified race/ethnicity categories ‘white, non-Hispanic’, ‘black, non-Hispanic’, and ‘Hispanic’ are hereafter referred to as white, black, and Hispanic, respectively. This study complied with the Helsinki Declaration, was approved by institutional review boards for each site, and participants gave written informed consent.
Identifying genetic polymorphisms
This clinical trials population has undergone whole genome analysis for a project related to immune control of HIV-1 replication [29]. A total of 1739 individuals received genome-wide genotyping. Genotyping was conducted in three phases, where the first phase utilized the Illumina 650Y Beadchip and the latter two phases, the Illumina Human-1M Duo Beadchip (Illumina Inc., San Diego, California, USA). For statistical analyses we used only those polymorphisms that were common to both platforms and were successfully genotyped. The NIH grant that supported A5202 was submitted to NIH before 2008, and support for genome-wide genotyping was not from NIH, so this study is not subject to NIH policy that requires submission of genome-wide association study data to the database of Genotypes and Phenotypes (dbGaP). The ACTG has well-established procedures to facilitate data and specimen sharing with non-ACTG investigators.
Genome-wide data were complemented by additional targeted genotyping in a subset of 891 participants. A total of 73 polymorphisms in CYP2B6 (n=47), CYP2A6 (n=21), CYP3A4 (n=1), CYP3A5 (n=1), and ABCB1 (n=3) were genotyped by MassARRAY iPLEX Gold (Sequenom Inc., San Diego, California, USA). The Sequenom assay design was as follows: We tagged the entire CYP2B6 gene using SeattleSNPs [30], including 5 kB in each 5′ and 3′ untranslated regions (UTR), using a cosmopolitan strategy across populations (Yoruba, Asian, African-American, European-American, and Hispanic) with a 5% allelic frequency cutoff, a 0.80 threshold for r2, 85% data convergence for tagging polymorphisms, and 70% data convergence for clustering. Additional polymorphisms of interest (but that were not extremely infrequent) were added based on previous reports [12,13]. We also added polymorphisms with at least 5% allelic frequency in 20 kB of the 5′ UTR identified using Ensembl Genome Browser [31], as well as upstream polymorphisms possibly associated with CYP2B6 expression based on a previous report [28]. (Final Sequenom assay design available upon request.) Genotypes were confirmed by visual inspection of plots. The MassARRAY iPLEX Gold were merged with the genome-wide data for statistical analyses. In addition, a custom MassARRAY iPLEX Gold assay was designed for test whether selected polymorphisms from whole genome analyses would replicate in a separate group of individuals.
Laboratory personnel with no knowledge of clinical data performed genotyping. Quality control of genotype data was performed using PLINK [32] and EIGENSTRAT [33]. Samples which were potential sex misclassifications, population outliers according to principal components analysis projected onto HapMap Phase III data [29,34], individuals with estimated IBD greater than or equal to 0.25, and those with exceptionally high or low heterozygosity values (|F|>0.1) were excluded. Subsequently, genetic markers were excluded if the genotyping completion rate was lower than 98% or if minor allele frequency was below 1%.
Pharmacokinetic endpoint
Plasma efavirenz concentrations were assayed by high-performance liquid chromatography at treatment weeks 1, 4, 12, and 24. Sampling times were not prespecified, and the time of prior dose was by patient report. For association analyses, efavirenz Cmin values were estimated based on measured efavirenz concentrations at times in the dosing interval before Cmin, adjusted for dosing time using simulated efavirenz concentration–time curve percentiles, and generated by pharmacokinetic model simulations [35]. On the basis of dosing time, each observed concentration was assigned to a percentile. For each individual the summary statistic for analysis was median of observed time-adjusted percentiles over up to four time points for each individual. To minimize confounding by nongenetic factors and to assure steady state, we excluded individual efavirenz concentrations if beyond 24 h postdose or before at least 2 weeks of efavirenz, individuals with only one evaluable efavirenz value, and those with greater than 30-percentile difference between any two concentrations. For each participant, the median efavirenz percentile was used to estimate a 24-h postdose plasma efavirenz concentration (Cmin), which was then used in all statistical analyses.
Statistical analysis
Linear regression was used to test for association between each genetic variant and efavirenz estimated Cmin. Before regression analysis, estimated Cmin values were log-transformed because of non-normality. Next, the residual from regression with the top 10 principal component vectors was taken to adjust for potential confounding by ancestry. The number of statistically significant principal components was computed using twstats in the EIGENSOFT software package [33,36]. Linear regression was conducted first using this residual phenotype and then adjusting for rs3745274 (CYP2B6 516G→T), then for rs3745274 and rs28399499 (CYP2B6 983T→C), and then for rs3745274, rs28399499, and rs4803419 (CYP2B6 15582C→T). Linear regression was performed using PLINK, whereas residuals were taken using the lm command in the R stats package [37]. In the replication analysis, the linear regression process was repeated adjusting for ethnicity and the three single-nucleotide polymorphisms as categorical variables instead of computing residuals from the phenotype. Manhattan plots were generated using the R stats package [37].
Results
Genome-wide association analyses of efavirenz estimated Cmin
Initial analyses comprised 856 individuals (16% women, 50% self-reported white, 33% self-reported black, 18% self-reported Hispanic). Associations between genetic polymorphisms and efavirenz estimated Cmin values by linear regression, correcting for the top 10 principal components, are shown in Fig. 1a. There is a strong genome-wide significant (P<5×10 −8) peak of polymorphisms involving the CYP2B6 locus on chromosome 19. The lowest P-value represented CYP2B6 516G→T (rs3745274, P=8.5×10 − 41). To investigate associations beyond this peak, we repeated this linear regression while additionally adjusting for 516G→T. This analysis showed significant associations that included CYP2B6 983T→C (rs28399499, P=9.9×10 − 11) and CYP2B6 15582C→T (rs4803419, P=3.4×10 − 11). In addition, there were apparent associations with polymorphisms on chromosome 8 (rs7818576, P=7.2×10 − 9) and on chromosome 18 (rs288982, P=1.5×10 − 8; rs2959521, P=2.2×10 − 8), as shown in Fig. 1b. We continued to investigate associations by again repeating linear regression, this time adjusting for both CYP2B6 516G→Tand 983T→C. (This polymorphism took priority over rs4803419 because of the former’s strong a-priori evidence of association with efavirenz pharmacokinetics.) This analysis continued to show a significant association with CYP2B6 15582 C→T (P=4.4×10 − 15), along with variants on chromosomes 8 and 18 (Fig. 1c). In a final analysis adjusted for 516G→T, 983T→C, and rs4803419, there appeared to be a continued significant association with rs7818576 on chromosome 8 (P=1.1×10 − 9), as well as an association with rs2288657 on chromosome 2 (P=3.6×10 − 8). The polymorphisms on chromosome 18 no longer exceeded the threshold for genome-wide significance (Fig. 1d). Of note, after adjusting for 516G→T, 983T→C, and rs4803419 there was no association detected between efavirenz estimated Cmin values and CYP2B6 rs2279343 (P=0.49). The latter polymorphism defines CYP2B6*4, and was recently reported to be associated with increased plasma clearance of the CYP2B6 substrate nevirapine among HIV-infected Cambodians [38].
Fig. 1.
Genome-wide associations with efavirenz estimated Cmin values by multiple linear regression analysis. Manhattan plots of associations between genetic polymorphisms and efavirenz log-transformed estimated Cmin values. Linear regression was used. Each analysis includes the top 10 principal component vectors to adjust for potential confounding by ancestry. The – log10 of P-values are shown. Note the different y-axis scale on each panel. (a) Adjusting only for principal component vectors. (b) Also adjusting for CYP2B6 516G→T. (c) Also adjusting for CYP2B6 516G→T and 983TC. (d) Also adjusting for CYP2B6 516G→T, 983T→C, and rs4803419 C→T. Arrows indicate non-CYP2B6 polymorphisms that achieved (or nearly achieved) P=5×10−8 genome-wide significance in the final model.
Replication or genotype–phenotype associations
In the clinical trials studied herein there were 240 individuals with efavirenz pharmacokinetic data, but who were not included in genome-wide association analyses. We used this group to attempt replication of the above associations with CYP2B6 polymorphisms as well as with 18 other polymorphisms with P-value less than 10− 5 in the final genome-wide analysis above (Table 1). Of note, none of the 18 polymorphisms were close to any gene with clear plausibility for affecting efavirenz disposition. Each polymorphism was examined for association with efavirenz estimated Cmin individually, and with correction for CYP2B6 516G→T. Self-identified race/ethnicity (rather than principal components because genome-wide genotype data were not available) was used to adjust for systematic ancestry differences that might constitute population stratification. In the analysis unadjusted for CYP2B6 genotype, 516G→Tremained strongly associated with efavirenz estimated Cmin (P=6.1×10− 14). A chromosome 5 polymorphism (rs1897833) was also nominally associated (P=0.004), however, its effect was in the opposite direction as in the genome-wide analysis. After adjusting for 516G→T, CYP2B6 rs4803419 C→T was significantly associated with efavirenz estimated Cmin (P=2.3×10− 5), with a trend toward significance for CYP2B6 983T→C (P=0.070). In a final analysis that adjusted for both CYP2B6 516G→T and rs4803419 C→T, CYP2B6 983T→C was nominally associated with efavirenz estimated Cmin (P=0.017). Thus, all three CYP2B6 polymorphisms replicated, whereas no polymorphism beyond CYP2B6 replicated.
Table 1.
Associations between genetic polymorphisms with lowest P-values and efavirenz estimated Cmin values in genome-wide analysis cohort and in replication cohort
| Primary analysisa
|
Replication unadjusted
|
Replication 516G-T adjusted
|
|||||
|---|---|---|---|---|---|---|---|
| Chromosome | Polymorphism | β | P-value | β | P-value | β | P-value |
| 19 | CYP2B6 516G→T | 0.63 | 8.5 × 10− 41 | 0.68 | 6.1 × 10− 14 | NA | NA |
| 19 | CYP2B6 983T→C | 0.79 | 9.9 × 10− 11 | 0.52 | 0.444 | 1.09 | 0.070 |
| 19 | rs4803419 | 0.34 | 4.4 × 10− 15 | 0.03 | 0.769 | 0.41 | 2.3 × 10− 5 |
| 1 | rs2034525 | − 0.33 | 3.8 × 10− 6 | 0.10 | 0.509 | 0.05 | 0.695 |
| 2 | rs13386913 | − 0.68 | 2.7 × 10− 6 | 0.12 | 0.798 | 0.15 | 0.727 |
| 2 | rs2288657 | − 0.44 | 3.6 × 10− 8 | 0.24 | 0.224 | 0.14 | 0.423 |
| 3 | rs7625304 | − 0.42 | 9.0 × 10− 6 | − 0.40 | 0.129 | − 0.38 | 0.101 |
| 3 | rs17070662 | − 0.45 | 2.70 × 10− 6 | 0.61 | 0.055 | 0.43 | 0.130 |
| 3 | rs17295791 | − 0.28 | 6.0 × 10− 6 | 0.03 | 0.846 | 0.03 | 0.823 |
| 4 | rs16999951 | − 0.37 | 6.0 × 10− 6 | − 0.17 | 0.413 | − 0.30 | 0.118 |
| 5 | rs1897833 | − 0.23 | 1.5 × 10− 6 | 0.33 | 0.003 | 0.29 | 0.004 |
| 8 | rs11988660 | − 0.53 | 6.9 × 10− 6 | 0.69 | 0.149 | 0.54 | 0.207 |
| 8 | rs7818576 | − 0.47 | 1.1 × 10− 9 | − 0.01 | 0.954 | − 0.06 | 0.780 |
| 8 | rs10086443 | − 0.23 | 2.3 × 10− 6 | − 0.02 | 0.895 | 0.02 | 0.879 |
| 11 | rs10834309 | − 0.23 | 7.1 × 10− 6 | − 0.05 | 0.660 | − 0.05 | 0.636 |
| 11 | rs4556569 | − 0.23 | 6.0 × 10− 6 | 0.18 | 0.180 | 0.08 | 0.523 |
| 13 | rs2439613 | − 0.42 | 6.3 × 10− 6 | 0.50 | 0.103 | 0.15 | 0.588 |
| 18 | rs288982 | − 0.63 | 8.2 × 10− 8 | 0.16 | 0.633 | − 0.01 | 0.971 |
| 18 | rs4800112 | − 0.63 | 8.9 × 10− 8 | 0.51 | 0.080 | 0.26 | 0.314 |
| 18 | rs2959521 | − 0.87 | 1.3 × 10−7 | 0.45 | 0.412 | 0.30 | 0.543 |
| 20 | rs6050259 | 0.18 | 1.8 × 10− 6 | 0.02 | 0.799 | 0.02 | 0.853 |
NA, not available.
Results for the three CYP2B6 polymorphisms reflect the β coefficient and P-value from the analysis in which they were significant.
Linkage disequilibrium with other CYP2B6 variants
To better understand relationships between efavirenz estimated Cmin and the three CYP2B6 polymorphisms that were independently associated in genome-wide analyses, we considered their linkage disequilibrium (LD) with other polymorphisms. In the first genomewide analyses, without adjusting for any CYP2B6 polymorphism, two groups of polymorphisms were associated with efavirenz estimated Cmin (Fig. 2, blue markers), the first comprising 13 polymorphisms in strong LD with CYP2B6 516G→T and with minor alleles associated with higher efavirenz estimated Cmin values (i.e. positive β values), and the second comprising 13 polymorphisms in weaker LD with CYP2B6 516G→Tand with minor alleles associated with lower efavirenz estimated Cmin values (i.e. negative β values). In the second analyses, which adjusted for CYP2B6 516G→T, only four of the above 26 polymorphisms remained associated with efavirenz estimated Cmin values, whereas eight additional polymorphisms became significant (Fig. 2, red markers). All but CYP2B6 983T→C were all in strong LD with rs4803419 C→T. In the third analyses, which adjusted for CYP2B6 516G→Tand 983T→C, only rs4803419 C→Tand seven polymorphisms in strong LD with rs4803419 were associated with efavirenz estimated Cmin values (Fig. 2, orange markers).
Fig. 2.
Association between polymorphisms in the CYP2B6 locus and efavirenz estimated Cmin values, considering linkage disequilibrium (LD). Synthesis-View plots [40] of associations for the chromosome 19 region of interest are shown. At the top are chromosome positions, below which are panels showing – log10 P-values, β values, and minor allele frequencies (MAF). At the bottom shown LD between polymorphisms. Blue markers: Adjusting only for principal component vectors; Red markers: Also adjusting for CYP2B6 516G→T; Orange markers: Also adjusting for CYP2B6 516G→T and 983T→C. The blue, red, and orange markers correspond to Fig. 1a–c, respectively. Only polymorphisms with P≤5×10−8 are shown.
The rs4803419 polymorphism had not been previously associated with efavirenz pharmacokinetics. We therefore used 1000 Genomes data to further characterize LD with rs4803419 C→T in different populations. For polymorphisms within 200 kB of rs4803419 and that are in LD at r2 values of at least 0.5 among individuals of African, Asian, European, and Hispanic ancestry, r2 values are shown in Online Supplemental Materials (Supplementary digital content 1, http://links.lww.com/FPC/A523). Of note, the many polymorphisms in strong LD with rs4803419 included rs7251950 (r2=0.88 in Asians, r2=0.68 in Europeans, r2=1.00 in Hispanics). As reported elsewhere in this journal issue, rs7251950 was associated with decreased plasma nevirapine clearance among HIV-1-infected patients in Cambodia [38].
Associations with efavirenz estimated Cmin values
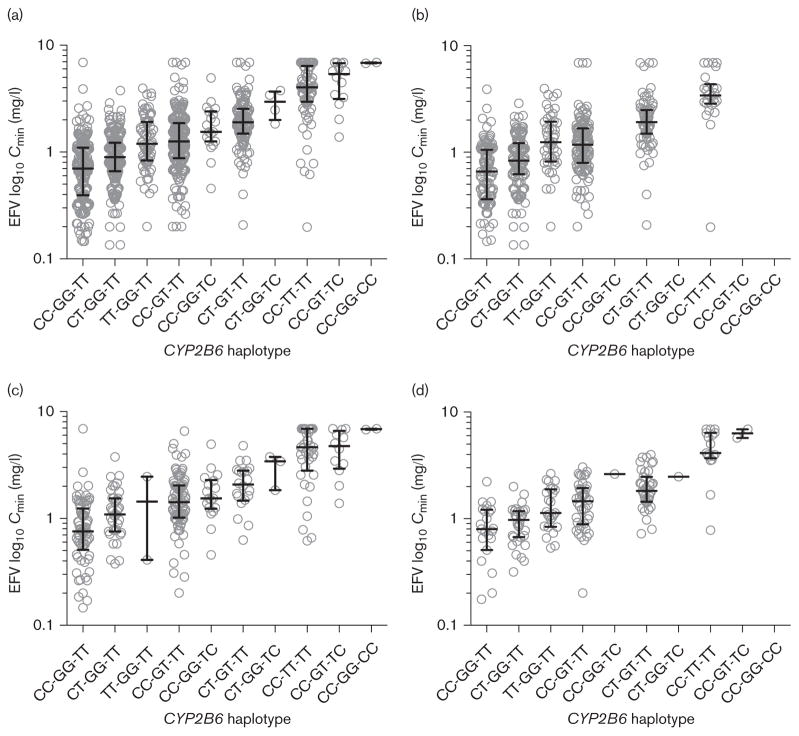
Relationships between each of the three significant CYP2B6 polymorphisms and efavirenz estimated Cmin values among all evaluable study participants are shown in Fig. 3. Homozygosity for any one of these polymorphisms appears to preclude the presence of the other two, suggesting that CYP2B6 516T, 983C, and rs4803419T may be on mutually exclusive haplotypes. Both CYP2B6 516G→T and 983T→C were associated with markedly increased median efavirenz estimated Cmin values. The median estimated Cmin among individuals homozygous for CYP2B6 516 TTwas 3.98 μg/ml, 5.4 times higher than for individuals lacking variant alleles at all three loci (0.74 μg/ml). This ratio was 7.1 among individuals who were concomitantly heterozygous for both 516Tand 983C (5.38 μg/ml). In contrast, compared with individuals lacking variant alleles at all three loci, homozygosity for rs4803419 TT was associated with only 1.7 times higher median estimated Cmin values (1.24 μg/ml), comparable with CYP2B6 516 GT heterozygosity (1.27 μg/ml). Among individuals heterozygous for CYP2B6 516T or 983C, the concomitant presence of a rs4803419 T allele conferred somewhat higher median estimated Cmin values. In the genome-wide association analyses described above, the somewhat smaller P-value for rs4803419 C→T than for 983T→C is explained by the former’s greater allelic frequency, despite its lesser magnitude of effect.
Fig. 3.
Relationships with CYP2B6 variants and efavirenz estimated Cmin values. Efavirenz Cmin values were estimated as described in the Materials and Methods section. (a) All participants (self-identified white, black, and Hispanic); (b) Self-identified white participants; (c) Self-identified black participants; (d) Self-identified Hispanic participants. On x-axis, CYP2B6 haplotypes represent (in order) CYP2B6 rs4803419 C→T (CC, CT, TT), 516G→T (GG, GT, TT), and 983T→C (TT, TC, CC). The need to convert measured efavirenz concentrations to percentiles to derive estimated Cmin values creates a spurious plateau of concentrations at approximately 7 mg/l. Actual Cmin values for some individuals undoubtedly exceed this value.
In the genome-wide analysis cohort, a linear regression model that included the three significant CYP2B6 polymorphisms and the top 10 principal components vectors explained 34% of the variance in log-transformed Cmin. A model that considered the CYP2B6 polymorphisms without principal component vectors explained 33% of the variance, whereas in a model that considered the top 10 principal component vectors without CYP2B6 polymorphisms explained only 3% of the variance. In the replication cohort, the three significant CYP2B6 polymorphisms and self-identified race/ethnicity indicators (rather than principal component vectors) explained 31% of the variance in log-transformed Cmin.
Previously reported associations beyond CYP2B6
Previous reports have suggested associations between polymorphisms in CYP2A6 [20,21], UGT2B7 [21], CYP3A4 [22], CYP3A5 [4], and CAR [23] and increased plasma efavirenz exposure. Genotype results for six previously implicated polymorphisms were available in the present genome-wide dataset. After adjusting for CYP2B6 516G→T, 983T→C, and rs4803419, no association was detected between efavirenz estimated Cmin values and 28399433 (CYP2A6*9, CYP2A6*13, and CYP2A6*15, P=0.82), rs1801272 (CYP2A6*2, P=0.07), rs28399454 (CYP2A6*17, P=0.39), rs4646437 (CYP3A4*1B, P=0.91), rs776746 (CYP3A5*3, P=0.59), or rs2307424 (CAR, P=0.99).
Discussion
Here we present results of the first genome-wide association analyses of efavirenz pharmacokinetics. We readily show at genome-wide significance that both CYP2B6 516G→T and 983T→C are independently associated with efavirenz estimated Cmin values. After controlling for these two polymorphisms we identify an independent association with a third CYP2B6 variant (rs4803419), which replicated in a separate group of study participants. A multivariable model that included all three CYP2B6 polymorphisms and the top 10 principal components vectors explained approximately one-third of variance in efavirenz log-transformed estimated Cmin value. It is noteworthy that two of these three genomewide significant polymorphisms were already known to predict efavirenz pharmacokinetics based on prior candidate gene studies [5–8,12–14].
Several lines of evidence support the validity of association between rs4803419 and efavirenz Cmin. First, this association achieved genome-wide significance in our discovery cohort, and replicated in a separate group of clinical trials participants. Second, this polymorphism has recently been associated with increased plasma clearance of the CYP2B6 substrate nevirapine among patients in Cambodia, as reported elsewhere in this journal issue [38]. Third, in a previous ex-vivo analysis of human liver tissue samples, this polymorphism was associated with decreased CYP2B6 activity among women [39]. As this polymorphism is adjacent to an intron 3 splicing branch site, it has been speculated that this may cause aberrant splicing with resultant decreased expression of the functional enzyme [39]. However, Hofmann et al. [15] found no association between rs4803419 and splice variants in human liver. We cannot exclude the possibility of an indirect effect through LD with other polymorphisms. In this regard, Lang et al. [40] reported that rs4803419 resides on several haplotypes (CYP2B6*13B, *15A, and *15B) that include nonsynonymous polymorphisms, several of which (K139E, Q172H, I391N, and K262R) were associated with decreased CYP2B6 protein expression and/or decreased enzymatic activity in COS-1 cells and in ex-vivo liver samples. Our query of 1000 Genomes data showed rs4803419 to be in strong LD with many polymorphisms, although none in exons. The effect size for this polymorphism was modest in comparison with those for CYP2B6 516G→Tand 983T→C, and rs4803419 appears to reside on a haplotype(s) separate from the former two polymorphisms. The implication is that rs4803419 does not improve our ability to predict very high plasma efavirenz concentrations (e.g. >4.0 μg/ml) beyond 516G→T and 983T→C, because rs4803419 T cannot be present among individuals who already have two variant alleles at CYP2B6 516G→Tand/or 983T→C. The rs4803419 polymorphism does, however, improve considerably the overall description of efavirenz estimated Cmin values, especially at lower concentrations. Individuals homozygous for rs4803419 CC who also lacked 516T and 983C had the lowest median efavirenz estimated Cmin values (Fig. 3).
In multivariable analyses that controlled the three significant CYP2B6 polymorphisms, no additional CYP2B6 polymorphisms reached genome-wide significance. Although several polymorphisms beyond chromosome 19 achieved genome-wide significance in the genome-wide dataset, none of these replicated in the targeted analyses of a separate group of study participants. These polymorphisms were therefore almost certainly spurious. There may very well be other polymorphisms in the present cohort that are independently associated with substantially increased plasma efavirenz Cmin values. These must, however, be very infrequent, because among 383 individuals lacking variant alleles at both CYP2B6 516G→T and 983T→C only one (0.26%) had an estimated Cmin greater than 4.0 μg/ml (Fig. 3). Alternatively, other yet-to-be-identified variants (not assayed herein) may have modest effects alone, however, with much greater impact when combined with CYP2B6 516G→T or 983T→C.
There were limitations to this study. We largely studied individuals of European descent, African descent, and Hispanics. Different associations may be identified in other populations. We limited our statistical analyses to polymorphisms that were shared across the two genomewide assay platforms (plus additional targeted genotyping of CYP2B6 and selected other genes). More in-depth genotyping might identify additional polymorphisms associated with efavirenz pharmacokinetics, particularly if such polymorphisms are infrequent or were poorly tagged by our assays. Efavirenz was not directly quantified at the time of Cmin, but rather Cmin values were estimated by applying pharmacokinetic model simulations to assays obtained earlier in the dosing interval, and the time since previous dose was by self-report. Therefore, actual efavirenz Cmin values may differ from these estimated values. Several previous associations between plasma efavirenz exposure and polymorphisms in CYP2A6 [20,21], CYP3A4 [22], CYP3A5 [4], and CAR [23] did not replicate herein. However, genotype data were unavailable for the previously implicated CYP2A6*1H, *1J [20], and UGT2B7 [21] variants.
Conclusion
To date it has been difficult to replicate putative associations between human genetic variants and efavirenz efficacy and/or toxicity. Previous studies based on candidate gene approaches have consistently associated CYP2B6 516G→T [4,6–8] and CYP2B6 983T→C [12–14] with higher plasma efavirenz concentrations. Additional CYP2B6 polymorphisms suggested to affect CYP2B6 activity have been extremely infrequent [12,17,18] or have not predicted plasma efavirenz exposure [18,19], and data for associations beyond CYP2B6 are limited [4,20,21,23]. The present study provides a more comprehensive understanding of genetic determinants of efavirenz plasma exposure. These results should support continued work to define the potential utility of human genetic testing to inform the prescribing of efavirenz.
Supplementary Material
Acknowledgments
The authors are grateful to the many persons with HIV infection who volunteered for ACTG 384, A5095, A5097, A5202, and A5128. In addition, they acknowledge the contributions of study teams and site staff for protocols ACTG 384, A5095, A5097s, A5202, and A5128. They are also grateful to Danielle Richardson, Cara Sutcliffe, and the Vanderbilt DNA Resources Core for MassARRAY iPLEX Gold genotyping.
This work was supported in part by the AIDS Clinical Trials Group funded by the National Institute of Allergy and Infectious Diseases (AI-068636, AI-038858, AI-068634, and AI-038855). Grant support included AI-069439, TR-000011, AI-077505, and AI-54999 (D.W.H.), BRS-ACURE-06-00140-T001 (G.D.M.), AI-051966, AI-069419, and UL1 RR-024996 (R.M.G.), AI-069472 and AI-062435 (G.K.R.). Study drugs were provided by Bristol–Myers Squibb Company (Princeton, New Jersey, USA), Gilead Sciences (Gilead Sciences Inc., Foster City, California, USA), and GlaxoSmithKline Inc. (Research Triangle Park, North Carolina, USA).
Clinical Research Sites that participated in ACTG protocols ACTG 384, A5095, or A5202, and collected DNA under protocol A5128, were supported by the following grants from NIH National Institute of Allergy and Infectious Diseases (NIAID): AI-069532, AI-069484, AI-069432, AI-069450, AI-069495, AI-069434, AI-069424, AI-069439, AI-069467, AI-069423, AI-069513, AI-069477, AI-069465, AI-069419, AI-069502, AI-069474, AI-069472, AI-069501, AI-069418, AI-069494, AI-069471, AI-069511, AI-069452, AI-069428, AI-069556, AI-069415, AI-032782, AI-046376, AI-046370, AI-038858, AI-034853, AI-027661, AI-025859, AI-069470, AI-027675, AI-073961, AI-050410, AI-045008, AI-050409, AI-072626, AI-069447, AI-027658, AI-027666, AI-058740, and AI-025868, and by the following grants from NIH National Center for Research Resources (NCRR): RR-000046, RR-000425, RR-025747, RR-025777, RR-025780, RR-024996, RR-024160, RR-023561, RR-024156, RR-024160, and RR-024160.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Website (www.pharmacogeneticsandgenomics.com).
Conflicts of interest
Eric S. Daar: Research grants from Bristol–Myers Squibb, Abbott, Merck, Pfizer, ViiV, and Gilead and a consultant/advisor to Bristol–Myers Squibb, Merck, ViiV, and Gilead. Roy M. Gulick: Research Grants from Gilead, Janssen, Pfizer, and ViiV, ad-hoc consultant to Gilead, Glaxo-SmithKline, Janssen, and Koronis. Gregory K. Robbins: Research support from Gilead Sciences and Schering–Plough. Received royalties from Wolters Kluwer. David B. Clifford: Serves on Data Safety Boards for Biogen, Millennium, Genzyme, Genentech, and Pfizer. He has been a consultant to Genentech, Genzyme, Bristol–Myers Squibb, Millennium, Biogen Idec, Janssen, and Pfizer. He has received research support from Biogen Idec, NeurogesX, Tibotec, and Pfizer. David W. Haas: Research grants from Bristol–Myers Squibb, Boehringer Ingelheim, Merck, and Gilead Sciences. For the remaining authors there are no conflicts of interest.
References
- 1.Gulick RM, Ribaudo HJ, Shikuma CM, Lalama C, Schackman BR, Meyer WA, et al. Three- vs four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA. 2006;296:769–781. doi: 10.1001/jama.296.7.769. [DOI] [PubMed] [Google Scholar]
- 2.Clifford DB, Evans S, Yang Y, Acosta EP, Goodkin K, Tashima K, et al. Impact of efavirenz on neuropsychological performance and symptoms in HIV-infected individuals. Ann Intern Med. 2005;143:714–721. doi: 10.7326/0003-4819-143-10-200511150-00008. [DOI] [PubMed] [Google Scholar]
- 3.Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther. 2003;306:287–300. doi: 10.1124/jpet.103.049601. [DOI] [PubMed] [Google Scholar]
- 4.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–2400. [PubMed] [Google Scholar]
- 5.Tsuchiya K, Gatanaga H, Tachikawa N, Teruya K, Kikuchi Y, Yoshino M, et al. Homozygous CYP2B6*6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319:1322–1326. doi: 10.1016/j.bbrc.2004.05.116. [DOI] [PubMed] [Google Scholar]
- 6.Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbe L, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult AIDS Clinical Trials Group study. J Infect Dis. 2005;192:1931–1942. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 7.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Novoa S, Barreiro P, Rendon A, Jimenez-Nacher I, Gonzalez-Lahoz J, Soriano V. Influence of 516G > T polymorphisms at the gene encoding the CYP450-2B6 isoenzyme on efavirenz plasma concentrations in HIV-infected subjects. Clin Infect Dis. 2005;40:1358–1361. doi: 10.1086/429327. [DOI] [PubMed] [Google Scholar]
- 9.dbSNP. [Accessed 5 January 2012];Short genetic variations. Available at: http://www.ncbi.nlm.nih.gov/projects/SNP/
- 10.Barrett JS, Joshi AS, Chai M, Ludden TM, Fiske WD, Pieniaszek HJ., Jr Population pharmacokinetic meta-analysis with efavirenz. Int J Clin Pharmacol Ther. 2002;40:507–519. doi: 10.5414/cpp40507. [DOI] [PubMed] [Google Scholar]
- 11.Pfister M, Labbe L, Hammer SM, Mellors J, Bennett KK, Rosenkranz S, Sheiner LB, et al. Population pharmacokinetics and pharmacodynamics of efavirenz, nelfinavir, and indinavir: Adult AIDS Clinical Trial Group Study 398. Antimicrob Agents Chemother. 2003;47:130–137. doi: 10.1128/AAC.47.1.130-137.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Sonnerborg A, Rane A, Josephson F, Lundgren S, Stahle L, Ingelman-Sundberg M, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genomics. 2006;16:191–198. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 13.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, Brockmeyer NH, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61:914–918. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribaudo HJ, Liu H, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202:717–722. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann MH, Blievernicht JK, Klein K, Saussele T, Schaeffeler E, Schwab M, Zanger UM. Aberrant splicing caused by single nucleotide polymorphism c. 516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325:284–292. doi: 10.1124/jpet.107.133306. [DOI] [PubMed] [Google Scholar]
- 16.Klein K, Lang T, Saussele T, Barbosa-Sicard E, Schunck WH, Eichelbaum M, et al. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genomics. 2005;15:861–873. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 17.Rotger M, Colombo S, Cavassini M, Furrer H, Elzi L, Ganthard H, et al. Genetic variability of CYP2B6 in individuals with extremely high efavirenz plasma concentrations. Presented at 13th Conference on Retroviruses and Opportunistic Infections; February 2006; Denver, CO. 2006. p. abstract 572. [Google Scholar]
- 18.Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Decosterd L, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81:557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 19.Rotger M, Saumoy M, Zhang K, Flepp M, Sahli R, Decosterd L, Telenti A, et al. Partial deletion of CYP2B6 owing to unequal crossover with CYP2B7. Pharmacogenet Genomics. 2007;17:885–890. doi: 10.1097/FPC.0b013e3282ef5cd1. [DOI] [PubMed] [Google Scholar]
- 20.Di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics. 2009;19:300–309. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 21.Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS. 2009;23:2101–2106. doi: 10.1097/QAD.0b013e3283319908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arab-Alameddine M, Di Iulio J, Buclin T, Rotger M, Lubomirov R, Cavassini M, et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Ther. 2009;85:485–494. doi: 10.1038/clpt.2008.271. [DOI] [PubMed] [Google Scholar]
- 23.Wyen C, Hendra H, Siccardi M, Platten M, Jaeger H, Harrer T, et al. Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J Antimicrob Chemother. 2011;66:2092–2098. doi: 10.1093/jac/dkr272. [DOI] [PubMed] [Google Scholar]
- 24.Robbins GK, De GV, Shafer RW, Smeaton LM, Snyder SW, Pettinelli C, et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349:2293–2303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafer RW, Smeaton LM, Robbins GK, De Gruttola V, Snyder SW, D’Aquila RT, et al. Comparison of four-drug regimens and pairs of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349:2304–2315. doi: 10.1056/NEJMoa030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sax PE, Tierney C, Collier AC, Fischl MA, Mollan K, Peeples L, et al. Abacavir-lamivudine versus tenofovir-emtricitabine for initial HIV-1 therapy. N Engl J Med. 2009;361:2230–2240. doi: 10.1056/NEJMoa0906768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daar ES, Tierney C, Fischl MA, Sax PE, Mollan K, Budhathoki C, et al. Atazanavir plus ritonavir or efavirenz as part of a 3-drug regimen for initial treatment of HIV-1. Ann Intern Med. 2011;154:445–456. doi: 10.1059/0003-4819-154-7-201104050-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas DW, Wilkinson GR, Kuritzkes DR, Richman DD, Nicotera J, Mahon LF, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 29.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.SeattleSNPs. [Accessed 5 January 2012];Variation discovery resource. Available at: http://pga.gs.washington.edu/
- 31. [Accessed 5 January 2012];Ensembl Genome Browser. Available at: http://useast.ensembl.org/index.html.
- 32.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Weale ME. Quality control for genome-wide association studies. Methods Mol Biol. 2010;628:341–372. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 35.Acosta EP, King JR. Methods for integration of pharmacokinetic and phenotypic information in the treatment of infection with human immunodeficiency virus. Clin Infect Dis. 2003;36:373–377. doi: 10.1086/345993. [DOI] [PubMed] [Google Scholar]
- 36.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RDC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 38.Bertrand J, Chou M, Richardson DM. Multiple genetic variants predict steady-state nevirapine clearance in HIV-infected Cambodians. Pharmacogenet Genomics. 2012 doi: 10.1097/FPC.0b013e32835a5af2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamba V, Lamba J, Yasuda K, Strom S, Davila JC, Hancock M, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR expression. J Pharmacol Exp Ther. 2003;307:906–922. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 40.Lang T, Klein K, Richter T, Zibat A, Kerb R, Eichelbaum M, et al. Multiple novel nonsynonymous CYP2B6 gene polymorphisms in Caucasians: demonstration of phenotypic null alleles. J Pharmacol Exp Ther. 2004;311:34–43. doi: 10.1124/jpet.104.068973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.