Abstract
The classical Mendelian genetic perspective has failed to adequately explain the biology and genetics of common metabolic and degenerative diseases. This is because these diseases are primarily systemic bioenergetic diseases, and the most important energy genes are located in the cytoplasmic mitochondrial DNA (mtDNA). Therefore, to understand these “complex” diseases, we must investigate their bioenergetic pathophysiology and consider the genetics of the thousands of copies of maternally inherited mtDNA, the more than 1,000 nuclear DNA (nDNA) bioenergetic genes, and the epigenomic and signal transduction systems that coordinate these dispersed elements of the mitochondrial genome.
Origins and implications of human mitochondrial genetics
The application of Mendelian genetic principles, using the most sophisticated technologies, has failed to adequately explain the genetics or pathophysiology of many common metabolic and degenerative diseases. This shortcoming can now be understood through the discovery that mutations in the maternally inherited mtDNA can cause many of the symptoms associated with “complex” diseases and that the mtDNA codes for the central genes of the mitochondrial energy–generating process oxidative phosphorylation (OXPHOS). Therefore, the common metabolic and degenerative diseases must be bioenergetic in origin and non-Mendelian in inheritance.
The central player in bioenergetics is the mitochondrion. Mitochondria produce about 90% of cellular energy, regulate cellular redox status, produce ROS, maintain Ca2+ homeostasis, synthesize and degrade high-energy biochemical intermediates, and regulate cell death through activation of the mitochondrial permeability transition pore (mtPTP). The mitochondrial genome consists of thousands of copies of the maternally inherited mtDNA plus between 1,000 and 2,000 nDNA genes. mtDNA codes for 13 OXPHOS polypeptides, plus the 22 transfer RNAs (tRNAs) and the 12S and 16S rRNAs necessary for the bacteria-like mitochondrial protein synthesis. mtDNA polypeptides encompass seven of the 45 polypeptides of OXPHOS complex I (ND1, ND2, ND3, ND4, ND4L, ND5, and ND6), one of the 11 polypeptides of complex III (cytochrome b), three of the 13 polypeptides of complex IV (COI, COII, and COIII), and two of the approximated 17 polypeptides of complex V (ATPase6 and ATPase8). Complexes I, III, and IV constitute the electron transport chain (ETC), which oxidizes the reducing equivalents (hydrogen-derived electrons) from food with the oxygen we breathe. As the electrons flow sequentially through complexes I, III, and IV, protons are pumped out across the mitochondrial inner membrane through these complexes to generate an electrochemical gradient. This mitochondrial capacitor is the vital force and can be used to drive many biological processes, including the condensation of ADP and Pi to form ATP via complex V. Thus oxidation is coupled with phosphorylation in OXPHOS. The 1,000–2,000 nDNA mitochondrial genes, scattered across the chromosomes, code for the remaining approximately 80 OXPHOS subunits, the intermediary metabolism enzymes, and the mitochondrial biogenesis proteins (1–3).
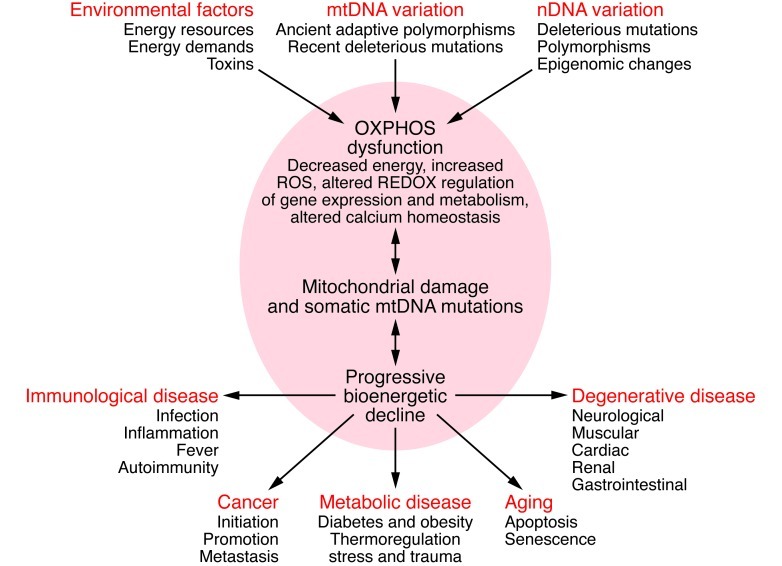
Three factors can perturb mitochondrial bioenergetics and result in disease: variation in the mtDNA sequence, variation in the sequences of nDNA-coded mitochondrial genes or in the expression of these genes, or variation in environmental calories and the caloric demands made on the organism. Since different tissues rely on mitochondrial energy to different extents, partial systemic energy deficiency can result in tissue-specific symptoms. The brain, which represents only 2% of the body weight but consumes 20% of the oxygen, is the organ most sensitive to subtle energy diminution. Other high-energy demand tissues include the heart, muscle, kidney, and endocrine system, the organs commonly affected in metabolic and degenerative diseases (Figure 1).
Figure 1. Bioenergetic paradigm for metabolic and degenerative diseases, cancer, and aging.
Mitochondrial OXPHOS can be perturbed by nDNA genetic alterations and/or epigenomic regulation, by mtDNA ancient adaptive of recent deleterious mutations, or by variation in the availability of calories and in caloric demands. Alterations in mitochondrial structure and function can impair OXPHOS, which in turn can reduce energy production, alter cellular redox state, increase ROS production, deregulate Ca2+ homeostasis, and ultimately activate the mtPTP, leading to apoptosis. These and other consequences of OXPHOS perturbation can destabilize mtDNA. This results in progressive accumulation of somatic mtDNA mutations and decline of mitochondrial function, which accounts for aging and the delayed-onset and progressive course of degenerative diseases. As energy output declines, the most energetic tissues are preferentially affected, resulting in degenerative diseases of the central nervous system, heart, muscle, and kidney. Aberrant mitochondrial caloric metabolism also leads to metabolic deregulation, endocrine dysfunction, and symptoms such as diabetes, obesity, and cardiovascular disease. The release into the blood stream of mtDNA mutant N-formylmethionine polypeptides plus the mtDNA can initiate the inflammatory response, contributing to autoimmune diseases (e.g., multiple sclerosis and type I diabetes) and possibly also to the inflammatory component of late-onset degenerative diseases. Finally, cancer cells must manage energy resources to permit rapid replication (95). Figure adapted with permission from Cold Spring Harbor Press (55).
mtDNA mutations
Since the first report of an inherited mtDNA disease mutation 25 years ago (4), hundreds of clinically relevant mtDNA mutations have been identified. These can either be polypeptide mutations or protein synthesis mutations, the latter altering the tRNA or rRNA genes (3, 5). There are three clinically relevant classes of mtDNA mutations: recent deleterious mutations that result in matrilineal disease, ancient adaptive variants that predispose to the common diseases, and somatic mutations that accumulate in tissues with age and provide the aging clock (Figure 1).
Recent deleterious mutations and maternally inherited diseases.
A prime example of a pathogenic mtDNA polypeptide missense mutation is the NADH dehydrogenase subunit 4 (ND4) nt 11,778 G>A mutation (histidine 340 to arginine), or ND4 G11778A (R340H), that causes Leber hereditary optic neuropathy (LHON) (4). LHON is characterized by acute or subacute midlife blindness that is two to five times more likely to affect males than females, even though all maternal relatives generally have close to 100% mutant mtDNAs (homoplasmic) (6). mtDNA also accumulates mutations within tissues with age. mtDNA deletions that occur early in development can become widely disseminated throughout the body and cause spontaneous mitochondrial myopathy (24 ).
The two most common mtDNA protein synthesis mutations cause myoclonic epilepsy and ragged red fiber disease (MERRF) (tRNALys A8344G) (7, 8) and mitochondrial encephalomyopathy and stroke-like episodes (MELAS) (tRNALeu(UUR) A3243G) (9). These more severe mtDNA mutations produce multisystem neuromuscular diseases, and the mutant mtDNAs are generally mixed within the cell with normal (wild-type) mtDNAs (heteroplasmic) (3, 5). As a heteroplasmic cell replicates, the percentages of mutant and normal mtDNAs are randomly distributed into the daughter cells (replicative segregation). Consequently, the mtDNA genotype can drift during both meiotic and mitotic cell division. Meiotic replicative segregation can cause maternal relatives to harbor different percentages of mutant mtDNAs, have different degrees of the energetic defect, and manifest widely different phenotypes. For example, when the MELAS-causing tRNALeu(UUR) A3243G mutant is present in 10%–30% of the mtDNAs, the individual will develop type I or type II diabetes mellitus, but when the mutant is present at higher percentages, myopathy, cardiomyopathy, and stroke-like episodes will develop. Mitotic segregation can give rise to an individual with significantly different percentages of mutant mtDNAs in different tissues when derived from a heteroplasmic oocyte, further contributing to phenotypic variability (3, 5, 7, 8).
The milder mtDNA variants can affect caloric metabolism and result in metabolic abnormalities such as diabetes and obesity and/or affect the most energy-demanding organs such as the brain and lead to late-onset degenerative diseases, such as psychiatric disorders, Parkinson disease (PD), and Alzheimer disease (AD). The more severe mtDNA mutations, like MERRF and MELAS, cause progressive multisystem diseases, frequently resulting in premature death. The most severe mtDNA mutations can lead to lethal childhood diseases, such as Leigh syndrome (3).
Ancient adaptive mtDNA variants: common variants that predispose to common diseases.
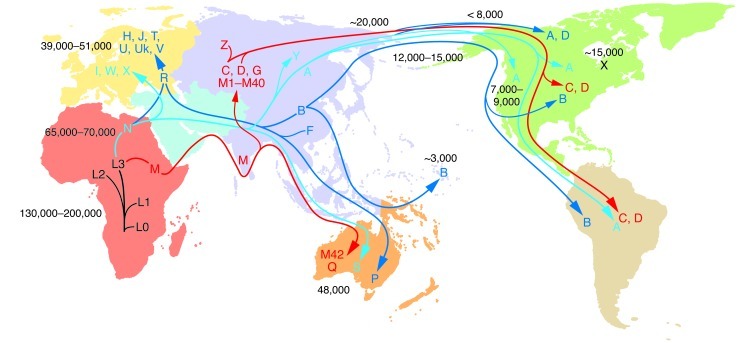
Population-specific mtDNA polymorphisms have been linked to predisposition to a broad range of metabolic and degenerative diseases (3, 10, 11). These variants are generally ancient, having accumulated along radiating maternal lineages during the human expansion out of Africa. By superimposing the human mtDNA mutational tree on the geographic locations of indigenous populations that harbor the various mtDNA types, mtDNA polymorphisms have been used to reconstruct the origins and ancient migrations of women (Figure 2).
Figure 2. Radiation of human mtDNA as women migrated out of Africa to colonize Eurasia, Australia, and the Americas.
The uniparentally inherited mtDNA can only change by sequential accumulation of mutations along radiating female lineages. Therefore, the mtDNA mutational tree and ancient migrations of women were reconstructed by sequencing mtDNAs from indigenous populations and correlating their regional clusters of related haplotypes (haplogroups) with the population’s geographic location. The haplogroups are regional because they were founded by regionally adaptive functional variants. The mtDNA tree originates in Africa, and all African mtDNAs are classified together as macrohaplogroup L. From haplogroup L3, two mtDNA lineages, M and N, arose in Ethiopia and successfully left Africa to colonize the rest of the world about 65,000–70,000 YBP. The founder mtDNA of macrohaplogroup N harbored two mtDNA missense mutations, ND3 G10398A (A114T) and ATP6 G8701A (A59T), whereas the founder of macrohaplogroup M did not harbor major functional mutations (3, 15). Early M and N emigrants from Africa moved through Southeast Asia, ending in Australia (96, 97). N mtDNAs also moved north from Africa into the Middle East to generate submacrohaplogroup R and European-specific haplogroups H, J, T, U, Uk, and V (from R) and I, W, and X (from N). N and R gave rise to Asian haplogroups A+Y and B+F, respectively. M moved north out of Southeast Asia to colonize Asia, generating haplogroups C and D and multiple M haplogroups. Haplogroups A, B, C, D, and X subsequently migrated to the Americas. The mtDNA mutation rate is 2.2%–2.9% per million years (numbers within the figure denote YBP). Figure adapted with permission from MITOMAP (5).
mtDNA has a very high mutation rate, yet codes for the most important mitochondrial energy genes. This should be a lethal combination. However, mammalian females have evolved an intraovarian selection system that destroys the proto-oocytes with the most severe mitochondrial defects (12, 13), a process that may account for atresia. Therefore, mild mtDNA variants are continuously being introduced into populations with minimum genetic load. Those variants can be beneficial for a population within a specific regional energetic environment and be selectively enriched in that regional population. Since different mtDNA variants are adaptive in different environments, different mtDNAs became enriched in different populations. The subsequent accumulation of random mtDNA mutations on the founding adaptive mtDNA produced regional clusters of related mtDNA haplotypes, known as haplogroups.
Because of the complexity of mitochondrial physiology, a mtDNA variant might be beneficial early in reproductive life, but deleterious postreproductively. Such variants are said to be antagonistically pleiotropic. For example, increased ROS production might help fight infection in the young but cause lifelong chronic oxidative stress that predisposes to development of age-related degenerative diseases.
The human mtDNA tree originated in Africa about 130,000–200,000 years before present (YBP) and gave rise to a series of African-specific haplogroups, which in aggregate form African macrohaplogroup L (Figure 2). Of all the African mtDNAs, only two mtDNA lineages, macrohaplogroups M and N, successfully left Africa (about 65,000–70,000 YBP) and colonized the rest of the world. In one migration, M and N left Africa and traveled along the tropical Southeast Asian coast, ultimately reaching Australia.
Macrohaplogroup N mtDNAs also moved north into the Middle East and radiated to create submacrohaplogroup R. Both N and R lineages spread into Europe to generate the eight to nine European-specific haplogroups.
In Asia, macrohaplogroup N radiated to form haplogroups A and Y, and the N-derived R lineage generated haplogroups B and F. From Southeast Asia, macrohaplogroup M moved northward to form an array of Asian-specific mtDNA haplogroups (C, D, and M1∼M40). Ultimately, haplogroups A, B, C, D, and X migrated into the Americas to found the Native American populations (Figure 2).
A major environmental barrier to the migration of sub-Saharan Africans into Eurasia must have been the cold of the northern latitudes. To survive, early humans would have needed to produce more core body heat. This might have been achieved by mtDNA mutations that decreased the “coupling efficiency” of OXPHOS. This would require that more calories be burned to generate the same amount of ATP. Since a calorie is a unit of heat, reduced coupling efficiency would increase the core body temperature and resistance to cold, but would necessitate a higher-calorie diet (2, 14).
Prior to moving out of Ethiopia, the founding macrohaplogroup N mtDNA acquired two functional variants, ND3 G10398A (A114T) and ATP6 G8701A (A59T) (3, 15), which changed the mitochondrial membrane potential and Ca2+ metabolism (ref. 16 and Figure 3). These mutations likely contributed to cold resistance in the temperate zone. Macrohaplogroup M, in contrast, stayed in the tropics, so cold-adaptive variants were not initially fixed in this lineage.
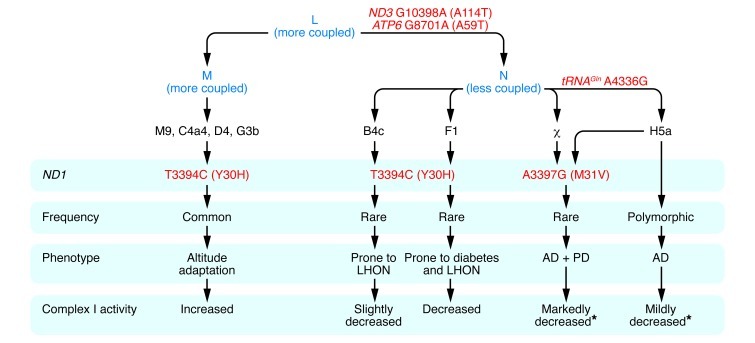
Figure 3. Relationship between ancient adaptive mtDNA variants and predisposition to metabolic and degenerative diseases.
African haplogroup L3 gave rise to macrohaplogroups M and N, which colonized Europe and Asia. N differed from M in harboring the ND3 G10398A (A114T) and ATP6 G8701A (A59T) variants. In Europe, N gave rise to haplogroup H, and H acquired the tRNAGln A4336G variant to generate H5a, which predisposes to AD, PD, and both AD and PD (17). The ND1 A3397G (M31V) missense mutation arose twice in Europeans, once on H5a and once independently, and in both cases was associated with predisposition to both AD and PD (17). Both tRNAGln A4336G and ND1 A3397G (M31V) mutations are likely to reduce mitochondrial complex I activity, augmenting the founding N variants. The ND1 T3394C (Y30H) mutation, which is adjacent to the ND1 M31 codon, arose on N and M mtDNAs. When arising on N haplogroups B and F, the ND1 T3394C (Y30H) variant is associated with complex I deficiency and increased penetrance of the primary LHON mutations. However, complex I activity is also modulated by N haplogroup background, with haplogroup F mtDNAs having lower complex I activity than haplogroup B mtDNAs, consistent with haplogroup F predisposition to diabetes (23). The ND1 T3394C (Y30H) mutation has arisen on several M mtDNAs, with all haplogroup M9 mtDNAs having the 3394C allele. Both M9 and 3394C mtDNAs increase in frequency with altitude in Tibet. Finally, M9 complex I activity is equal to or greater than that of any of the N haplogroups with the wild-type T3394 allele (20). Asterisks indicate that the stated complex I activity is predicted, based on the known genotype and complex I activities determined for cell lines harboring ND1 T3394C (Y30H)–containing mtDNAs.
The most common European N-R–derived mtDNA haplogroup is H. In Europe, haplogroup H acquired additional functional variants, one of which arose between 8,500 and 17,000 YBP in the tRNAGln gene (tRNAGln A4336G), creating subhaplogroup H5a. Today, this variant is found in only 0.4% of the general European population, but is present in 3.3% of AD patients, 5.3% of PD patients, and 6.8% of patients with both AD and PD (ref. 17 and Figure 3). The retention of this variant in the European population may be an example of antagonistic pleiotropy. A second European mtDNA variant observed in patients with both AD and PD is a missense mutation in the ND1 gene: ND1 A3397G (M31V). This variant arose twice, once in the tRNAGln A4336G lineage, and once independently (ref. 17 and Figure 3).
A related ND1 variant, ND1 T3394C (Y30H), changes the amino acid adjacent to M31. When arising on macrohaplogroup N mtDNAs, this variant is associated with increased penetrance of the milder primary LHON mtDNA mutations (18, 19). Yet when this same ND1 T3394C (Y30H) variant arose on macrohaplogroup M mtDNAs, it became enriched in high-altitude Tibetans. Haplogroup M9 mtDNA with the ND1 T3394C (Y30H) variant is present at less than 2% at sea level, but increases to about 35% of the mtDNAs in the highest Tibetan villages (ref. 20 and Figure 3).
The ND1 T3394C (Y30H) variant results in a 15%–28% reduction in complex I–specific activity when arising on macrohaplogroup N mtDNAs, which explains its enhancement of LHON mutation penetrance. However, the specific activity of complex I can also vary among different macrohaplogroup N haplogroups by up to 30%, independent of their 3394 allele. Even more surprising, the 3394C allele, when present in the M9 haplogroup mtDNA, is associated with complex I activity equal to or greater than any of the macrohaplogroup N mtDNAs with the wild-type T3394 (Y30) variant (ref. 20 and Figure 3).
Similar physiological differences have been documented between European haplogroup H and Uk mtDNAs (11, 21). Therefore, the metabolic consequences of a particular mtDNA nucleotide variant can be strongly influenced by genetic and environmental context.
The clinical relevance of mtDNA haplogroup variation has been repeatedly demonstrated through case-control studies on a wide variety of metabolic and degenerative diseases, cancer, and aging (3, 10, 11, 22), and the physiological associations are being elucidated. For example, haplogroup F mtDNAs are associated with low complex I activity (20) and predilection to diabetes (23). Hence, ancient mtDNA variants and haplogroups are likely the long-sought common variants that predispose to common diseases.
Somatic mtDNA mutations.
mtDNA also accumulates mutations within tissues with age. mtDNA deletions that occur early in development can become widely disseminated throughout the body and cause spontaneous mitochondrial myopathy (24). However, mtDNA deletions (25–28) and base substitutions (29, 30) can arise in tissues throughout life, and their accumulation has been shown to modulate aging and longevity (31–33). Therefore, the accumulation of somatic mtDNA mutations may be the aging clock.
The rate of accumulation of somatic mtDNA mutations can be modulated by nuclear or cytoplasmic genetic variants and by environmental factors (Figure 1). For example, factors that increase mitochondrial ROS production would increase the mtDNA mutation rate and lead to premature organ failure. Increased mtDNA somatic mutation levels have been documented in ischemic heart disease (34), AD brains (35–37), PD brains (38, 39), Huntington disease brains (40), and Down syndrome with dementia (DSAD) brains (37). In AD and DSAD brains, elevated somatic mtDNA base substitution mutations have been correlated with reduced mtDNA copy number and ND6 transcript levels (37).
As somatic mtDNA mutations accumulate, they change the amino acid sequences of the mtDNA-coded mitochondrial N-formylmethionine–initiated polypeptides. These bacterial-like variant polypeptides can then be seen as foreign and initiate an inflammatory response. This may contribute to the inflammation frequently observed in late-stage degenerative diseases (Figure 1 and refs. 38, 41–43).
nDNA mitochondrial gene mutations
Bioenergetic disease can also result from mutations in any one of the hundreds of nDNA genes that code for mitochondrial proteins. Mutations in more than 200 nDNA gene loci have already been reported to cause mitochondrial bioenergetic dysfunction (3, 44).
Structural and metabolic nDNA-coded mitochondrial genes.
In addition to mutations in the nDNA-coded enzymes of mitochondrial intermediary metabolism, which exhibit classical Mendelian transmission, pathogenic mutations have been identified in multiple nDNA-coded OXPHOS structural and assembly factor genes. When both copies of a chromosomal gene are mutated, severe OXPHOS defects can occur and cause devastating pediatric disease, the most commonly recognized phenotype being Leigh syndrome (3, 44).
Diseases of nDNA-mtDNA interactions.
Mutations in the nDNA-coded mtDNA biogenesis genes can cause degenerative diseases by destabilizing mtDNA biogenesis, resulting in multiple mtDNA deletions and/or mtDNA depletion (3, 44). Pathogenic mutations have been reported in mtDNA polymerase γ (POLG) (45), Twinkle helicase (46), mitochondrial deoxyguanosine kinase and thymidine kinase 2 (47, 48), cytosolic thymidine phosphorylase (49), and the heart-muscle adenine nucleotide (ADP/ATP) translocator (ANT1) (50, 51), to name a few (3). Mitochondrial disease can also result from the incompatible interaction of two otherwise nonpathogenic nDNA and mtDNA genetic variants (52).
Alterations in nDNA-coded mitochondrial gene expression and the epigenome.
While mtDNA mutations have permitted humans to adapt to stable regional environmental energetic differences, many energy resources and demands fluctuate cyclically, for example, seasonal changes in temperature and food supply. Adaptation to this type of energetic variation is accomplished by changes in the levels of mitochondrially generated high-energy intermediates, such as acetyl-CoA and ATP, and in the mitochondrial modulation of the cellular redox state. As these mitochondrial bioenergetic parameters fluctuate with the environment, they drive posttranslational modification of the proteins of the epigenome and the signal transduction pathways. In this way, the expression of the hundreds of nDNA-coded bioenergetic genes is coupled to environmental fluctuations through mitochondrial energy flux (53–55).
The nDNA and mtDNA genetics of “complex” diseases
This new bioenergetic perspective provides a framework to reevaluate the genetics and pathophysiology of “complex” diseases, such as PD, AD, autism spectrum disorders (ASDs), and psychiatric disorders. In PD, numerous nDNA loci that have been linked to developing movement disorders are directly involved in modulating mitochondrial integrity and function (38). For example, mutations in parkin (PARK2) and PTEN-induced kinase 1 (PINK1; also known as PARK6) impede mitochondrial autophagy (mitophagy), which leads to the accumulation of mitochondrial damage (38, 56, 57). The resulting increased mtDNA somatic mutation rate degrades the mtDNA in the basal ganglion and substantia nigra, ultimately resulting in neuronal dysfunction, cell death, and movement disorders (26, 39, 58).
In AD, Aβ toxicity is generally assumed to be the cause, yet systemic mitochondrial defects have been repeatedly reported (1). At high concentrations, Aβ oligomerizes and is toxic (59), specifically inhibiting mitochondrial function (38, 60). However, at low concentrations, monomeric Aβ is protective of mitochondrial function (38). Therefore, Aβ appears to be bifunctional. From this perspective, under normal conditions, Aβ functions to protect the mitochondria and associated neurons and synapses and is induced in response to mitochondrial stress, possibly mitochondrial ROS production. However, when mitochondrial dysfunction becomes so severe that neuronal function is irreversibly impaired, Aβ induction becomes excessive and leads to Aβ oligomerization. The oligomerized Aβ then inhibits mitochondrial function, activates the mtPTP, and destroys the neuron with the defective mitochondria, thus eliminating noise from the neuronal information network.
Early-onset AD, then, is the result of mutations in APP or the presenilins, which aberrantly increase Aβ levels, leading to premature Aβ aggregation and destruction of potentially repairable mitochondria, neurons, and synapses. Late-onset AD, in contrast, is the result of chronic mitochondrial stress, perhaps mediated by ROS toxicity, Ca2+ overload, or other factors. Increased chronic mitochondrial oxidative stress can result from a variety of factors that partially inhibit OXPHOS, including the tRNAGln A4336G and ND1 A3397G (M31V) variants. Excessive mitochondrial Ca2+ exposure can also increase mitochondrial ROS production and activate the mtPTP. Mitochondrially destined Ca2+ is released from the endoplasmic reticulum within the mitochondria-associated membranes (MAMs), and MAMs harbor the presenilin complexes (61, 62). Inappropriate MAM Ca2+ regulation can cause chronic mitochondrial stress by increasing mitochondrial ROS production and Ca2+ activation of the mtPTP. These and other stressors could cause the premature accumulation of neuronal somatic mtDNA mutations, bioenergetic decline, mutant mtDNA peptide–induced inflammation, synaptic loss, and dementia (38).
ASDs have also proven enigmatic when viewed from a classical Mendelian perspective. Yet the genetics and pathophysiology of ASDs are fully consistent with the expectations for mild mitochondrial dysfunction. Like LHON (6), in ASDs, males are four times more likely than females to be affected (63, 64). Mitochondrial metabolic defects have been repeatedly reported in ASD patients, and mtDNA mutations have been found in several ASD pedigrees (63, 65–67).
Elevated Ca2+ levels have also been observed in ASD brains, and the excess Ca2+ could activate the neuronal aspartate/glutamate carrier of the mitochondrial NADH shuttle system (68) and the tricarboxylic acid cycle dehydrogenases (69, 70). Both of these effects would drive excessive reducing equivalents into the ETC, stimulating mitochondrial ROS production, oxidative stress, mitochondrial damage, and synaptic loss. Mutations in the CACNA1C Ca2+ channel gene have been shown to cause the syndromic ASD Timothy syndrome (71), and mutations in the CACNA1F Ca2+ channel gene have been reported in ASD patients (72).
Copy number variants (CNVs) are also increased in number in autism patients (73, 74), and CNVs that remove a copy of the PARK2 or ubiquitin protein ligase E3A (UBE3A) genes have been observed repeatedly (75, 76). Since loss of PARK2 would impair mitochondrial quality control (77), and mutations in UBE3A are associated with hippocampal mitochondrial defects in Angelman syndrome, another syndromic ASD (78), these observations also implicate bioenergetics in ASDs.
Since there are more than 1,000 nDNA mitochondrial genes, and partial mitochondrial defects can be sufficient to cause neurodegenerative disease, random CNVs that delete one copy of a nDNA mitochondrial gene could be sufficient to predispose to the neurological symptoms of ASD. Indeed, in one study, an ASD subject with one CNV had near-normal OXPHOS function, while another patient with 13 CNVs had a severe OXPHOS defect (67).
Another surprise from the Mendelian perspective has been that genetic elements linked to ASDs are also associated with other neuropsychiatric disorders (79, 80). However, this would be predicted if neuropsychiatric disorders share a common bioenergetic pathophysiological mechanism. Mitochondrial dysfunction has been documented in psychiatric disorders (81), evidence of matrilineal bias in transmission has been reported (82, 83), and mtDNA haplogroups and brain mtDNA somatic mutations have been observed in patients with psychiatric conditions (21, 28).
Mouse mtDNA mutants cause neuromuscular disease
To prove that mitochondrial defects cause metabolic and degenerative diseases, mitochondrial gene mutations have been introduced into the mouse, and metabolic and degenerative disease phenotypes have been observed (84). The introduction of mtDNA mutations into the mouse germline has proven particularly instructive.
To introduce a mtDNA mutation into the mouse, an appropriate mtDNA mutation must be isolated in a cultured mouse cell line and the mutant mtDNA transferred into the mouse female germline, most commonly mediated by mouse female embryonic stem cell (mfESC) transmitochondrial cybrids (85). Introduction into the mouse of a mtDNA harboring a 12S rRNA chloramphenicol resistance (CAPR) mutation (86, 87) resulted in chimeric CAPR mice with cataracts, retinal dysfunction, and optic nerve hamartomas. Homoplasmic CAPR transgenic mice had stunted growth, mitochondrial myopathy, and cardiomyopathy and died prematurely (87).
Introduction of mtDNAs harboring a homoplasmic mtDNA with a COI T6589C (V421A) missense mutation that were also heteroplasmic for a ND6 13886 insertion C frameshift mutation resulted in animals that rapidly and directionally lost the frameshift mtDNA within three generations. This observation revealed the existence of the prefertilization ovarian selective system, which eliminates proto-oocytes with the most deleterious mtDNA mutations (12, 88). The mice that remained after segregation of the ND6 13886 insertion C frameshift mtDNA were homoplasmic for the COI T6589C (V421A) missense mutation. These animals had a 50% reduction in complex IV activity and developed mitochondrial myopathy and cardiomyopathy (12).
Introduction into the mouse of an ND6 G13997A (P25L) mtDNA mutation, which is functionally equivalent to the human ND6 G14600A (P25L) mutation reported to cause optic atrophy when heteroplasmic and Leigh syndrome when homoplasmic (89), resulted in animals with all of the anatomically possible physiological and pathological features of LHON. The physiological effects of the mutation on neurons were analyzed using synaptosomes from the mouse LHON model brain. This revealed that the optic atrophy mutation did not diminish synaptic ATP levels, but instead chronically increased mitochondrial ROS production. If this is the case for other LHON mutations, it suggests that the delayed onset of acute vision loss may be the result of cumulative oxidative damage (90).
A heteroplasmic mtDNA rearrangement mutation has also been introduced into the mouse. This resulted in mice with complex IV–negative (COX-negative) muscle and heart myofibers, renal dysfunction (91), and infertility (92). Therefore, both base substitution and mtDNA rearrangement mutations are sufficient to cause degenerative diseases.
To determine the consequences of much milder mitochondrial defects, mice were created in which two normal but different mouse mtDNAs were mixed within the female germline, thus subverting maternal inheritance. The mtDNAs were from NZB and 129 mice and differed at 91 nt positions encompassing 15 missense mutations, 5 tRNA mutations, 7 rRNA mutations, and 11 control region mutations. All mice were maintained on the C57BL/6J nuclear background (93). As previously observed (94), the heteroplasmy levels within the tissues of individual animals segregated, with NZB mtDNAs predominating in liver and kidney, and 129 mtDNAs in spleen and pancreas. However, the tail, muscle, heart, and brain heteroplasmy levels remained relatively stable. In mating experiments, the NZB mtDNAs were progressively lost from the maternal lineage, with the rate of segregation being greatest when the mtDNAs were at relatively equal percentages.
By random segregation and selective breeding, the heteroplasmic mice were used to derive three mouse lines: homoplasmic NZB, homoplasmic 129, and heteroplasmic NZB-129. These three different mtDNA genotype strains were then examined for behavioral alterations. While the homoplasmic NZB and homoplasmic 129 mice were essentially the same and phenotypically normal, the heteroplasmic NZB-129 mice were markedly different. The heteroplasmic NZB-129 animals were hypoactive during the normally active dark period, in association with reduced food intake and respiratory exchange ratio, but were hyperexcitable under stress conditions. Even more remarkably, the heteroplasmic mice showed a striking learning defect, being slow to learn and quick to forget (93).
Extensive biochemical studies of the heteroplasmic NZB-129 mice have failed to detect a significant OXPHOS defect. Therefore, even extremely subtle bioenergetic dysfunction is sufficient to cause neuropsychiatric symptoms. This can account for why maternal inheritance of the mtDNA is strictly imposed throughout most of the eukaryotic kingdom and might explain why it has been so difficult to determine the pathophysiological basis of neuropsychiatric disorders.
Conclusion
Elucidation of the novel genetics of mtDNA and the demonstration of its central role in bioenergetics has provided a new set of genetic rules and physiological parameters for understanding the intraspecific genetic variation of relevance to human health and disease. This bioenergetic perspective not only provides a coherent theory for the etiology of the “complex” metabolic and degenerative diseases, it suggests powerful new approaches for their presymptomatic diagnoses, reliable prognosis, and effective treatment and prevention (10). However, reaping the benefits of these new insights will require a major redirection of the way we think about medical genetics and origin of disease. If we can change, the bioenergetic perspective promises to reduce the burden of chronic diseases and markedly improve global health span.
Acknowledgments
The author thanks Marie Lott for assistance. This work was supported by NIH grants NS21328, NS070298, AG24373, and DK73691 and by Simon Foundation grant 205844 awarded to D.C. Wallace.
Footnotes
Conflict of interest: The author’s research has received some support from Glaxo-Smith Kline.
Citation for this article:J Clin Invest. 2013;123(4):1405–1412. doi:10.1172/JCI61398.
References
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace DC. Why do we have a maternally inherited mitochondrial DNA? Insights from Evolutionary Medicine. Annu Rev Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 3. Wallace DC, Lott MT, Procaccio V. Mitochondrial medicine: the mitochondrial biology and genetics of metabolic and degenerative diseases, cancer, and aging. In: Rimoin DL, Pyeritz RE, Korf BR, eds.Emery and Rimoin’s Principles and Practice of Medical Genetics. Philadelphia, Pennsylvania, USA: Churchill Livingstone Elsevier; 2013. [Google Scholar]
- 4.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Pesini E, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35(database issue):D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadun AA, La Morgia C, Carelli V. Leber’s Hereditary Optic Neuropathy. Curr Treat Options Neurol. 2011;13(1):109–117. doi: 10.1007/s11940-010-0100-y. [DOI] [PubMed] [Google Scholar]
- 7.Wallace DC, et al. Familial mitochondrial encephalomyopathy (MERRF): Genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55(4):601–610. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 8.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. . Cell. 1990;61(6):931–937. doi: 10.1016/0092-8674(90)90059-N. [DOI] [PubMed] [Google Scholar]
- 9.Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. . Nature. 1990;348(6302):651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC. Mitochondria as chi. Genetics. 2008;179(2):727–735. doi: 10.1534/genetics.104.91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez-Duran A, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19(17):3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 12.Fan W, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319(5865):958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9(9):657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 14.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A. 1994;91(19):8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238(1):211–230. doi: 10.1016/S0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 16.Kazuno AA, et al. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2(8):e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoffner JM, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17(1):171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 18.Brown MD, Torroni A, Reckord CL, Wallace DC. Phylogenetic analysis of Leber’s hereditary optic neuropathy mitochondrial DNA’s indicates multiple independent occurrences of the common mutations. Hum Mutat. 1995;6(4):311–325. doi: 10.1002/humu.1380060405. [DOI] [PubMed] [Google Scholar]
- 19.Liang M, et al. Leber’s hereditary optic neuropathy is associated with mitochondrial ND1 T3394C mutation. Biochem Biophys Res Commun. 2009;383(3):286–292. doi: 10.1016/j.bbrc.2009.03.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji F, et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci U S A. 2012;109(19):7391–7396. doi: 10.1073/pnas.1202484109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rollins B, et al. Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PLoS One. 2009;4(3):e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khusnutdinova E, et al. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson’s disease. Ann N Y Acad Sci. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- 23.Fuku N, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. 2007;80(3):407–415. doi: 10.1086/512202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331(6158):717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 25.Cortopassi G, Arnheim N. Mitochondrial DNA in Human Pathology. 1992. Accumulation of mitochondrial DNA mutation in normal aging brain and muscle. In: DiMauro S, Wallace DS, eds. pp. 125–136. New York, New York, USA: Raven Press; [Google Scholar]
- 26.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 27.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992;2(4):318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 28.Sequeira A, et al. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet. 2012;3:103. doi: 10.3389/fgene.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286(5440):774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 30.Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res. 2000;28(21):4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 32.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 33.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308(5730):1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 34.Corral-Debrinski M, Stepien G, Shoffner JM, Lott MT, Kanter K, Wallace DC. Hypoxemia is associated with mitochondrial DNA damage and gene induction. Implications for cardiac disease. JAMA. 1991;266(13):1812–1816. doi: 10.1001/jama.1991.03470130092035. [DOI] [PubMed] [Google Scholar]
- 35.Corral-Debrinski M, et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23(2):471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 36.Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101(29):10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coskun PE, et al. Systemic mitochondrial dysfunction and the etiology of Alzheimer’s disease and down syndrome dementia. J Alzheimers Dis. 2010;20(suppl 2):S293–S310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coskun P, et al. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2011;1820(5):553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 40.Horton TM, et al. Marked increase in mitochondrial DNA deletion levels in the cerebral cortex of Huntington’s Disease patients. Neurology. 1995;45(10):1879–1883. doi: 10.1212/WNL.45.10.1879. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Q, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krysko DV, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32(4):157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Oka T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485(7397):251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. 2012;366(12):1132–1141. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 45.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28(3):211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 46.Spelbrink JN, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28(3):223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 47.Mandel H, et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet. 2001;29(3):337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- 48.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29(3):342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 49.Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283(5402):689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 50.Kaukonen J, et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289(5480):782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 51.Palmieri L, et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet. 2005;14(20):3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- 52.Potluri P, et al. A novel NDUFA1 mutation leads to a progressive mitochondrial complex I-specific neurodegenerative disease. Mol Genet Metab. 2009;96(4):189–195. doi: 10.1016/j.ymgme.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10(1):12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallace DC. Bioenergetic origins of complexity and disease. Cold Spring Harb Symp Quant Biol. 2011;76:1–16. doi: 10.1101/sqb.2011.76.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107(26):11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 59.Glabe CG, Kayed R. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 2006;66(2 suppl 1):S74–S78. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 60.Crouch PJ, et al. Copper-dependent inhibition of human cytochrome c oxidase by a dimeric conformer of amyloid-beta1-42. J Neurosci. 2005;25(3):672–679. doi: 10.1523/JNEUROSCI.4276-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schon EA, Area-Gomez E. Is Alzheimer’s disease a disorder of mitochondria-associated membranes? J Alzheimers Dis. 2010;20(suppl 2):S281–S292. doi: 10.3233/JAD-2010-100495. [DOI] [PubMed] [Google Scholar]
- 62.Contreras L, Drago I, Zampese E, Pozzan T. Mitochondria: the calcium connection. Biochim Biophys Acta. 2010;1797(6–7):607–618. doi: 10.1016/j.bbabio.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 63. Filipek PA. Medical aspects of autism. In: Volkmar FR, Klin A, Paul R, Cohen DJ, eds.Handbook of Autism and Pervasive Developmental Disorders. New York, New York, USA: John Wiley and Sons; 2005:534–578. [Google Scholar]
- 64.Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. Am J Biochem Biotech. 2008;4(2):198–207. [Google Scholar]
- 65.Giulivi C, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389–2396. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chauhan A, et al. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem. 2011;117(2):209–220. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith M, et al. Mitochondrial and ion channel gene alterations in autism. Biochim Biophys Acta. 2012;1817(10):1796–1802. doi: 10.1016/j.bbabio.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palmieri L, et al. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15(1):38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- 69.McCormack JG, Denton RM. A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex and NAD+-isocitrate dehydrogenase from a variety of sources. Biochem J. 1981;196(2):619–624. doi: 10.1042/bj1960619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCormack JG, Halestrap AP, Denton RM. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev. 1990;70(2):391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- 71.Splawski I, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci U S A. 2005;102(23):8089–8096. doi: 10.1073/pnas.0502506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myers RA, et al. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet. 2011;7(2):e1001318. doi: 10.1371/journal.pgen.1001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104(31):12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gai X, et al. Rare structural variation of synapse and neurotransmission genes in autism. Mol Psychiatry. 2012;17(4):402–411. doi: 10.1038/mp.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Su H, et al. Mitochondrial dysfunction in CA1 hippocampal neurons of the UBE3A deficient mouse model for Angelman syndrome. . Neurosci Lett. 2011;487(2):129–133. doi: 10.1016/j.neulet.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Daniels JL, et al. Parental psychiatric disorders associated with autism spectrum disorders in the offspring. Pediatrics. 2008;121(5):e1357–e1362. doi: 10.1542/peds.2007-2296. [DOI] [PubMed] [Google Scholar]
- 80.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gardner A, Boles RG. Beyond the serotonin hypothesis: mitochondria, inflammation and neurodegeneration in major depression and affective spectrum disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35(3):730–743. doi: 10.1016/j.pnpbp.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 82.McMahon FJ, Stine OC, Meyers DA, Simpson SG, DePaulo JR. Patterns of maternal transmission in bipolar affective disorder. Am J Hum Genet. 1995;56(6):1277–1286. [PMC free article] [PubMed] [Google Scholar]
- 83.Verge B, et al. New evidence for the involvement of mitochondrial inheritance in schizophrenia: results from a cross-sectional study evaluating the risk of illness in relatives of schizophrenia patients. J Clin Psychiatry. 2012;73(5):684–690. doi: 10.4088/JCP.10m06718. [DOI] [PubMed] [Google Scholar]
- 84.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23(15):1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. MacGregor GR, Fan WW, Waymire KG, Wallace DC. Generating animal models of human mitochondrial genetic disease using mouse ES cells. In: Notarianni E, Evans MJ, eds.Embryonic Stem Cells. New York, New York, USA: Oxford University Press; 2006:72–104. [Google Scholar]
- 86.Marchington DR, Barlow D, Poulton J. Transmitochondrial mice carrying resistance to chloramphenicol on mitochondrial DNA: developing the first mouse model of mitochondrial DNA disease. Nat Med. 1999;5(8):957–960. doi: 10.1038/11403. [DOI] [PubMed] [Google Scholar]
- 87.Sligh JE, et al. Maternal germ-line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc Natl Acad Sci U S A. 2000;97(26):14461–14466. doi: 10.1073/pnas.250491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6(1):e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malfatti E, et al. Novel mutations of ND genes in complex I deficiency associated with mitochondrial encephalopathy. Brain. 2007;130(pt 7):1894–1904. doi: 10.1093/brain/awm114. [DOI] [PubMed] [Google Scholar]
- 90.Lin CS, et al. A mouse mtDNA mutant model of Leber’s Hereditary Optic Neuropathy. Proc Natl Acad Sci U S A. 2012;109(49):20065–20070. doi: 10.1073/pnas.1217113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inoue K, et al. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet. 2000;26(2):176–181. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- 92.Nakada K, et al. Mitochondria-related male infertility. Proc Natl Acad Sci U S A. 2006;103(41):15148–15153. doi: 10.1073/pnas.0604641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sharpley MS, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151(2):333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16(1):93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 95.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Holst Pellekaan SM, Ingman M, Roberts-Thomson J, Harding RM. Mitochondrial genomics identifies major haplogroups in Aboriginal Australians. Am J Phys Anthropol. 2006;131(2):282–294. doi: 10.1002/ajpa.20426. [DOI] [PubMed] [Google Scholar]
- 97.Hudjashov G, et al. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci U S A. 2007;104(21):8726–8730. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]