Abstract
Centromeres direct faithful chromosome inheritance at cell division but are not defined by a conserved DNA sequence. Instead, a specialized form of chromatin containing the histone H3 variant, CENP-A, epigenetically specifies centromere location. We discuss current models where CENP-A serves as the marker for the centromere during the entire cell cycle in addition to generating the foundational chromatin for the kinetochore in mitosis. Recent elegant experiments indicate that engineered arrays of CENP-A-containing nucleosomes are sufficient to serve as the site of kinetochore formation and for seeding centromeric chromatin that self-propagates through cell generations. Finally, recent structural and dynamic studies of CENP-A-containing histone complexes—before and after assembly into nucleosomes—provide models to explain underlying molecular mechanisms at the centromere.
Introduction
All eukaryotes share a common mechanism to achieve equal partitioning of genetic information at cell division. Paired sister chromosomes are held together by protein tethers called cohesins, and attach to a microtubule-based spindle via a locus called the centromere [1,2]. Centromeres, except in some budding yeasts (e.g. S. cerevisiae and K. lactis), are not defined by a specific DNA sequence. However, particular repeat sequences (e.g. α-satellites in H. sapiens) can be common at the centromeres in a given species but are dispensable for centromere function and identity [3]. In addition, while the centromere is the locus that confers stable inheritance of all the conserved genes on the chromosome, centromere DNA sequences are not well conserved between species [4]. In fact, centromere DNA is one of the most rapidly evolving parts of the chromosome [5]. One aspect of centromeres that is now clear is that their location on the chromosome is specified epigenetically.
The histone H3 variant, CENP-A (called Cse4 in yeast, CID in flies, and CenH3 in plants) is the key to the epigenetic specification of centromeres [6]. In cases of chromosome fusion creating a pseudo-dicentric (or centromere relocation on an intact chromosome; i.e. creating a neo-dicentric), CENP-A is found at the active centromere but not at the location that undergoes inactivation [7–9]. In this review, we discuss recent experiments that provide compelling evidence that the presence of CENP-A is sufficient to form a functional centromere de novo. This is a major advance for a key question in cell biology and genetics: understanding how the chromosome directs its own inheritance. These findings also add important insight into the epigenetic basis of centromere specification, since the new studies support the notion that, once seeded, CENP-A-containing chromatin directs its own propagation in perpetuity in a manner that is independent of DNA sequence. In addition, we will focus on other recent studies that have yielded major insight into the physical basis for how CENP-A distinguishes centromeric chromatin from the rest of the chromosome, as well as efforts that have uncovered a conserved cell cycle-coupled pathway for delivery of newly expressed CENP-A to the centromere.
Two Roles for CENP-A: Persistent Centromere Marker and Chromatin Foundation of the Mitotic Kinetochore
During the entire course of the cell cycle, the presence of CENP-A maintains the location of the centromere [2,6] (Figure 1, top). CENP-A is also required in mitosis for nucleating the kinetochore (Figure 1, bottom), the proteinaceous mega-complex that mediates spindle microtubule attachment and generates a diffusible checkpoint signal to arrest cells if attachments are not made [10]. Centromeres define the location of final chromosome cohesion at metaphase and recruitment of the Aurora B-containing chromosome passenger complex that selectively destabilizes erroneous microtubule attachments [8,9]. One key question that remains unresolved is whether or not identical physical features of CENP-A (modifications and/or organization within the nucleosome) are used for epigenetic marking throughout the cell cycle and for its mitotic functions. Indeed, there are several proposed forms of CENP-A containing nucleosomes, from conservative models where two CENP-A molecules replace both copies of H3 within a nucleosome of conventional octameric stoichiometry [11–14], to models where it alters the stoichiometry of histone subunits [15–17], switches the handedness of DNA wrapping around the CENP-A-containing particle [18,19], or replaces H2A/H2B dimers with a non-histone protein (e.g. Scm3 in budding yeast; [20]). These diverse proposals have been discussed at length along with the idea that they could be reconciled by a model of cell cycle-coupled centromere maturation [21].
Figure 1. Epigenetic marking and kinetochore nucleating functions of CENP-A-containing nucleosomes.
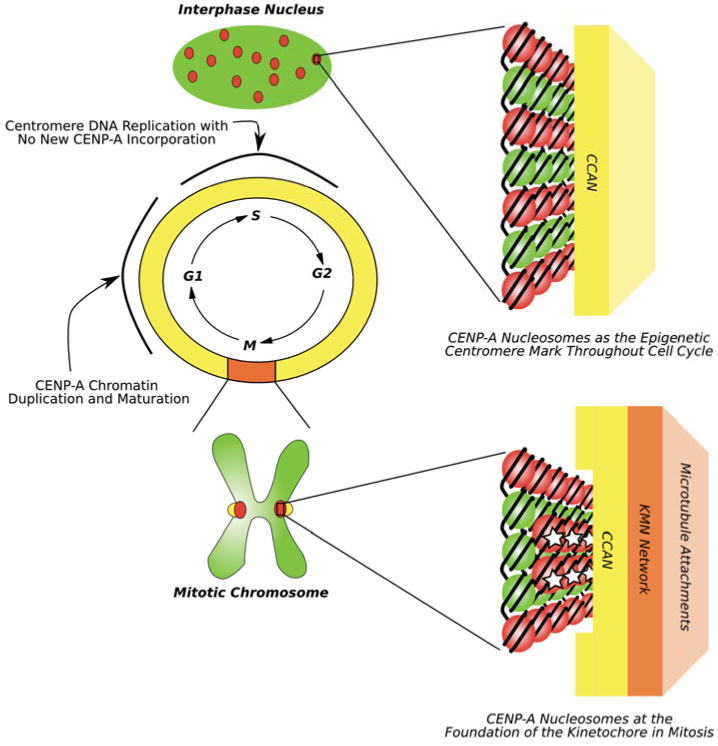
Diagram showing that CENP-A (red dots in the green interphase nucleus and on the mitotic chromosome) marks centromere location throughout the cell cycle. Two key points during interphase that are currently under investigation are S-phase when centromere DNA is replicated but no new CENP-A is deposited and early G1 when new CENP-A protein incorporates at centromeres and results in doubling the number of CENP-A nucleosomes. Throughout the cell cycle, CENP-A nucleosomes are associated with the constitutive centromere-associated protein network (CCAN) (represented by a yellow rectangle). In mitosis, CENP-A nucleosomes are (re)organized on the face of the chromosome. Connections are made between the CCAN and the so-called KMN network (orange rectangle) that is directly involved in attaching to spindle microtubules [82]. As detailed in the text, one untested possibility is that specialized and somehow modified subset of CENP-A nucleosomes (denoted with white stars) serve as the key connection point, linking the chromosome to a spindle microtubule through the CCAN and KMN network.
An idea has been put forward that the kinetochore-forming function of centromeric chromatin in most eukaryotic species is represented by a minor, specialized population of CENP-A nucleosomes [20,22] (Figure 1, bottom, copies with a white star). Candidate organisms where a ‘special’ pool of CENP-A nucleosomes may be particularly useful include those like nematodes in which CENP-A nucleosomes are arranged in a subset of chromatin that somehow discontinuously spans the entire length of the chromosome (i.e. ‘holocentric’ chromosomes), but where only a tiny fraction of CENP-A nucleosomes are sites of kinetochore formation [23]. Even in more ‘regional’ centromeres (i.e. in mammals, birds, many plants, etc.), there are typically thought to be more CENP-A-containing nucleosomes than microtubule attachment sites [24,25]. Indeed, in human cells, reduction of 90-95% of CENP-A protein is required to eliminate the targeting of at least some kinetochore and inner centromere components [25,26]. Perhaps this requirement for extreme depletion of CENP-A indicates that, as in the worm, the CENP-A-containing nucleosomes are in stoichiometric excess to the kinetochore formation proteins, and further that they are the final pool to be removed upon knockdown of CENP-A expression. An equally attractive possibility is that CENP-A nucleosomes are all equivalent to each other but in excess of nucleation sites for microtubule attachments. In this scenario, the precise CENP-A-containing nucleosome at the chromatin foundation of each microtubule attachment site would then be determined stochastically. The “point” centromere of budding yeast is clearly a unique type of centromere where there is likely to be a simplification of CENP-A-containing chromatin. In these organisms, CENP-A does not mark centromere location but rather centromere specification is determined by a particular DNA sequence [27]. Clearly budding yeast CENP-ACse4 maintains its role at the foundation of the kinetochore [28,29], and by removing a role in epigenetic centromere specification it is possible that the physical properties imparted by Cse4 have evolved solely for the purpose of nucleating kinetochore components.
Generating New Centromeres From Scratch
Key data supporting the idea that the presence of an array of CENP-A-containing nucleosomes is sufficient to seed new centromeres, and thus acts as the epigenetic component that establishes centromere location, come from experiments where CENP-A is directed to chromosome arms (Figure 2a,b,d). One approach to direct CENP-A to chromosome arms is by its massive over-expression in mammalian tissue culture cells [11,30]. Not surprisingly, spreading CENP-A nucleosomes throughout the genome does not turn the entire chromosome into a centromere [30,31]. After an initial strong pulse of CENP-ACID expression in fly cells, CENP-ACID is removed from most genomic locations, but where it is retained, centromere activity (e.g. recruitment of centromere/kinetochore proteins and interactions with the mitotic spindle) is occasionally detected [32,33] (Figure 2a). The genomic locations where CENP-ACID is retained are not random, showing a distinct preference for junctions between heterochromatic and euchromatic regions of the chromosome [33]. This junction is similar to the telomere-proximal locations of new centromere (neocentromere) formation in fission yeast that follow the engineered deletion of endogenous centromeres [34]. It should be noted, however, that at the naturally-occurring human neocentromeres that have been examined to date there is little or no detectable classical heterochromatin at adjacent chromatin location [35].
Figure 2. Summary of various experimental approaches used to seed a new epignetic centromere (a-d), to generate an ectopic kinetochore (e-f) and a synthetic kinetochore (g).
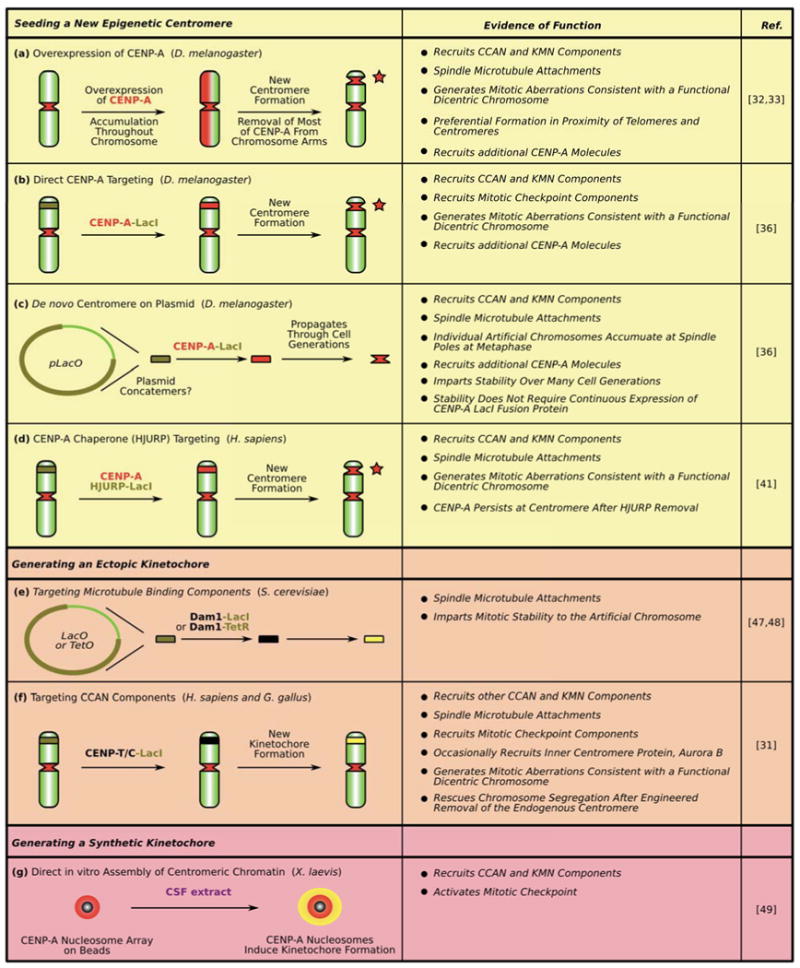
See text for details. Dark green segments with black line boundaries on chromosome arms denote the location of integrated LacO arrays. Red stars denote location of the new centromere in panels a, b and d. Yellow denotes kinetochore activity in panels e-g.
Directed targeting of CENP-ACID to a specific genomic location, by fusing CENP-ACID to the Lac repressor (LacI) to bring it to an array of Lac operator (LacO) sites is sufficient to drive new centromere formation [36] (Figure 2b). This approach also extends to CENP-ACid-LacI recruited to LacO-containing plasmids that gain centromere activity [36] (Figure 2c). Importantly, following an initial pulse of CENP-ACID-LacI targeting to the chromosomal array, endogenous CENP-ACID is recruited locally, perpetuating centromeric chromatin that spreads to locations adjacent to the LacO sites [36]. This observation shows that once seeded with CENP-A, the artificial centromere successfully (self)propagates through many cell cycles. Another attractive mode of targeting CENP-A to a new location in the genome is to exploit its dedicated chaperone, HJURP (called Scm3 in yeast) [20,37–40]. Indeed, initial targeting to a LacO array by an HJURP-LacI fusion protein is sufficient to recruit endogenous CENP-A [41] (Figure 2d). After HJURP-LacI is removed from the locus by addition of the LacI allosteric effector molecule, IPTG, CENP-A is retained, presumably integrated in the chromatin. Taken together these experiments provide strong evidence that CENP-A chromatin is sufficient to form a new centromere.
In budding yeast, de novo centromere experiments have been performed for more than three decades, a feature that is routinely exploited by investigators of budding yeast because the small (∼125 bp) CEN sequence confers mitotic stability to plasmids [27,42]. While biochemical evidence strongly argues for CENP-ACse4-containing chromatin that is restricted to the genetically defined ∼125 bp budding yeast centromere [29,43,44], recent fluorescence-based approaches indicate additional copies of CENP-ACse4 [45,46]. Regardless of the size of centromeric chromatin in this species, CENP-ACse4 is required but not sufficient for (neo)centromere formation and function in budding yeast. Nonetheless, de novo kinetochore activity that is sufficient to direct chromosome segregation can be generated by tethering the Dam1 outer kinetochore complex to an ectopic chromosome locus [47,48] (Figure 2e). Unlike the de novo centromere experiments involving CENP-A or HJURP (Figure 2a-d), these approaches require the persistent, engineered tethering of a Dam1 component to the locus, that, in turn, recruits other components of the spindle microtubule binding machinery (Figure 2e). Likewise, two members of the so-called constitutive centromere associated network (CCAN), CENP-C and CENP-T, are sufficient in mammalian and avian cells to confer robust kinetochore function to a LacO array site by recruiting other constitutive centromere components that nucleate the assembly of kinetochore components in mitosis [31] (Figure 2f).
Another approach for nucleating new kinetochores is biochemical. The nucleosome arrays with sites available for 19 nucleosomes containing human CENP-A are sufficient, when attached to beads, to nucleate the formation of functional kinetochores in frog egg extracts [49] (Figure 2g). It is important to emphasize that these functional arrays are entirely comprised of recombinant components assembled through traditional nucleosome reconstitution approaches, supporting the notion that the octameric nucleosome form underlies the kinetochore formation function of CENP-A.
Recent Progress in Defining the Physical Features of Octameric CENP-A-containing Nucleosomes
While proposals for diverse alternative forms of CENP-A nucleosomes are now being considered, as mentioned above, there has been nonetheless recent major progress in the detailed understanding of the octameric form (i.e. [CENP-A/H4/H2A/H2B]2) where CENP-A replaces the two chains of canonical H3 (Figure 3). Human (CENP-A/H4)2 heterotetramers that spontaneously form upon co-expression in bacteria [50] self-assemble with H2A/H2B dimers and DNA into octameric nucleosomes that wrap DNA in a left-handed manner [13] (the strategy used to form the functional arrays [Figure 2g; [49]]) and similar findings have been reported with CENP-A proteins from other species [12,51,52]. It is also notable that CENP-ACID/CENP-ACID is readily crosslinked through opposing cysteine residues [53], and in both fly and human versions of CENP-A, mutations that disrupt the CENP-A/CENP-A interface eliminate centromere targeting [53,54], supporting models where CENP-A replaces both histone H3 chains in an octameric nucleosome.
Figure 3. Unique physical properties of the mammalian CENP-A-containing nucleosome.
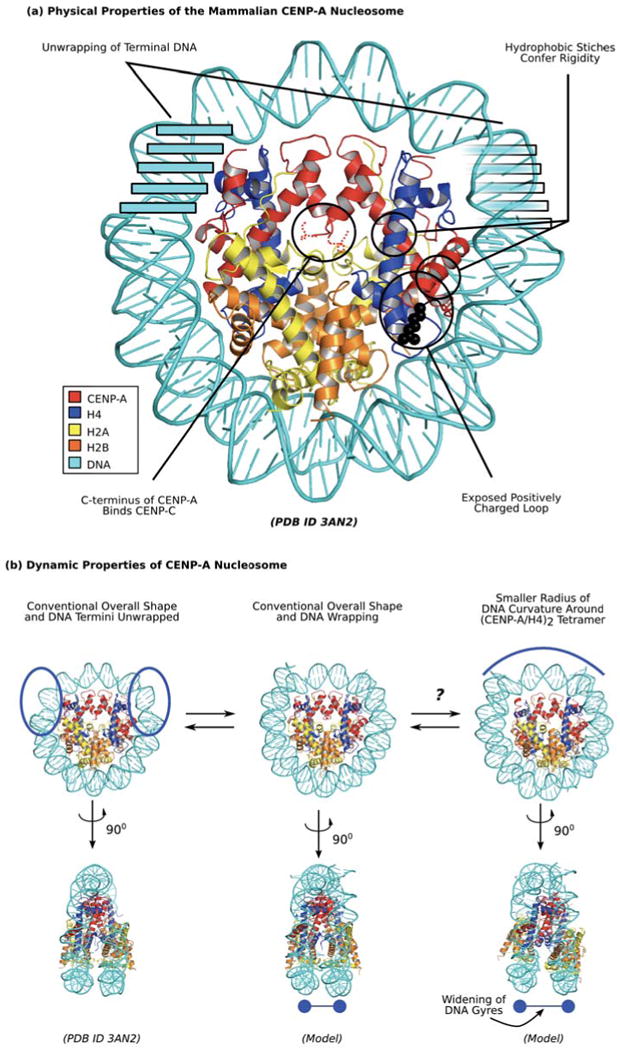
(a) Distinguished physical properties of CENP-A nucleosome are highlighted in black circles. Clockwise from the top left, these include transient unwrapping/flexibility of the final helical turn of DNA at each nucleosome terminus [14,57,58], hydrophobic stitches that rigidify the CENP-A/H4 interface [13,50,54,56], a bulged loop L1 that is of opposite charge as on H3 in the conventional nucleosome [13,14], and the unstructured C-terminus that mediates recognition of CENP-A nucleosomes by CENP-C [83]. (b) Dynamic properties of CENP-A nucleosome indicate potential sampling between distinct conformations. Mammalian CENP-A containing nucleosomes wrap ∼145 bp of DNA at steady-state (center; [13,58]), but sample unwrapping at the terminal DNA to a greater extent than conventional nucleosomes with canonical H3 [57,58]. The crystal form of the nucleosome includes only 120 bp of DNA that is visible in the structure (left; [14]), supporting the notion of increased flexibility at nucleosome termini. Another potential alternate form under investigation is one where rotation at the CENP-A/CENP-A is maintained as in the isolated sub-nucleosomal (CENP-A/H4)2 heterotetramer [13], leading to a smaller radius of curvature of DNA wrapping near the nucleosomal dyad (right, top) and wider spacing of H2A/H2B heterodimers and the two strands of nucleosomal DNA (right, bottom).
The first high-resolution structure of CENP-A from any species came in the form of the human (CENP-A/H4)2 heterotetramer [13]. One clear structural feature coming from this analysis and unique to CENP-A, is a bulged loop L1 where local side-chains generate a positive surface charge [13]. This is in contrast to the negative surface charge that exists at this location on H3-containing nucleosomes [55]. Another observed feature, unique to (CENP-A/H4)2 heterotetramers, is a rotated CENP-A/CENP-A interface that makes them ∼10 Å more compact at their largest dimension than (H3/H4)2 heterotetramers. This compaction is clearly measured in solution relative to the flexible and ‘open’ (H3/H4)2 heterotetramers by small-angle x-ray scattering (SAXS) experiments [13]. The crystal structure also identified hydrophobic stitches, patches of highly hydrophobic residues on α2 helix and loop L1 unique to CENP-A that directly contact hydrophobic residues on H4 [13], providing an explanation for how CENP-A confers conformational rigidity to centromeric nucleosomes [56].
The structure of a nucleosome containing CENP-A was recently reported [14] and revealed four important features (Figure 3a):
It confirmed the earlier reports [13,51,52] of left-handed DNA wrapping.
The αN helix of CENP-A is shorter [14] than the corresponding helix in H3 in the conventional nucleosome [55]. In both types of nucleosomes this helix is juxtaposed to the final turn of DNA at each nucleosome terminus (also called the nucleosome ‘entry/exit’ DNA). As opposed to the conventional nucleosome, where the entire ∼146 bp of nucleosomal DNA is visible in the crystal structure [55], the terminal 13 bp of DNA are not visible in the CENP-A-containing nucleosome structure [14]. While nucleosomes containing human CENP-A were previously reported to wrap ∼146 bp of DNA [13], this apparent ‘disorder’, resulting in more open conformations (Figure 3b, left), is likely due to increased flexibility in the interaction of CENP-A with the terminal DNA fragment, consistent with previous results from the topological analysis of CENP-A mononucleosomes assembled on DNA mini-circles [57] as well as the analysis of CENP-A polypeptide backbone dynamics in folded nucleosome arrays [58]. The flexibility is probably consequence of a substitution of a single Arg→Lys at the position corresponding to Arg49 in conventional H3 [57,58]. The side-chain of H3R49 intercalates the terminal DNA in conventional nucleosomes [55] in a manner that is not possible with a Lys side-chain. Interestingly, this same position in budding yeast CENP-ACse4 is a Tyr, which would be predicted to highly disrupt local DNA binding, and, indeed, octameric nucleosomes assembled with CENP-ACse4 wrap only ∼120 bp of DNA with no apparent ability to fully wrap the conventional 145 bp [51,52].
It confirmed that positively-charged loop L1, [13] is bulged and exposed on the surface of the nucleosome [14].
In the nucleosome crystal structure, the CENP-A/CENP-A four-helix bundle rotates to a conventional rotation so that the sub-nucleosomal (CENP-A/H4)2 heterotetramer and the entire nucleosome has very similar overall shape and dimensions to canonical nucleosomes [14].
This final finding is in contrast to (our) earlier proposal that the CENP-A/CENP-A interface would maintain its rigid, ‘closed’ conformation after incorporation into nucleosomes and concomitantly force the sub-nucleosomal H2A/H2B heterodimers to rotate away at the H4/H2B four-helix bundle, generating a differently shaped nucleosome with altered radius of DNA resulting in more exposed histone H2A and wider separated DNA gyres [13] (Figure 3b, right). Since this does not occur in the CENP-A-containing nucleosome crystal structure [14], it calls into question if, or to what extent (Figure 3b), CENP-A uses its rotated CENP-A/CENP-A interface that is an intrinsic feature of isolated (CENP-A/H4)2 heterotetramers. On the other hand, the C-terminal ∼45 a.a. of the H2A subunit of CENP-A-containing polynucleosome arrays undergoes rapid hydrogen/deuterium exchange relative to in arrays assembled with conventional H3 [58]. Whether or not this increased local flexibility accompanies a structural rearrangement of H2A/H2B dimers in CENP-A polynucleosome arrays—the functional context of centromeric chromatin of most eukaryotes [49,58,59]—awaits further investigation. The so-called CENP-A targeting domain, or CATD, is comprised of 22 amino acids substitutions relative to histone H3 in its loop L1 and α2 helix within its histone fold domain [50]. In some biological contexts the CATD appears to be functionally important before and after nucleosome assembly [25,60,61], while other studies have led to the proposal that the CATD may only be relevant for steps preceding nucleosome incorporation [49,62]. While further functional tests are clearly warranted, the CATD harbors both the bulged L1 that is exposed before and after nucleosome assembly [13,14] and the residues important for the CENP-A/CENP-A rotation that has only been directly observed in the heterotetramer context [13]. The CATD also contains six buried residues at the H4 interface [13] in nearly identical orientation before and after nucleosome assembly [13,14], that generate hydrophobic stitches and confer rigidity to CENP-A-containing nucleosome [50,56]. Hydrophobic stitching seems to be a required feature of centromeric nucleosomes. Indeed, a version of CENP-A in which the six residues that constitute the stitches are swapped with their counterpart residues in conventional H3 is unable to accumulate at centromeres [54]. The apparent functional importance of the rigid interior of CENP-A nucleosomes reveals a potentially rich area of future investigation to examine the role of such deviations in sub-nucleosomal dynamics to generate local, physical diversity within the chromatin fiber of eukaryotic chromosomes.
Propagating Centromere Identity Every Cell Cycle: HJURPScm3 and CENP-A Chromatin Assembly
A particular challenge to propagating centromeric chromatin is the delivery of newly expressed CENP-A to centromeres each cell cycle so that it is not diluted through replication of the underlying DNA. The recent discovery of HJURPScm3 [20,37–40] and its subsequent characterization as the chromatin assembly factor specific for generating new CENP-A nucleosomes [41,52,63,64] represent landmark achievements for the field in understanding this aspect of epigenetic centromere propagation. While HJURPScm3 is present at yeast centromeres for much [65] or all [22,66] of the cell cycle, it is present at mammalian centromeres for a small portion of the cell cycle consisting of several hours following mitotic exit and the subsequent G1 phase of the cell cycle [37,38]. This time window in the cell cycle overlaps with new CENP-A deposition at centromeres in insects and mammals [67,68]. The cell cycle timing of CENP-A deposition appears to be primarily dictated in mammalian and bird cells by conventional cell cycle machinery (i.e. Cdk1 and Cdk2) that restrict to mitotic exit a ‘centromere licensing’ event [69], presumably mediated by a complex containing Mis18 and associated factors [41,64,65,70]. Using purified components, HJURPScm3 assembles CENP-A-containing nucleosomes [41,52,71], and on the extremely AT-rich centromere sequences in budding yeast HJURPScm3 can co-assemble with CENP-A and H4 into non-nucleosomal particles [22]. In mammalian cells, HJURPScm3 is detectable at centromeres for several hours every cell cycle, so perhaps it forms a similar non-nucleosomal particle as an intermediate in a stepwise centromere maturation program that is coupled to the cell cycle [21].
Three atomic resolution structures are now available of the ternary HJURPScm3/CENP-ACse4/H4 complex, two fungal complexes (from S. cerevisiae [72] and K. lactis [62]) and one H. sapiens complex [73] (Figure 4). The K. lactis and H. sapiens structures are from crystallographic studies, and in both cases binding of HJURPScm3 occurs along a surface of an otherwise intact CENP-ACse4/H4 histone fold [62,73]. The S. cerevisiae structure from NMR studies, however, showed major perturbations of the histone fold, notably the separation of the long α2 helices of CENP-ACse4 and H4 by the insertion of the αA helix from HJURPScm3, as well as breakage of the C-terminal ∼one-third of the CENP-A α2 helix [72]. Cho and Harrison have called the NMR structure into question, chiefly on the basis that the construct lacked the α1 helix of H4 [62]. Given the intertwined helical fold of the histones and the essential contacts therein by all three helices of each histone [55], this criticism must be strongly considered. Bai and colleagues have countered that there is no strong evidence that the α1 helix is structured in solution prior to nucleosome formation, and that the αB helix of HJURPScm3, absent in the construct used for the K. lactis crystal likely takes the place of an unstructured H4 α1 helix [74].
Figure 4. Structures of HJURPScm3/CENP-ACse4/H4 ternary complexes.
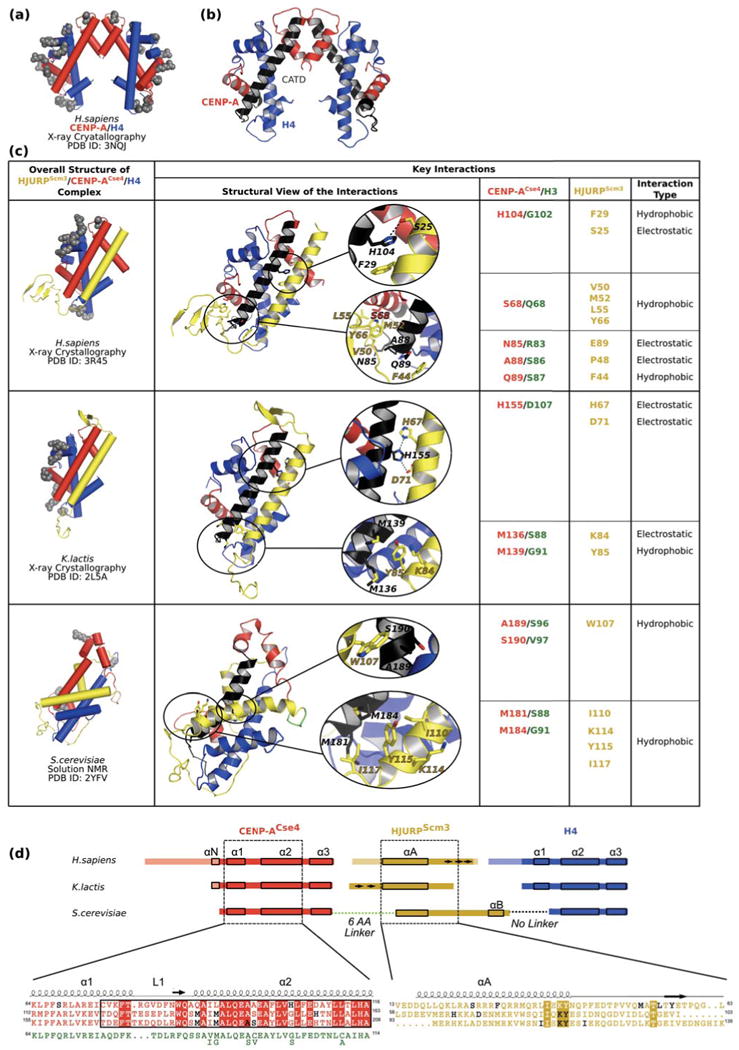
(a) The sub-nucleosomal (CENP-A/H4)2 structure [13] is shown with cylindrical helices as a reference point for how CENP-A and H4 are affected by binding to HJURPScm3. Positively charged residues making DNA contacts [14] are depicted as grey spheres. (b) Ribbon diagram of CENP-A/H4 tetramer (PDB ID 3NQJ) with the CATD of CENP-A colored black [13] shown for easier comparison with complexes in panel c. (c) Structure of HJURPScm3 (yellow)/CENP-ACse4 (red)/H4 (blue) complexes [62,72,73] with cylindrical helices (right column) and details of specific interactions in each of the structures (middle and right columns). The middle column includes ribbon diagrams of complexes with sites of specific interactions circled in black and accompanying enlargements. For easier comparison between each other and the tetramer in panel b, all structures were overlaid using residues within α2 helix of CENP-A (labeled black). (d) Linear diagram of constructs used in the three HJURPScm3/CENP-ACse4/H4 ternary complex structural studies. In constructs used for crystal structures, the residues present in the construct but lacking order in the corresponding structure are made transparent. The shared contact surfaces in each structure are contained within the dashed boxed regions of CENP-ACse4 (red) and HJURPScm3 (yellow), with α-helices indicated by small solid boxes and β-strands indicated by arrows. The S. cerevisiae study used a single-chain fused polypeptide with a 6 a.a. linker sequence between CENP-ACse4 and HJURPScm3 (green) and no linker between HJURPScm3 and H4. Sequence overlays are presented underneath the scheme with conserved residues between all structures labeled with boxes of solid colors (red for CENP-ACse4 and yellow for HJURPScm3). The CATD of CENP-A is boxed with a black rectangle. The interacting residues (listed in panel c) proposed as determinants of specificity between CENP-ACse4 and HJURPScm3 are shown in black. For easier analysis, the corresponding region of human histone H3 sequence (green) is placed underneath CENP-ACse4 sequences. Six residues differ between human H3.1 and fungal H3 proteins and the matching amino acids in fungal H3 are shown below the human H3 sequence.
While each of the three studies were of proteins originating from different species, all of the resulting structures agree on several points (Figure 4c):
HJURPScm3 binds CENP-ACse4/H4 dimer and prevents (CENP-ACse4/H4)2 heterotetramer formation as observed in the absence of HJURPScm3 [13] and within nucleosomes [14].
HJURPScm3 obstructs DNA binding of CENP-ACse4 as it occurs within the nucleosome [14]. On the other hand, some of the positively charged DNA-binding ridges remain accessible (Figure 4a and 4c, grey spheres), leaving open the possibility that the ternary complex could bind to DNA in a non-nucleosomal form. Indeed, this type of binding could account for HJURPScm3/CENP-ACse4/H4 complexes that bind to yeast CEN sequences, and are proposed to represent the centromeric CENP-ACse4-containing particle at the foundation of the budding yeast kinetochore [22].
HJURPScm3 uses as a major binding surface electrostatic and hydrophobic contacts with CATD region of CENP-ACse4.
A central question in understanding the propagation mechanism that replenishes CENP-A at the centromere each cell cycle is how it is recognized and sorted away from bulk H3 destined for other locations in the genome. For example, HJURPScm3 binds CENP-ACse4/H4 in a dimer form generating trimeric 1:1:1 complex of chaperone and two histones, the same stoichiometry as the ternary complex of the histone chaperone Asf1 with conventional H3 and H4 [75,76]. Since the contact region on H3/H4 for Asf1 [75,76] is not strongly distinguished from the corresponding portion of CENP-A/H4 [13], bulk histone chaperones may not be excluded from binding to CENP-A/H4. Indeed, RbAp46/48 and nucleophosmin can co-purify with CENP-A/H4 out of mammalian cells [37,38], the former presumably through its interactions with the N-terminus of H4 [77,78]. Thus, it seems most likely that HJURP uses distinguishing features encoded by CENP-A to sort it away and sequester it from bulk H3.
While the atomic-level structural studies have added important insight to HJURP binding to CENP-A/H4, divergent findings from the three structures [62,72,73] did not generate a unified view as of how is specificity actually achieved. The fungal complex structures have partially overlapping residues proposed to be important for CENP-ACse4 binding specificity (Figure 4c,d) [62,72]. For instance, two Met residues within the N-terminal portion of the CENP-ACse4 α2 helix were highlighted in both studies [62,72]. The K. lactis structure finds another key interaction towards the C-terminal end of the CENP-ACse4 α2 helix: His155 of the CENP-ACse4 interacts electrostatically with His67 and Asp71 on HJURPScm3 [62]. The S. cerevisiae structure, however, does not involve capture of this amino acid contact due to bending of αA helix of HJURPScm3 away form C-terminal end of the α2 helix of CENP-ACse4 [72]. Instead, the authors highlight residues in the central portion of α2 helix of CENP-ACse4 (Ala189 and Ser190), as they were identified as important in other functional studies for growth defects (e.g. mutation of Met184, Ala189 and Ser190 in CENP-ACse4 to the corresponding H3 residues [39]) and cell viability (e.g. Ile110Asp and Ile117Asn mutations [79]). However, it is not immediately clear from the structure how these residues provide specificity over analogous residues in conventional H3 histone (Figure 4c). In H. sapiens CENP-A, there is a histidine (His104), located one turn up the α2 helix relative to His155 in K. lactis CENP-ACse4 [73]. The authors of the human ternary complex study have concluded, however, that neither His104 nor any other residues encoded by the CATD of CENP-A (i.e. in CENP-A loop L1 and α2 helix) are likely candidates to confer specificity by CENP-A. Instead, they proposed Ser68 within the α1 helix of CENP-A as the primary specificity determinant for HJURP-binding [73]. This view should be tempered, though, by functional approaches in cells as well as experiments to monitor HJURP/CENP-A/H4 ternary complex formation by size exclusion chromatography where the critical specificity determinants map to surface residues within the CATD, and not to Ser68 [54]. Indeed, as few as three contact residues within the CATD suffice for HJURP recognition [54].
Concluding Remarks and Future Challenges
Over the last ∼two years, the centromere field has seen major progress in two key areas: tests that confirm the notion that arrays of CENP-A-containing nucleosomes are sufficient to generate functional centromeric chromatin [31-33,36,41,47–49] and structural findings that finally give atomic resolution snapshots [13,14,62,72,73] of the histone variant ∼25 years after its initial discovery [80,81]. Major questions remain, however, and effort is needed now to define the precise nature of the CENP-A-containing particle at natural centromeres, as well as the nature of potential intermediate forms accompanying CENP-A chromatin assembly reactions. In addition, it will be important to test whether or not the interphase epigenetic centromere specifying function of CENP-A is mediated by a nucleosomal form that is the same as the kinetochore-building form.
Acknowledgments
We thank K. Gupta for assistance with modeling. Many important contributions could not be cited due to space constraints, and we apologize to our colleagues for any relevant exclusions. Work in the Black Lab is supported by a grant from the National Institutes of Health (GM082989), a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and a Rita Allen Foundation Scholar Award. N.S. is supported in by a postdoctoral fellowship from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cleveland DW, et al. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichler EE. Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff S, et al. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 5.Murphy WJ, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 6.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Warburton PE, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 8.Amor DJ, et al. Human centromere repositioning “in progress”. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassett EA, et al. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelby RD, et al. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekulic N, et al. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 15.Williams JS, et al. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalal Y, et al. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krassovsky K, et al. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci USA. 2012;109:243–248. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavelle C, et al. Right-handed nucleosome: myth or reality? Cell. 2009;139:1216–1217. doi: 10.1016/j.cell.2009.12.014. author reply 1217-1218. [DOI] [PubMed] [Google Scholar]
- 20.Mizuguchi G, et al. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao H, et al. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddox PS, et al. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro SA, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Liu ST, et al. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 28.Stoler S, et al. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 29.Meluh PB, et al. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 30.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 31.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olszak AM, et al. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- 34.Ishii K, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 35.Alonso A, et al. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendiburo MJ, et al. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 37.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 38.Foltz DR, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camahort R, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Barnhart MC, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hieter P, et al. Functional selection and analysis of yeast centromeric DNA. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 43.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henikoff S, Henikoff JG. “Point” Centromeres of Saccharomyces Harbor Single CenH3 Nucleosomes. Genetics. 2012 doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coffman VC, et al. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawrimore J, et al. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiermaier E, et al. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1109–1115. doi: 10.1038/ncb1924. [DOI] [PubMed] [Google Scholar]
- 48.Lacefield S, et al. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1116–1120. doi: 10.1038/ncb1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guse A, et al. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 51.Kingston IJ, et al. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dechassa ML, et al. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, et al. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bassett EA, et al. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–62. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 56.Black BE, et al. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conde e Silva N, et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 58.Panchenko T, et al. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci USA. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blower MD, et al. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranjitkar P, et al. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreno-Moreno O, et al. The F Box Protein Partner of Paired Regulates Stability of Drosophila Centromeric Histone H3, CenH3CID. Current Biology. 2011;21:1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 62.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shuaib M, et al. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernad R, et al. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol. 2011;192:569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pidoux AL, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luconi L, et al. The CENP-A chaperone Scm3 becomes enriched at kinetochores in anaphase independently of CENP-A incorporation. Cell Cycle. 2011;10:3369–3378. doi: 10.4161/cc.10.19.17663. [DOI] [PubMed] [Google Scholar]
- 67.Schuh M, et al. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 68.Jansen LET, et al. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silva MCC, et al. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 70.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Shivaraju M, et al. Scm3 is a centromeric nucleosome assembly factor. J Biol Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng H, et al. Structure of the budding yeast Saccharomyces cerevisiae centromeric histones Cse4-H4 complexed with the chaperone Scm3. Proc Natl Acad Sci USA. 2011;108:E596. doi: 10.1073/pnas.1109548108. author reply E597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.English CM, et al. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 77.Murzina NV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song JJ, et al. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keith KC, et al. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palmer DK, et al. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 82.Gascoigne KE, Cheeseman IM. Kinetochore assembly: if you build it, they will come. Curr Opin Cell Biol. 2011;23:102–108. doi: 10.1016/j.ceb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carroll CW, et al. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]


