Abstract
Skin secretion of Hypsiboas punctatus is the source of a complex mixture of bioactive compounds where peptides and small proteins prevail, similarly to many other amphibians. Among dozens of molecules isolated from H. punctatus in a proteomic based approach, we report here the structural and functional studies of a novel peptide named Phenylseptin (FFFDTLKNLAGKVIGALT-NH2) that was purified as two naturally occurring D- and L-Phes configurations. The amino acid epimerization and C-terminal amidation for both molecules were confirmed by a combination of techniques including reverse-phase UFLC, ion mobility mass spectrometry, high resolution MS/MS experiments, Edman degradation, cDNA sequencing and solid-phase peptide synthesis. RMSD analysis of the twenty lowest-energy 1H NMR structures of each peptide revealed a major 90° difference between the two backbones at the first four N-terminal residues and substantial orientation changes of their respective side chains. These structural divergences were considered to be the primary cause of the in vitro quantitative differences in antimicrobial activities between the two molecules. Finally, both molecules elicited equally aversive reactions in mice when delivered orally, an effect that depended entirely on peripheral gustatory pathways.
Introduction
According to Stahl [1], multifunctional drugs are agents with more than one therapeutic mechanism and it has been observed that almost all existing drugs have more than one known pharmacological target when higher doses exceeding the remedial recommendations are administrated. Fine-tuned with Paracelsuś legendary quote, the multifunctional concept of a drug depends upon a given concentration to reveal its multiple pharmacological activities. Among many others, a classical example of this principle is Aspirin [2], the well-known drug used for more than a century as analgesic, antipyretic and anti-inflammatory, in the end 1960`s was also confirmed experimentally to be an effective inhibitor of platelet aggregation [3], therefore an important agent for ischemic stroke prevention [4]. Pondering these facts and more recent ones [5], it is reasonable to ask: why the multifunctional drug principle should be confined only to small molecules, natural products and/or their synthetic analogs? Would peptides and proteins also show similar properties under comparable experimental conditions? How many different structures, functional sites and therefore biological activities a single polypeptide chain would display after an extensive investigation?
A noteworthy indication towards a more comprehensive answer to these questions was given by Brandenburg and co-authors [6] in their recent review on antimicrobial peptides, when the multifunctional drug attributes of this class of molecules and a possible range of different applications were underlined.
The other end of the same spectrum show us that in principle, from the organism perspective, any exogenous substance alone may represent a potential hazardous to its integrity and/or survival. Living beings are equipped with numerous barriers, defense strategies and metabolic mechanisms to deal with physical, chemical and biological threats. In all that, small and macromolecules may be found acting in both ways, as belligerent and defense agents depending on the situation. It is interesting to note that animals may engender complex multimodal signals combined with volatile and toxic substances that play critical roles in defense strategies as well as in social interactions of a number of animal species [7]–[10]. In plants, it was demonstrated that homoterpene could also be released from damaged aerial tissues induced by herbivore attacks revealing a signaling mechanism similar to those in animals [11], [12]. In other words, many molecules may have multiple functions and activities depending on the metabolic target and relative concentration.
As for amphibians, it is widely known that they protect themselves against various biotic hazards using an intricate collection of strategies including noxious secretions, aposematism, deimatic behavior and evasive tactics [13]. The granular glands in anurans skin store extraordinary quantities of toxic compounds [14]–[17], which are also the most evident molecular ingredients of some aversive chemosensory signals released against potential predators to reduce the chances of future attacks [18], [19], as exemplified by the fact that some amphibian eggs and larvae are perceived as repulsive by some fish and birds [20]–[22]. Amongst the great variety of substances secreted by these glands, several of them are antimicrobial peptides [15], [23]–[28] typically characterized by basic, linear and amphiphilic primary structures of 30–35 residues in a single-chain [27], [29]–[31]. Although their protecting roles as microbicidal agentes have been thoroughly investigated during previous decades, little is known about other biological activities that antimicrobial peptides may display [32], [33]. In the present work, we report the 1H NMR structures and functional studies of a novel antimicrobial peptide that also displays aversive gustatory properties in mice models. The peptide naturally occurring in two enantiomeric configurations was isolated from Hypsiboas punctatus skin secretion and sequenced by a combination of ion mobility mass spectrometry, Edman degradation and cDNA methodologies.
Materials and Methods
1. Amphibian Skin Secretion
Adult specimens of H. punctatus were collected in Palmas, Tocantins, Brazil. All procedures in this study were approved and performed are under the license IBAMA 0637/91 AC. The frog skin glands were subjected to mild electrical stimulation (6 V during 1 min) and skin secretion was collected in ice-cold Milli-Q H2O, filtered, and freeze-dried and stored at −80°C, as described by Prates, et al 2004 [34], [35].
2. Peptide Purification
The crude extract was dissolved in aqueous 0.1% trifluoroacetic acid (TFA), then fractionated by RP-HPLC (Class VP Shimadzu Co., Japan) using a Vydac® 218TP510 semi-preparative C18 column (Grace Vydac® TP, USA). The fractions were eluted in a 120 minutes linear gradient of Solvent A (0.1% TFA in Milli-Q H2O) and Solvent B (0.1% TFA in acetonitrile) using a 2.5 mL ˙min−1 flow rate. Chromatographic fractions containing peptides of interest were submitted to further purification steps on analytical columns Vydac® 218TP54 C18 (Grace Vydac® TP, USA), Source™ 5 RPC ST (GE Healthcare, WI), and Shim-Pack-XR-ODS (30.0 mm×2.0 mm) C18 column on an Ultra-Fast-Liquid-Chromatographer (UFLC Prominence System Shimadzu Co., Japan). All the experiments were monitored at 216 and 280 nm (or 254 nm when phenylalanine residues were present), collected manually, frozen, lyophilized, and stored at −20°C.
3. Mass Spectrometry Analysis and Peptide Sequencing
The accurate molecular mass values of the peptides were determined by ESI-micrOTOF-Q II (Bruker Daltonics, Germany). The purity of each chromatographic fraction was evaluated by MALDI-TOF/MS (UltraFlex III, Bruker Daltonics, Germany) using close external calibration under reflector mode. Approximately 20 nM of lyophilized peptide was dissolved in Milli-Q H2O, mixed to a saturated solution of α-cyano-4-hydroxycinnaminic acid, spotted on a MALDI sample plate, and dried at room temperature. Peptide fragmentation was obtained by MALDI-TOF MS/MS experiments and the resulting data were analyzed manually using both Pepseq [36] and Flex Analysis 3.0 (Bruker Daltonics) softwares. The synthetic peptides were analyzed with a high-capacity ion trap LC/MSn system HCT Ultra ETD (Bruker Daltonics, Germany) equipped with a standard ESI ion source, with full scan data in UltraScan mode between m/z 100–3000, in positive mode, and with a fixed accumulation time of 30 ms. Additionally, amino acid sequencing was performed by the automated Edman degradation method on a PPSQ-23 protein peptide sequencer (Shimadzu Co., Japan).
4. Structural Studies on Ion Mobility Mass Spectrometry
Experiments were performed on a both MALDI Synapt G1 and ESI G2 HDMS instrument (Quadrupole Ion Mobility High-Definition mass spectrometry – Waters Co. MA, USA) equipped with nano-electrospray ionization source. All spectra were acquired by direct infusion of 1 µL·min−1 at a range of 300 up to 2000 m/z. Cross section calculations of Phenylseptin were conducted as described [37].
5. Nucler Magnetic Resonance (NMR)
The sample was prepared dissolving the peptide at 2 mM in a 60% TFE/D2O (v/v) solution. The NMR experiments were conducted at 20°C on a Bruker Avance III spectrometer (Bruker DRX-800) operating at 800 MHz for the 1H frequency. The Total Correlation Spectroscopy (TOCSY) spectra were acquired using the MLEV-17 pulse sequence. The spectral width was determined as 6,900 Hz, and the 512 t1 increments were collected with eight transients of 4,096 points. NOESY spectra were acquired using mixing times of 80, 100, 120, 140 and 160 ms. For this experiment, the spectral width was 6,900 Hz and the 512 t1 increments were collected with 16 transients of 4,096 points for each F1. The 1H–13C HSQC spectra were acquired with F1 and F2 spectral widths of 27,160 and 8,993 Hz respectively. The 400 t1 increments were collected with 56 transients of 1,024 points. The acquisitions were carried out in an edited mode allowing CH and CH3 correlations could show a positive phase and CH2 correlations, a negative phase. The 1H–15N HSQC spectra were acquired with F1 and F2 spectral widths of 27,160 and 8,993 Hz, respectively. Eighty t1 increments were collected (with 400 transients of 1,024 points), for each free induction decay. NMR spectra were analyzed using NMRVIEW, version 5.0.3. NOE intensities obtained at 120 ms mixing times were semi-quantitative converted into distance restraints using the calibration described by [38]. The highest intensity peaks corresponded to restraints with upper limit of 2.8 Å, the medium intensity with 3.4 Å, and the weakest peaks, with 5.0 Å. The backbone atoms chemical shift values were used to generate the dihedral angle restraints, using the TALOS+ software [39]. Structures were calculated (200) using these restraints on the Xplor-NIH v.2.17 software [40] by simulated annealing protocol. The structure calculation started with an extended model, with 14000 steps at high temperature, 6000 steps during cooling and a time step of 0.005 ps. The generated structures were then submitted to a refinement procedure, using again a simulated annealing calculation, using the Internal Variable Module [41] available in Xplor-NIH, with the Hydrogen-Bonding Database, Ramachandran Torsion-Angle Database and the Carbon Chemical Shift potentials. Among the refined structures, the 20 lowest-energy ones were selected to represent the model ensemble and then validated on Procheck on-line software [42].
6. Gene Cloning, cDNA Sequencing and Analysis
Total RNA from H. punctatus was extracted using the Trizol reagent according to a previously described protocol [23]. Reverse transcription of total RNA (1 µg) was performed with d(T)-anchor primer and the Superscript Reverse Transcriptase kit™ (Invitrogen), according to the manufacturer’s instructions. PCR reactions were performed using degenerate primers designed based on conserved signal peptide [24]. The amplified cDNA fragments were cloned into pGEM-T easy® (Promega, Maldison, WI, USA) and sequenced in both strands using an ABI Prism 3700 DNA Analyzer system with BigDye™ terminator and POP-5™ Polymer (Applied Biosystem Perkin Elmer). Comparisons of the cloned cDNA sequence with other GenBank™ database sequences were performed using the BLASTp software [43] from the NCBI databank (http://www.ncbi.nlm.nih.gov). The Conserved Domain Database search (CDD-Search) from the NCBI site was used to compare motif identity and similarity with known conserved domains [44]. Sequence alignments were obtained by CLUSTAL W software [45] and were edited using BIOEDIT [46]. The calculated molecular mass of deduced proteins and sequenced peptides were determined by the Protein Machine software available at the Expasy website (http://us.expasy.org/tools/).
7. Solid Phase Peptide Chemical Synthesis
The peptides L-Phes, D-Phes and one synthetic short-analog L-Phes.1 were manually synthesized by solid phase using the Fmoc/t-butyl strategy. An Fmoc-PAL-PEG-polystyrene resin was used for synthesis of the amidated C-terminal segment. Cleavage and final deprotection were conducted with a TFA:thioanisole:ethanedithiol:triisopropylsilane (91.5∶5∶2.5∶1, v:v:v:v) solution for 1 hour at room temperature. Peptide purification was performed through RP-HPLC with a Vydac 218TP1022 preparative column and purity was assessed by MALDI-TOF/MS.
8. Antimicrobial Assays
The antimicrobial activity of the peptides L-Phes and D-Phes were investigated using the bacterial strains Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 43300, and Escherichia coli ATCC 25992. In addition, we investigated the antimicrobial activity of the peptides towards a phytopathogenic bacterium Xanthomonas. axonopodis pv glycines ISBF 327 obtained from Embrapàs microorganism collection. All microorganisms were grown in stationary culture at 37°C and after that were transferred to Mueller-Hinton liquid medium, according to the National Committee for Clinical and Laboratory Standards (NCCLS) to perform the bioassays (NCCLS Institute protocols). The peptide was dissolved up to 8-fold in Mueller-Hinton liquid broth. The highest peptide concentration used in the assay was 131 µM in an initial inoculum of 2.5.105 cfu·mL−1 (colony-forming units/mL). The final volume was 100 µL per well, 50 µL of the peptide and 50 µL of the inoculum. The experiment was carried out in stationary culture at 37°C, and the spectrophotometer readings were performed 12 h after incubation. The minimal inhibitory concentration (MIC) was determined based on three independent measurements, using the optical density parameter (A595 nm). Conventional antibiotics (ampicillin and chloramphenicol) had their minimum inhibitory concentrations determined against the three experimental bacterial strains.
9. Behavioral Gustatory Assays
Male, adult (8–10 weeks old) mice on a C57BL/6 background were used for behavioral assays. Animals used in this study included littermates generated from mice heterozygous for a partial deletion of the Trpm5 gene, as previously described [47], originally bred from mice generously donated by C. S. Zuker (UCSD, San Diego, CA). Genotype was confirmed by PCR amplification using specific primers for Trpm5 gene (forward 5′-ATTCTAGAGCCCACCCGCCCCATC-3′ and reverse 5′-TTCACCTGCCCAGCCCTCATCTAC-3′) and for PGK-neor cassette that replaced exons 15 to 19 encoding the first five transmembrane domains of Trpm5 in mutant animals (forward 5′-TTGCACGCAGGTTCTCCGGC-3′ and reverse: 5′-TAGAAGGCGATGCGCTGCGA-3′). Heterozygous mice were identified by amplification of both the Trpm5 gene and PGK-neor cassette; knockout mice were identified by the absence of Trpm5 gene but amplification of PGK-neor cassette; and wild-type animals by absence of PGK-neor cassette but positive Trpm5 gene amplification. Behavioral experiments were conducted in two mouse behavior chambers enclosed in a ventilated and sound-attenuating cubicle (Med Associates Inc., St. Albans, VT). Each chamber was equipped with two slots for sipper tubing placement in symmetrical locations of one of the walls. All slots were equipped with licking detection devices (contact lickometers, Med Associates Inc., St. Albans, VT, USA) performing with a 10 ms resolution. All experiments were conducted in accordance with the J.B. Pierce Laboratory and Yale University regulations on usage of animals in research. All procedures performed in this study were approved by the Animal Care and Use Committee of The J. B. Pierce Laboratory.
9.1 Controls and stimulus concentrations
As a positive control, well known plant bitter alkaloids were used: caffeine, theophylline, and theobromine. Stimulus concentrations were based on previous studies, which reported, in rats, that the threshold for integrated cranial nerve responses is 10 mM for the glossopharyngeal nerve and approximately 10–30 mM for the chorda tympani [48]. Accordingly, we used 10–20 mM (as well as the sub-threshold concentration of 5 mM) in our studies to evaluate the role of Trpm5 in generating behavioral responses to caffeine. Finally, to allow for comparisons between behavioral responses across controls on a molar basis, we have used these same concentrations in theophylline and theobromine experiments. As a negative control we used free amino acid mixtures corresponding to the peptide primary structures. Stimulus concentrations were selected based on experiments conducted by Maehashi and Huang, 2009, where it was shown that a 10–20 mM threshold of free-amino acid is able to display bitter response in mice and rats. For the peptides L-Phes, D-Phes, and L-Phes.1 we considered the same concentrations as starting points.
9.2. Short-term two-bottle preference tests
Once habituated to the behavioral chamber, each animal was presented with two bottles, contained either water or the stimulus solution prepared in distilled water. Animals were given free access to both bottles during the experiment (10 minutes). The number of licks for each sipper was recorded and used to calculate the preference ratios (see below). To reduce confounds associated with side-biases, mice were tested in each condition for four consecutive days with daily inversion of bottle positions. The average preference ratio across testing days was then calculated for each animal. Animals had been water-deprived for 16 hours previous to the beginning of each session.
9.3. Data analysis
Results from data analyses were expressed as mean ± SD. Analyses of behavioral data were performed with the custom software Origin Professional (v8, OriginLab, Northampton, MA) using 2-way or 1-way ANOVAs followed by two-sample or independent one-sample t-tests. Bonferroni corrections for multiple comparisons were performed whenever appropriate.
9.4. Lick response and preference measures
All two-bottle preference tests were analyzed by calculating the preference ratios as:
Where n(.) denotes the total number of licks counted on a given sipper during a session. These values were subjected to a 2-way ANOVA genotype × stimulus concentration model and tested against 0.5, which is the reference value meaning indifference with respect to water.
Results
1. Phenylseptins on H. punctatus Skin Secretion
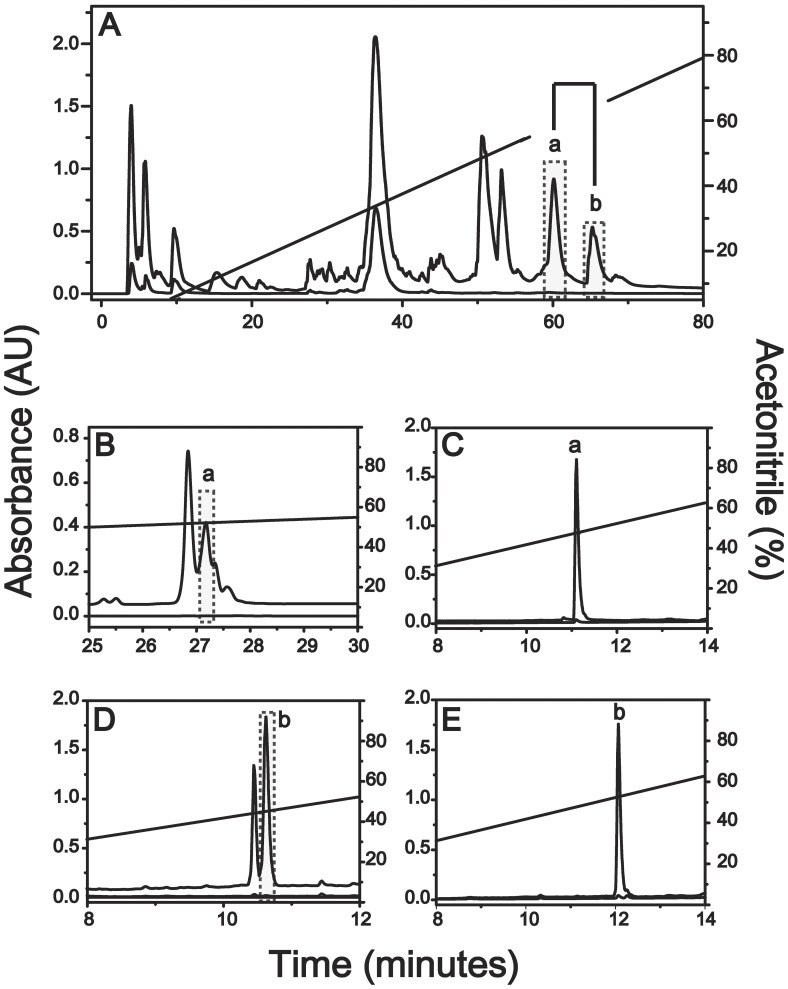
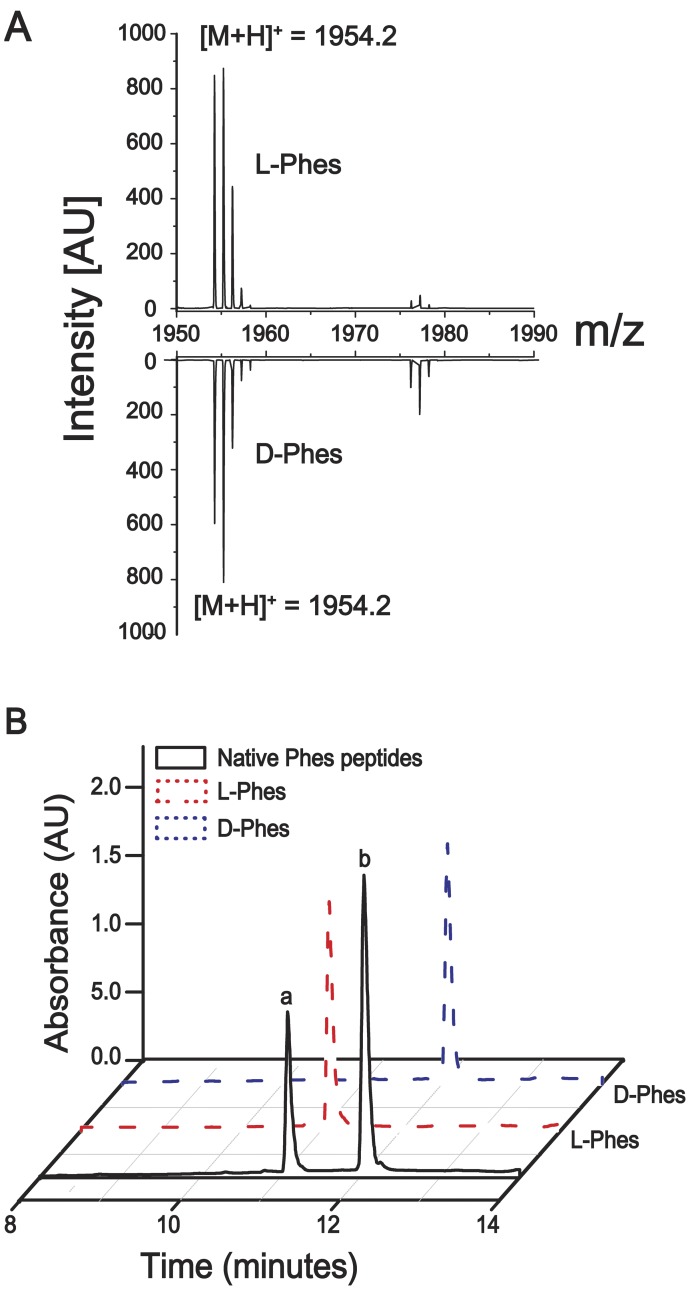
The crude skin secretion was fractionated by semi-preparative reverse-phase HPLC and mass analyzed by both MALDI TOF-TOF/MS and ESI Q-TOF/MS systems yielding about 20 major fractions (Figure 1A) from which, components “a” and “b” eluted at 61.0 and 64.3 minutes, respectively, showed identical molecular masses and MS/MS spectra. Each of the fractions was submitted to further purification steps on analytical scale to attain homogeneous peptide samples (Figure 1B–1E), confirmation of their distinct retention times, MALDI-TOF/MS monoisotopic molecular mass analyses (M+H+ = 1954.20 Da, Figure 2A) and primary structure elucidation from their corresponding MS/MS data (Figure S1). For both components (a and b), de novo sequencing based on the resulting sets of undistinguishable daughter ion series revealed 100% similarity between the amino acid sequences of the two components (see Table S1) bearing the following 18 residue polypeptide chain: FFFDTLKNLAGKVIGALT-NH2. Edman degradation of the intact components and their respective tryptic fragments excluded typical MS/MS data ambiguities regarding leucine, isoleucine, lysine and glutamine residues (Table S2). Moreover, the final sequence confirmation of both peptide and Thr18 C-terminal amidation was achieved by cDNA sequencing as shown in Figure 3 and ESI Q-TOF/MS exact mass of M+2H+ = 977.561 m/z (1.0 ppm) determinations (Figure S2).
Figure 1. Phenylseptin peptides present on H. punctatus skin secretion.
Chromatographic profile of H. punctatus crude extract and Phenylseptin isolation. (A) The crude extract was eluted with 0.1% TFA (Solvent A) and 95% acetonitrile containing 0.1% TFA (Solvent B), under a linear gradient of solvent B for 120 minutes in a semi-preparative C18 column. The identified Phenylseptin peptides ‘a’ and ‘b’ (B and D, respectively) were further purified by reverse-phase polystyrene/divinyl benzene chromatography using a 5 RPC ST 4.6 mm/150 mm column under optimized gradient of acetonitrile at a flow rate of 1.0 mL·min-1. After these two separation steps, final purification of the peptides ‘a’ and ‘b’ (C and E, respectively) was obtained using Ultra Fast Liquid Chromatography using a Shimpack-XR-ODS column under a linear gradient of acetonitrile at a flow rate of 0.4 mL·min-1. In all steps of purification the absorbance was measured at 216 nm, and when necessary at 254 or 280 nm.
Figure 2. UFLC analysis and molecular mass determination of Phenylseptin mixture.
(A) The accurate molecular masses and purity of Phes peptides were determined by MALDI-TOF/MS and the observed molecular mass was 1954.2 Da for both molecules. (B) Analytical chromatographic profile of natural (black line) and synthetic L-Phes (red dash line) and D-Phes (blue dash line). The two peptides were mixed in similar molar concentrations and load into an Ultra Fast Liquid Chromatography using a Shimpack-XR-ODS column under a linear gradient of acetonitrile at a flow rate of 0.4 mL·min-1. The two distinct fractions eluted around 11 and 12 minutes corresponded to L-Phes and D-Phes, respectively.
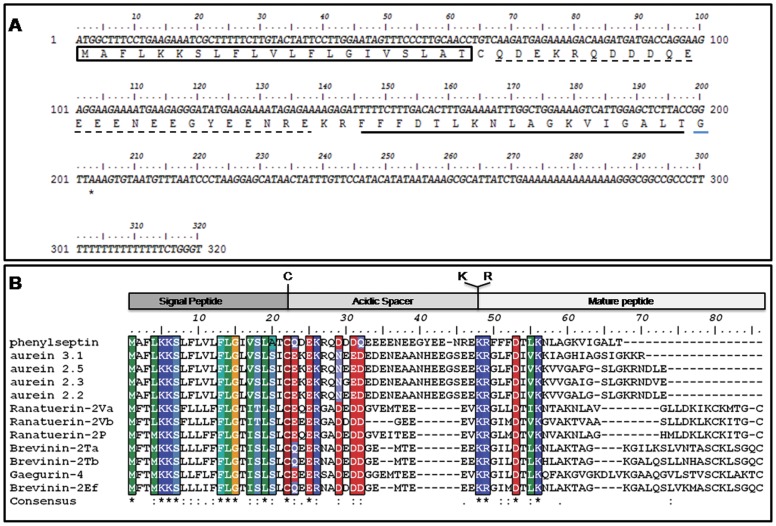
Figure 3. One single gene encoding Phes peptide.
(A) Nucleotide sequences of clone encoding precursor of selected Phenylseptin peptides. The putative signal peptide (in box), acidic spacer (dash underline) and mature peptide (bold underline), C-terminal codon for Glycine (blue underline) and stop codon (asterisk) are indicated. The nucleotide sequences were deposited in the NCBI Nucleotide Sequence Database under HQ012497 annotated accession code. (B) Predicted amino acid sequence alignment of Phenylseptin with previously sequenced Hylidae peptides aurein, ranateurin, brevinin and gaegurin. Sequence alignments were done using CLUSTAL W software and were edited with the BIOEDIT software.
2. One Single Gene Encoded Peptide
RT-PCR amplification produced various 320-bp fragments that revealed one single nucleotide sequence encoding a canonical antimicrobial pre-pro-peptide precursor (18) containing a deduced 18 residues peptide sequence (Figure 3A) identical to the previously obtained by mass spectrometry and Edman degradation. The pre-pro-peptide precursor also showed primary structure identity with antimicrobial sequences previously deposited in GenBank™ from three different Ranidae species, Rana rugosa (37% gaegurin), Rana temporaria (48% brevinin-2Ta and 46% 2Tb), Rana esculenta (50% brevinin-2Ef), and from one Hylidae specie, Litorea aurea (54% aurein 3.1; 56% aurein 2.5 and aurein 2.3; 57% aurein 2.2) (Figure 3B). The unusual triple phenylalanine residues at the N-terminal of the two putative antimicrobial peptides, their discrepancies in retention times by RP-HPLC and the confirmed complete sequence identity brought us to the hypothesis of a possible epimerization of at least one amino acid residue along the polypeptide chain, hence the name Phenylseptins for the two peptide analogs. The gene sequence was deposited under the genebank (gb|) accession number HQ012497.
3. D-phenylalanine is Present in Native D-Phes
Reverse-phase UFLC on a Shim-Pack-XR-ODS (2.0 mm i.d. × 30.0 mm) C18 column analyses determined that Phes ‘a’ was associated with a retention time of 11 minutes, and Phes ‘b’ with a retention time of 12 minutes (Figure 2B – black line). After enzymatic digestion with immobilized trypsin, UFLC analyses were performed to determine the retention times of all fragments from of these peptides (Figure S3 and Table S2). These analyses demonstrated that the fragments 1–7, corresponding to Phes ‘b’ N-terminal region (FFFDTLK; M+H+ = 917.55 Da) showed almost one minute difference in retention time when compared with the same fragment in Phes ‘a’. In order to investigate the presence of a D-phenylalanine residue at the second N-terminal position as suggested by previous related works [49]–[52], analogs containing [D-Phe2]-Phes and [L-Phe2]-Phes (henceforth D-Phes and L-Phes, respectively) were synthesized. After RP-UFLC native Phes ‘a’ co-eluted with L-Phes, while native Phes ‘b’ co-eluted with D-Phes (Figure 2B). In order to verify the presence of a D-phenylalanine residue at the N-terminal fragment, three possible analogues have been synthetized: [D-Phe1]-Phes, [D-Phe2]-Phes and [D-Phe3]-Phes. The three analogues (M+3H+977.7) were analyzed by RP-UFLC-MS and the individual retention time for each peptide was confirmed demonstrating that D-Phe2 analogue has similar retention time (over 12 min.) as the naturally occurring D-Phes peptide. (Figure S4).
4. Conformational Studies of Phenylseptin Analogs by Ion Mobility Mass Spectrometry (IM-MS)
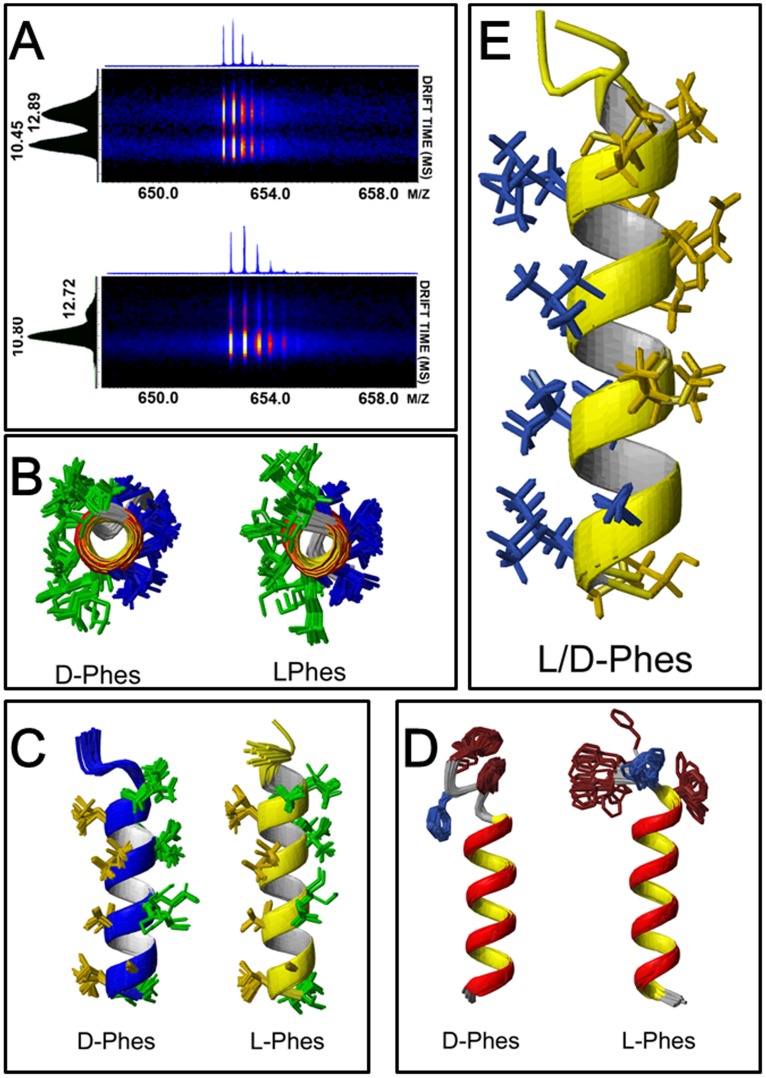
In order to investigate possible conformational attributes of both Phenylseptins, these molecules were submitted to IM-MS analyses revealing at least two conformers (M+3H+ = 652.04 m/z) with comparable intensities for L-Phes and a major one for D-Phes with determined drift times of 10.45, 12.89 and 10.80 ms, respectively (Figure 4A). These results suggest higher degrees of freedom for the L-Phes structure in the gas phase in comparison to the D-Phes molecule.
Figure 4. Structural studies on L- and D- Phenylseptin peptide isomers by Ion Mobility Mass Spectrometry (IM-MS) and Nuclear Magnetic Resonance (NMR).
(A) L-Phes and D-Phes were individually analyzed showing that each one (M+3H+ = 652.04 m/z) can assume at least two major conformations with distinct amounts of each type. L-Phes conformations at 10.45 and 12.89 ms and D-Phes conformations at 10.80 and 12.72 ms. D-Phes has its major conformation at 10.80 ms. Experiments were performed on a Synapt HDMS instrument (Quadrupole Ion Mobility High-Definition mass spectrometry – Waters Co. MA, USA) equipped with nano-electrospray ionization. All spectra were acquired with a direct infusion of 1 µL·min-1 of in a range m/z 300 up to 2000. Precursor charge state: 3. Tolerance: 0.1 Da. (B) and (C) The 20 lowest-energy structures for both peptides. The hydrophobic residues are represented in gold yellow, the hydrophilic residues in green. (D) The lowest-energy Phenylseptins showing Phenylalanine enatiomerization, the aromatic phenylalanine are in dark red and (E) The alignment of lowest-energy L- and D-Phes structure in the presence of 60% TFE viewed along the helix axis and from the side.
5. NMR Structures of the Phenylseptins
Both NMR structures of L- and D-Phenylseptins at 60% TFE were produced using total sets of 361 and 373 NOEs, respectively (see Table 1 for details). NOEs correlations at the peptides three phenylalanine N-terminal region indicated significant structural differences between L- and D-Phes most likely due to Phe2 enantiomerization (Figure 4). Nevertheless strong NOEs correlations of the sequential dNN(i,i+1) and dαN(i,i+3) obtained for the rest of the molecule (residues 4–18) indicated classical α-helical conformations at that region for both peptides (Figure 4F–4G). The Root Mean Square Deviation (RMSD) analyses of all backbone atoms for individual structures were 0.53±0.21 for L-Phes and 0.38±0.18 for D-Phes, and no distance violations greater than 11 was observed. The RMSD values for segment 4–17 were similar between L-Phes and D-Phes (0.22±0.09 and 0.22±0.08, respectively), while L-Phes showed 0.40±0.22 for the1Phe-3Phe segment and D-Phes showed 0.08±0.06. In the calculated D-Phes structure it was observed that Phe1 and Phe3 aromatic rings have significant interactions whilst Phe2 share NOEs with the peptide side chain with Leu6 producing a more structured N-terminal than in L-Phes (Figure S5).
Table 1. Summary of the structural restraints and statistical analysis of the calculated structures of L/D-Phes in TFE/D2O 60:40 v/v.
| Structural Statisticsa | L-Phes | D-Phes |
| Experimental restraints b | ||
| Distance restrains | ||
| Intraresidue (i–j = 0) | 185 | 184 |
| Sequential (|i–j| = 1) | 80 | 82 |
| Medium range (2≤|i–j|≤4) | 66 | 78 |
| Long range (|i–j|≥5) | 0 | 1 |
| Dihedral restrains | ||
| Torsion angle (phi/psi) | 30 | 28 |
| Total number of restrains | 361 | 373 |
| Restrains statistics | ||
| NOE violations >0.5 Å | 11 | 8 |
| Dihedral violations >5° | 3 | 1 |
| CNS energies (Kcal/mol) | ||
| Etotal | 238.54±5.59 | 37.66±4.56 |
| RMSD from average for residues 1–18 (Å) c | ||
| Backbone N, CA, C′ | 0.51±0.17 | 0.44±0.12 |
| Heavy atoms | 1.55±0.40 | 1.01±0.10 |
| RMSD from average for residues 1–3 (Å) c | ||
| Backbone N, CA, C′ | 0.47±0.22 | 0.24±0.18 |
| RMSD from average for residues 4–17 (Å) c | ||
| Backbone N, CA, C′ | 0.20±0.08 | 0.20±0.06 |
| Ramachandran plot d | ||
| Most favored regions (%) | 85.7 | 99.6 |
| Additional allowed regions (%) | 0 | 0.4 |
| Generously allowed regions (%) | 8.9 | 0 |
| Disallowed regions (%) | 5.4 | 0 |
The statistics was obtained from an ensemble of 20 lowest-energy vacuum-refined structures for both peptides (L/D-Phes).
Restraint statistics reported for unique, unambiguous assigned NOEs.
Coordinate precision is given as pair-wise Cartesian coordinate over the ensemble Root Mean Square Deviations from the average structure.
Values obtained from the PROCHECK-NMR analysis over all residues including the first seven highly flexible residues.
6. Phenylseptins Display Different Antimicrobial Activities
In vitro antimicrobial tests revealed that both peptides exhibited distinct levels of activities against pathogenic S. aureus (ATCC 29313), E. coli (ATCC 25922) and P. aeruginosa (ATCC 27853). Overall, D-Phes was more effective than L-Phes and Magainin (classical positive control) [15] against these pathogenic bacteria but less active than DS01 [53], which was used as the most potent antimicrobial peptide (our groups positive control) (Table 2). When tested against the soybean phytopathogen, Xanthomonas axonopodis pv glycines, D-Phes was found to be 8-fold more effective than L-Phes (4.1 and 32.7 mM, respectively).
Table 2. Antimicrobial activity of L/D-Phes.
| Microorganism | MIC (µM)a | |||||
| L-Phes | D-Phes | Mg | DS01 | Amp. | Chloram. | |
| S. aureus ATCC 29313 | 65.5 | 32.7 | ND | 26,5 | <11 | ND |
| E. coli ATCC 25922 | 65.5 | 65.5 | 79 | 6,6 | 46 | 25 |
| P. aeruginosa ATCC 27853 | >130 | 130 | 157.8 | NT | 25 | 25 |
| X. axonopodis pv glycines ISBF 327 a | 32.7 | 4.1 | 13 | <1.42 | ND | ND |
MIC -minimal peptide concentrations required for total inhibition of cell growth.
ND - antimicrobial activity not detectable;
Phytophatogenic bacteria; Amp and Chloram- ampicillin and chloramphenicol; Mg and DS01 - control peptides Magainin (Zasloff, et al and Brand, et al).
7. Bitterness Motif Hypothesis
Analyses of Phenylseptin sequences revealed that their Phe-rich N-terminals share sequence similarities with the bitter Phe-Phe-Phe motifs shown in Figure 5 [54]. Therefore, we hypothesized that the triple-phenylalanine N-terminal sequences found in the two peptides would also produce similar aversive behavioral responses to those produced upon stimulation of oral bitter taste sensors.
Figure 5. Phenylseptin Phe-Phe-Phe motif.
Phenylseptin N-terminal sequence was compared to other peptides previously described on the literature as being bitter. Sequence alignments were edited with the BIOEDIT software. On the right side a bitterness scale based on Kim, et al., 2006 analysis.
8. Gustatory Preference Tests Involving Prototypical Bitters in Trpm5 KO and WT Mice
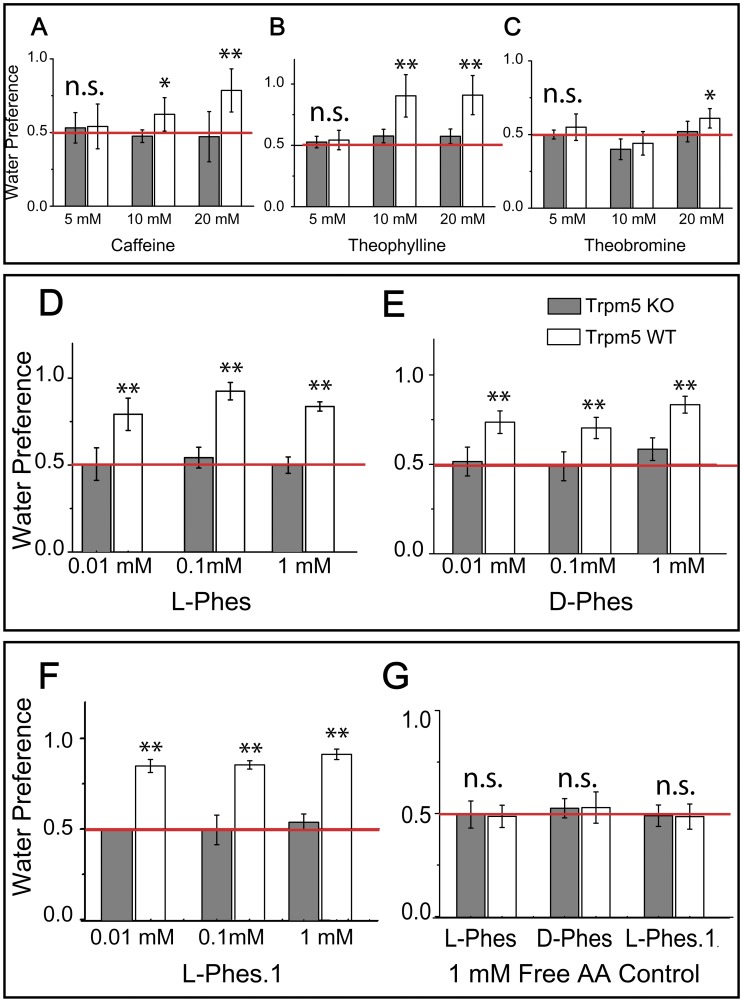
The methylxanthines caffeine, theophylline and theobromine are prototypical bitter tastants and were used as positive controls for bitter detection in WT animals and bitter-insensitivity in KO animals at 5, 10 and 20 mM concentrations. For the caffeine experiments, we designed a mixed-model two-way stimulus concentration × genotype ANOVA × which revealed, as expected, a robust effect of genotype (p<0.0003) on preference ratios. Similar analyses performed for theophylline revealed equally robust effects of genotype on preference ratios (p<0.001). In addition, these ANOVAs revealed significant stimulus concentration × genotype interactions (p<0.02 and p<0.008; caffeine and theophylline, respectively). The interaction effects prompted repeated measures one-way ANOVA models for each genotype separately (Table S3), which revealed a significant main effect of stimulus concentration on preference ratios in WT (p<0.02 and p<0.01, caffeine and theophylline, respectively), but not in KO mice (p = 0.4 and p = 0.95, caffeine and theophylline, respectively). For theobromine this analysis revealed neither significant main effect of genotype (p = 0.17) nor interaction (p = 0.4) on preference rations, although it was the only stimuli to show a significant effect of stimulus concentration (p<0.02). One-way ANOVAs performed for each genotype separately confirmed a significant effect of theobromine concentrations in WT (p<0.04), but not in KO (p = 0.29). Post-hoc analyses of the individual preference ratios for each stimulus confirmed that, while WT mice displayed significantly higher preferences for water against 10–20 mM caffeine, 5-10-20 mM theophylline and 20 mM theobromine, KO mice were uniformly indifferent to all choices. Our findings so far show that, in 10 min-long two-bottle preference tests, Trpm5 KO mice are indifferent all concentrations of methylxanthines, while some concentrations (10–20 mM caffeine; 5–10–20 mM theophylline; 20 mM theobromine) are robustly avoided by WT mice. All control results are depicted in Figure 6A–6C and Table S3.
Figure 6. Trpm5 dependent bitter substances.
Short-term (10 min) (A) caffeine vs. water, (B) theophylline and (C) theobromine two-bottle preference tests. Trpm5 knockout (“KO”) mice displayed indifference to all choices. (D) The bitterness of Phenylseptin peptides is dependent on Trpm5 regulation. Short-term (10 min) (E) L-Phes vs. water, (F) D-Phes vs. water and (3) L-Phes.1 vs. water two-bottle preference tests. Trpm5 knockout (“KO”) mice displayed indifference to all choices. Values displayed as preference ratios for water. (G) Amino acid organization on peptide structure is fundamental to gustatory perception. Short-term (10 min) L-Phes mixture vs. water and D-Phes mixture vs. water. Wild-type and Trpm5 knockout (“KO”) mice were equally indifferent to all choices. Values displayed as preference ratios for water. Values displayed as preference ratios for water. Post-hoc (Bonferroni-corrected) one sample t-test against indifference ratio of 0.5: *p<0.05; **p<0.04; all other comparisons p>0.05. Red line denotes level of indifference (preference ratio of 0.5).
9. Gustatory Preference Tests Involving Phenylseptins in Trpm5 KO and WT Mice
After methodology validation, we tested both Phenylseptin peptides and one shorter analog. The analog L-Phes.1 was used as a control to investigate the importance of the full peptide sequence. We reasoned that the peptide N-terminal might be a crucial modulator of bitterness potency in these peptides. The peptides were tested at 0.01 mM, 0.1 mM and 1 mM in behavioral tests identical to those previously described. WT (N = 7) and KO (N = 7) mice were exposed to 10-min two-bottle preference tests where choice was given between a Phes solution and (distilled) water. Analysis of two-way genotype × peptide concentration ANOVA designed as above revealed a robust effect of genotype for L-Phes (F1,12 = 31.9, p<0.0002), D-Phes (F1,12 = 21.2, p<0.0006) and L-Phes.1 (F1,12 = 76.1, p<0.0005), on preference ratios. Conversely, no concentration effects were detected for L-Phes (F1,12 = 1.12, p = 0.31), D-Phes (F1,12 = 1.32, p = 0.27) or L-Phes.1 (F1,12 = 1.23, p = 0.30). Also, no significant peptide concentration × genotype interactions for L-Phes (F1,12 = 0.29, p = 0.75), D-Phes (F1,12 = 1.32, p = 0.27) or L-Phes.1 (F1,12 = 0.04, p = 0.84), on the preference ratios. Pairwise comparisons using post-hoc Tukey tests showed that even in low concentrations WT mice rejected the peptides, while KO mice remained indifferent. As observed for the control experiment with methylxanthines, post-hoc analyses on individual preference ratios of peptides confirmed that WT mice demonstrated high preference for water, while KO mice were indifferent to the choices given. Specifically, WT mice have shown significant higher aversion against all concentrations of L-Phes, D-Phes and L-Phes.1 (one-tailed t-tests, p<0.05; Figure 6D–6F). In summary, we attribute the aversive oral properties of the Phenylseptins tested entirely to their gustatory, possibly bitter-like, properties, given the strong impairments observed in mice lacking the taste ion channel TRPM5.
10. Peptide Primary Structure and Gustatory Perception
1 mM of each amino acid free present in L-Phes and D-Phes sequences were mixed and the resulting solutions were offered to the mice in two-bottle preference tests as above. The two-way ANOVA free-amino acid solution X genotype model showed no significant effect for L-Phes (p = 0.20), D-Phes (p = 0.19) and L-Phes.1 (p = 0.23), or interactions (L-Phes p = 0.37, D-Phes p = 0.41, and L-Phes.1 p = 0.35) on preference ratios for both preparations. One-way ANOVAs were performed separately for each genotype using the amino acid preparation as factor. There were no effects on preference ratios in either WT or KO mice. In fact, as expected for the tested concentration, both WT and KO mice were equally indifferent to 1 mM free amino acid solution (Figure 6G), indicating the importance of the primary structure for the activity.
Discussion
The present work reports a novel peptide named Phenylseptin (Phes), purified from the skin secretion of H. punctatus. This peptide naturally occurs in two different primary structures constituted by L-Phe2 and D-Phe2 (Figure 2 and S3) and C-terminal amidation at Thr18. Both sequences display distinct in vitro antimicrobial activities but similar sensorial aversive properties on mice. It was observed that the epimerization of the second N-terminal phenylalanine residue induced a 90° conformational change at the N-terminal of the D-Phes structure (Figure 4) and that could be responsible for the significant differences on the antimicrobial activities obtained for two peptides (Table 1). However, the N-terminal structural differences demonstrated for both molecules appear to cause no detectable response regarding micès gustatory perception to bitterness. These findings are consistent with previous works concerning amino acid epimerization in frog peptides such as dermorphins and deltorphins, as well as predators aversive taste behaviors to various amphibian skin secretions [16], [51], [55]–[58].
The Phenylseptins reported in this study were found to adopt at least two major structural conformations and so far, according to our cDNA library, both forms should be considered a single gene product synthesized as a highly conserved pre-pro-protein that yields the two mature polypeptides (L−/D-Phes) after enzymatic processing and corresponding post-translational modifications. Electrospray and MALDI (data not shown) ion mobility mass spectrometry experiments demonstrated the presence of multiple peptide ion (M+H+) conformers in the gas phase (Figure 4A), which were confirmed by 1H NMR in solution (Figure 4D). The amount of structural data that was unveiled in this occasion for such a small polypeptide demonstrates how subtle, complex and non-trivial peptide studies can be, and how easily one might be driven to ignore relevant structural and functional features, particularly when high throughput approaches tend to be overemphasized. Furthermore, the uncommon triple phenylalanine N-terminal of the Phenylseptins seems to share similar bitterness attributes observed in peptides released during food hydrolysis or aging process in fermented products [59]. The aversive gustative responses registered in normal mice caused by aqueous solutions with different molar concentrations of Phenylseptins are comparable to those constituted by the classical bitter alkaloids caffeine, theophylline and theobromine, when offered to the same animals (Figure 6). Therefore, it is plausible to infer that similar aversive activity could also be part of a general warning strategy against natural predators of H. punctatus, especially if one may ponder what was uttered with remarkable precision in De Anima by Aristotle centuries ago: “…taste also must be a kind of touch, because it is a sensation of that which is tangible and nutritive… Hence excess in tangible qualities destroys not only the sense-organ, but also the animal itself. For touch is the one sense that the animal cannot do without… It (the animal) has taste on account of what is pleasant and painful, to the end that it may perceive what is pleasant in food and feels desire and be impelled to movement.” (Aristotle, III. 13) [60].
These results strongly suggest that Phenylseptins may be part of a multifunctional pool of peptides and proteins produced by the skin glands of H. punctatus with at least two experimentally tested defensive roles: against pathogenic microorganism and a potential predator warning agent.
Supporting Information
MS/MS spectra assignment for L-Phes and D-Phes fragmented peptides. The observed molecular mass was 1954.2 Da. The peptides were fragmented by MALDI-TOF MS/MS experiments showing the same fragmentation profile. The resulting data were analyzed manually using both Pepseq (Waters Co.) and Flex Analysis 3.0 (Bruker Daltonics) programs. The primary sequence was confirmed by the automated Edman degradation method on a PPSQ-23 protein peptide sequencer (from Shimadzu Corp.). The loss of 0.98 Da on C-terminal threonine residue indicates the amidation.
(TIF)
Theoretical and experimental accurate molecular mass values of Phenylseptins with internal calibration determined by direct infusion on an ESI MicrOTOF-Q II mass spectrometer operating with a standard ESI probe. Mass accuracy of <1 ppm RMS error; M+2H+ = 977.651.
(TIF)
Enzymatic digestion of natural L-Phes and D-Phes with immobilized trypsin and UFLC analyses. (A) Analytical chromatographic profile of the digested peptides loaded onto an Ultra Fast Liquid Chromatography (UFLC-HPLC) using a Shimpack-XR-ODS column under a linear gradient of acetonitrile at a flow rate of 0.4 mL•min-1. L-Phes (red line) and D-Phes (blue line) generated 3 fragments each. Fragments 1/1′ and fragments 2/2′ were eluted at the same time, while fragments 3/3′ were eluted differently with a Δt = 0.9 min. (B) The molecular masses and purity of all fragments were determined by MALDI-TOF/MS (UltraFlex III, Bruker Daltonics, Germany) and the observed molecular mass was 524.1 Da for fragments 1/1′, 594.1 Da for fragments 2/2′ and 917.5 Da for fragments 3/3′.
(TIF)
UFLC analyses of natural D-Phes and its analogues. (A) Analytical chromatographic profile of the native (dash line) and synthetic (dark line) peptides loaded onto an Ultra Fast Liquid Chromatography (UFLC-HPLC) using a Shimpack-XR-ODS column under a linear gradient of acetonitrile at a flow rate of 0.4 m•min-1. L-Phes (red line) and D-Phes (blue line). (B) The molecular masses and purity of peptides were determined by ESI-HCT Ultra ETD (Iontrap, Bruker Daltonics, Germany) and the observed molecular mass was 977.7 Da for all analogues.
(TIF)
NOESY spectra acquired using mixing times of 160 ms from D-Phes showing Phe2 and Leu6 interaction. For this experiment, the acquisitions were carried out in 60% TFE/H2O (v/v).
(TIF)
(DOCX)
(DOCX)
(TIF)
Acknowledgments
The authors wish to express their special gratitude to Dr. M. P. Bemquerer, Dr. G. D. Brand and Dr. L.P. da Silva from Embrapa Recursos Genéticos e Biotecnologia, and Dr. Xueying Ren from the J. B. Pierce Laboratory for assistance in data collection, analysis and insightful discussions. MTQM was the recipient of a doctoral CNPq and The J.B.Pierce Foundation scholarship. E.coli, P. aeruginosa and S. aureus microorganisms were cultured at the Ecology and Physiology of Microorganisms Laboratory, Microbiology Department, Federal University of Minas Gerais (UFMG) and Centro Nacional de Ressonância Magnética Nuclear Jiri Jonas (CNRMN), Rio de Janeiro, Brazil.
Funding Statement
The J.B. Pierce Foundation scholarship and Conselho Nacional de Pesquisa e Desenvolvimento (CNPq). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Poyurovsky M, Weizman R, Weizman A (2011) Mirtazapine - a multifunctional drug: low dose for akathisia. CNS Spectr 16: 63. [DOI] [PubMed] [Google Scholar]
- 2. Sneader W (2000) The discovery of aspirin: a reappraisal. BMJ 321: 1591–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans G, Packham MA, Nishizawa EE, Mustard JF, Murphy EA (1968) The effect of acetylsalicyclic acid on platelet function. J Exp Med 128: 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trial TIS (1997) A randomised trial of aspirin, subcutaneous heparin, both, or neither among 19435 patients with acute ischaemic stroke. International Stroke Trial Collaborative Group. Lancet 349: 1569–1581. [PubMed] [Google Scholar]
- 5. Minnerup J, Schabitz WR (2009) Multifunctional actions of approved and candidate stroke drugs. Neurotherapeutics 6: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandenburg L-O, Merres J, Albrecht L-J, Varoga D, Pufe T (2012) Antimicrobial Peptides: Multifunctional Drugs for Different Applications. Polymers 4: 539–560. [Google Scholar]
- 7. Rowe C, Guilford T (1996) Hidden colour aversions in domestic chicks triggered by pyrazine odours of insect warning displays. Nature 383: 520–522. [Google Scholar]
- 8. Rowe C (1999) Receiver psychology and the evolution of multicomponent signals. Anim Behav 58: 921–931. [DOI] [PubMed] [Google Scholar]
- 9. Skelhorn J, Rowe C (2005) Frequency-dependent taste-rejection by avian predation may select for defence chemical polymorphisms in aposematic prey. Biol Lett 1: 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, et al. (2011) Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci U S A 108: 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Boer JG, Posthumus MA, Dicke M (2004) Identification of volatiles that are used in discrimination between plants infested with prey or nonprey herbivores by a predatory mite. J Chem Ecol 30: 2215–2230. [DOI] [PubMed] [Google Scholar]
- 12. Lee S, Badieyan S, Bevan DR, Herde M, Gatz C, et al. (2010) Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc Natl Acad Sci U S A 107: 21205–21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duellman WE, Trueb L (1986) Biology of amphibian. Baltimore and London: Johns Hopkins Paperbacks edition. 670 p.
- 14. Erspamer V, Vialli M (1951) Presence of enteramine in the skin of Amphibia. Nature 167: 1033. [DOI] [PubMed] [Google Scholar]
- 15. Zasloff M (1987) Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A 84: 5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kreil G, Barra D, Simmaco M, Erspamer V, Erspamer GF, et al. (1989) Deltorphin, a novel amphibian skin peptide with high selectivity and affinity for delta opioid receptors. Eur J Pharmacol 162: 123–128. [DOI] [PubMed] [Google Scholar]
- 17. Amiche M, Delfour A, Nicolas P (1998) Opioid peptides from frog skin. EXS 85: 57–71. [DOI] [PubMed] [Google Scholar]
- 18. Roper TJ, Redston S (1987) Conspicuousness of Distasteful Prey Affects the Strength and Durability of One-Trial Avoidance-Learning. Animal Behaviour 35: 739–747. [Google Scholar]
- 19. Darst CR, Cummings ME (2006) Predator learning favours mimicry of a less-toxic model in poison frogs. Nature 440: 208–211. [DOI] [PubMed] [Google Scholar]
- 20. D’Heursel A, Haddad CFB (1999) Unpalatability of Hyla semilineata tadpoles (Anura) to captive and free-ranging vertebrate predators. Ethology Ecology and Evolution 11: 339–348. [Google Scholar]
- 21.Manteifel YB, Reshetnikov AN (2002) Avoidance of noxious tadpole prey by fish and invertebrate predators: adaptivity of a chemical defence may depend on predator feeding habits. Stuttgart, ALLEMAGNE: Schweizerbart.
- 22. Gunzburger MS, Travis J (2005) Critical Literature Review of the Evidence for Unpalatability of Amphibian Eggs and Larvae. Journal of Herpetology 39: 547–571. [Google Scholar]
- 23. Brand GD, Leite JR, de Sa Mandel SM, Mesquita DA, Silva LP, et al. (2006) Novel dermaseptins from Phyllomedusa hypochondrialis (Amphibia). Biochem Biophys Res Commun 347: 739–746. [DOI] [PubMed] [Google Scholar]
- 24. Brand GD, Krause FC, Silva LP, Leite JR, Melo JA, et al. (2006) Bradykinin-related peptides from Phyllomedusa hypochondrialis. Peptides 27: 2137–2146. [DOI] [PubMed] [Google Scholar]
- 25. Leite JR, Silva LP, Rodrigues MI, Prates MV, Brand GD, et al. (2005) Phylloseptins: a novel class of anti-bacterial and anti-protozoan peptides from the Phyllomedusa genus. Peptides 26: 565–573. [DOI] [PubMed] [Google Scholar]
- 26. Simmaco M, Mignogna G, Barra D (1998) Antimicrobial peptides from amphibian skin: what do they tell us? Biopolymers 47: 435–450. [DOI] [PubMed] [Google Scholar]
- 27. Mor A, Hani K, Nicolas P (1994) The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J Biol Chem 269: 31635–31641. [PubMed] [Google Scholar]
- 28. Morikawa N, Hagiwara K, Nakajima T (1992) Brevinin-1 and -2, unique antimicrobial peptides from the skin of the frog, Rana brevipoda porsa. Biochem Biophys Res Commun 189: 184–190. [DOI] [PubMed] [Google Scholar]
- 29.Kreil G (1994) Antimicrobial peptides from amphibian skin: an overview. Ciba Found Symp 186: 77–85; discussion 85–90. [DOI] [PubMed]
- 30. Shai Y (1999) Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta 1462: 55–70. [DOI] [PubMed] [Google Scholar]
- 31. Apponyi MA, Pukala TL, Brinkworth CS, Maselli VM, Bowie JH, et al. (2004) Host-defence peptides of Australian anurans: structure, mechanism of action and evolutionary significance. Peptides 25: 1035–1054. [DOI] [PubMed] [Google Scholar]
- 32. Moni RW, Romero FS, Daly JW (1995) The amphiphilic peptide adenoregulin enhances agonist binding to A1-adenosine receptors and [35S]GTP gamma S to brain membranes. Cell Mol Neurobiol 15: 465–493. [DOI] [PubMed] [Google Scholar]
- 33. Li J, Zhang C, Xu X, Wang J, Yu H, et al. (2007) Trypsin inhibitory loop is an excellent lead structure to design serine protease inhibitors and antimicrobial peptides. FASEB J 21: 2466–2473. [DOI] [PubMed] [Google Scholar]
- 34. Prates MV, Sforca ML, Regis WC, Leite JR, Silva LP, et al. (2004) The NMR-derived solution structure of a new cationic antimicrobial peptide from the skin secretion of the anuran Hyla punctata. J Biol Chem 279: 13018–13026. [DOI] [PubMed] [Google Scholar]
- 35. Andrews EJ, Bennett BT, Clark JD, Houpt KA, Pascoe PJ, et al. (1993) 1993 Report of the AVMA Panel on Euthanasia. J Am Vet Med Assoc 202: 229–249. [PubMed] [Google Scholar]
- 36.Garrett RH, Grisham CM (1995) Biochemistry; Saunders College Publishing S, editor.
- 37. Ruotolo BT, McLean JA, Gillig KJ, Russell DH (2005) The influence and utility of varying field strength for the separation of tryptic peptides by ion mobility-mass spectrometry. Journal of the American Society for Mass Spectrometry 16: 158–165. [DOI] [PubMed] [Google Scholar]
- 38. Hyberts SG, Goldberg MS, Havel TF, Wagner G (1992) The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures. Protein Sci 1: 736–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clore GM, Schwieters CD (2006) Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. J Mol Biol 355: 879–886. [DOI] [PubMed] [Google Scholar]
- 41. Schwieters CD, Clore GM (2001) The VMD-XPLOR visualization package for NMR structure refinement. J Magn Reson 149: 239–244. [DOI] [PubMed] [Google Scholar]
- 42. Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067. [DOI] [PubMed] [Google Scholar]
- 43. Altschul SF, Lipman DJ (1990) Protein database searches for multiple alignments. Proc Natl Acad Sci U S A 87: 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marchler-Bauer A, Anderson JB, Cherukuri PF, Deweese-Scott C, Geer LY, et al. (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res 33: p.D192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2 3. [DOI] [PubMed]
- 46. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- 47. Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, et al. (2003) Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112: 293–301. [DOI] [PubMed] [Google Scholar]
- 48. Iwasaki Y, Satoh H, Ikeda H, Uchimura H, Ohkubo A, et al. (1981) An evaluation of factors which affect circulating thyroid hormone levels in liver diseases. Kaku Igaku 18: 601–608. [PubMed] [Google Scholar]
- 49. Kamatani Y, Minakata H, Kenny PT, Iwashita T, Watanabe K, et al. (1989) Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Ferussac containing a D-amino acid residue. Biochem Biophys Res Commun 160: 1015–1020. [DOI] [PubMed] [Google Scholar]
- 50. Ishida T, In Y, Doi M, Inoue M, Yasuda-Kamatani Y, et al. (1992) Crystal structure and molecular conformation of achatin-I (H-Gly-D-Phe-Ala-Asp-OH), an endogenous neuropeptide containing a D-amino acid residue. Int J Pept Protein Res 39: 258–264. [DOI] [PubMed] [Google Scholar]
- 51. Barra D, Mignogna G, Simmaco M, Pucci P, Severini C, et al. (1994) [D-Leu2]deltorphin, a 17 amino acid opioid peptide from the skin of the Brazilian hylid frog, Phyllomedusa burmeisteri. Peptides 15: 199–202. [DOI] [PubMed] [Google Scholar]
- 52. Mangoni ML, Papo N, Saugar JM, Barra D, Shai Y, et al. (2006) Effect of natural L- to D-amino acid conversion on the organization, membrane binding, and biological function of the antimicrobial peptides bombinins H. Biochemistry. 45: 4266–4276. [DOI] [PubMed] [Google Scholar]
- 53. Brand GD, Leite JR, Silva LP, Albuquerque S, Prates MV, et al. (2002) Dermaseptins from Phyllomedusa oreades and Phyllomedusa distincta. Anti-Trypanosoma cruzi activity without cytotoxicity to mammalian cells. J Biol Chem 277: 49332–49340. [DOI] [PubMed] [Google Scholar]
- 54. Min BS, Noh YJ, Shin JH, Baek SY, Min KI, et al. (2006) Assessment of the quantitative real-time polymerase chain reaction using a cDNA standard for human group A rotavirus. J Virol Methods 137: 280–286. [DOI] [PubMed] [Google Scholar]
- 55. Erspamer V, Erspamer GF, Severini C, Potenza RL, Barra D, et al. (1993) Pharmacological studies of ‘sapo’ from the frog Phyllomedusa bicolor skin: a drug used by the Peruvian Matses Indians in shamanic hunting practices. Toxicon 31: 1099–1111. [DOI] [PubMed] [Google Scholar]
- 56. Auvynet C, Seddiki N, Dunia I, Nicolas P, Amiche M, et al. (2006) Post-translational amino acid racemization in the frog skin peptide deltorphin I in the secretion granules of cutaneous serous glands. Eur J Cell Biol 85: 25–34. [DOI] [PubMed] [Google Scholar]
- 57. Erspamer V, Melchiorri P, Falconieri-Erspamer G, Negri L, Corsi R, et al. (1989) Deltorphins: a family of naturally occurring peptides with high affinity and selectivity for delta opioid binding sites. Proc Natl Acad Sci U S A 86: 5188–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Heck SD, Faraci WS, Kelbaugh PR, Saccomano NA, Thadeio PF, et al. (1996) Posttranslational amino acid epimerization: enzyme-catalyzed isomerization of amino acid residues in peptide chains. Proc Natl Acad Sci U S A 93: 4036–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim HO, Li-Chan EC (2006) Quantitative structure-activity relationship study of bitter peptides. J Agric Food Chem 54: 10102–10111. [DOI] [PubMed] [Google Scholar]
- 60.Aristotle (1907) De Anima (R. D. Hicks, Translation); Press CU, editor.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MS/MS spectra assignment for L-Phes and D-Phes fragmented peptides. The observed molecular mass was 1954.2 Da. The peptides were fragmented by MALDI-TOF MS/MS experiments showing the same fragmentation profile. The resulting data were analyzed manually using both Pepseq (Waters Co.) and Flex Analysis 3.0 (Bruker Daltonics) programs. The primary sequence was confirmed by the automated Edman degradation method on a PPSQ-23 protein peptide sequencer (from Shimadzu Corp.). The loss of 0.98 Da on C-terminal threonine residue indicates the amidation.
(TIF)
Theoretical and experimental accurate molecular mass values of Phenylseptins with internal calibration determined by direct infusion on an ESI MicrOTOF-Q II mass spectrometer operating with a standard ESI probe. Mass accuracy of <1 ppm RMS error; M+2H+ = 977.651.
(TIF)
Enzymatic digestion of natural L-Phes and D-Phes with immobilized trypsin and UFLC analyses. (A) Analytical chromatographic profile of the digested peptides loaded onto an Ultra Fast Liquid Chromatography (UFLC-HPLC) using a Shimpack-XR-ODS column under a linear gradient of acetonitrile at a flow rate of 0.4 mL•min-1. L-Phes (red line) and D-Phes (blue line) generated 3 fragments each. Fragments 1/1′ and fragments 2/2′ were eluted at the same time, while fragments 3/3′ were eluted differently with a Δt = 0.9 min. (B) The molecular masses and purity of all fragments were determined by MALDI-TOF/MS (UltraFlex III, Bruker Daltonics, Germany) and the observed molecular mass was 524.1 Da for fragments 1/1′, 594.1 Da for fragments 2/2′ and 917.5 Da for fragments 3/3′.
(TIF)
UFLC analyses of natural D-Phes and its analogues. (A) Analytical chromatographic profile of the native (dash line) and synthetic (dark line) peptides loaded onto an Ultra Fast Liquid Chromatography (UFLC-HPLC) using a Shimpack-XR-ODS column under a linear gradient of acetonitrile at a flow rate of 0.4 m•min-1. L-Phes (red line) and D-Phes (blue line). (B) The molecular masses and purity of peptides were determined by ESI-HCT Ultra ETD (Iontrap, Bruker Daltonics, Germany) and the observed molecular mass was 977.7 Da for all analogues.
(TIF)
NOESY spectra acquired using mixing times of 160 ms from D-Phes showing Phe2 and Leu6 interaction. For this experiment, the acquisitions were carried out in 60% TFE/H2O (v/v).
(TIF)
(DOCX)
(DOCX)
(TIF)