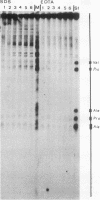
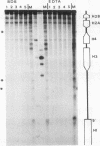
Abstract
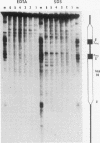
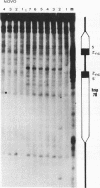
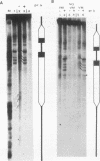
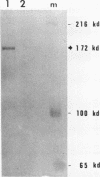
In the studies reported here we have used topoisomerase II as a model system for analyzing the factors that determine the sites of action for DNA-binding proteins in vivo. To localize topoisomerase II sites in vivo we used an inhibitor of the purified enzyme, the antitumor drug VM-26. This drug stabilizes an intermediate in the catalytic cycle, the cleavable complex, and substantially stimulates DNA cleavage by topoisomerase II. We show that lysis of VM-26 treated tissue culture cells with sodium dodecyl sulfate induces highly specific double-strand breaks in genomic DNA, and we present evidence indicating that these double-strand breaks are generated by topoisomerase II. Using indirect end labeling to map the cleavage products, we have examined the in vivo sites of action of topoisomerase II in the 87A7 heat shock locus, the histone repeat, and a tRNA gene cluster at 90BC. Our analysis reveals that chromatin structure, not sequence specificity, is the primary determinant in topoisomerase II site selection in vivo. We suggest that chromatin organization may provide a general mechanism for generating specificity in a wide range of DNA-protein interactions in vivo.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. L., Yang L., Rowe T. C., Halligan B. D., Tewey K. M., Liu L. F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Nov 10;259(21):13560–13566. [PubMed] [Google Scholar]
- DeLotto R., Schedl P. Internal promoter elements of transfer RNA genes are preferentially exposed in chromatin. J Mol Biol. 1984 Nov 15;179(4):607–628. doi: 10.1016/0022-2836(84)90158-x. [DOI] [PubMed] [Google Scholar]
- Desplan C., Theis J., O'Farrell P. H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988 Sep 23;54(7):1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Transcriptionally active chromatin is sensitive to Neurospora crassa and S1 nucleases. J Mol Biol. 1984 Nov 5;179(3):469–496. doi: 10.1016/0022-2836(84)90076-7. [DOI] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980 Aug;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Lee M. P., Sander M., Hsieh T. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. J Biol Chem. 1989 Dec 25;264(36):21779–21787. [PubMed] [Google Scholar]
- Liu L. F. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- Liu L. F. DNA topoisomerases--enzymes that catalyse the breaking and rejoining of DNA. CRC Crit Rev Biochem. 1983;15(1):1–24. doi: 10.3109/10409238309102799. [DOI] [PubMed] [Google Scholar]
- Muller M. T., Mehta V. B. DNase I hypersensitivity is independent of endogenous topoisomerase II activity during chicken erythrocyte differentiation. Mol Cell Biol. 1988 Sep;8(9):3661–3669. doi: 10.1128/mcb.8.9.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M., Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol Cell Biol. 1990 Jun;10(6):2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T. C., Wang J. C., Liu L. F. In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol Cell Biol. 1986 Apr;6(4):985–992. doi: 10.1128/mcb.6.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983 Jul 10;258(13):8421–8428. [PubMed] [Google Scholar]
- Sander M., Hsieh T., Udvardy A., Schedl P. Sequence dependence of Drosophila topoisomerase II in plasmid relaxation and DNA binding. J Mol Biol. 1987 Mar 20;194(2):219–229. doi: 10.1016/0022-2836(87)90370-6. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Maine E., Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985 Sep 20;185(2):341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P. Chromatin organization of the 87A7 heat shock locus of Drosophila melanogaster. J Mol Biol. 1984 Feb 5;172(4):385–403. doi: 10.1016/s0022-2836(84)80013-3. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Novel partitioning of DNA cleavage sites for Drosophila topoisomerase II. Cell. 1985 Apr;40(4):933–941. doi: 10.1016/0092-8674(85)90353-8. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Topoisomerase II cleavage in chromatin. J Mol Biol. 1986 Sep 20;191(2):231–246. doi: 10.1016/0022-2836(86)90260-3. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Worcel A., Gargiulo G., Jessee B., Udvardy A., Louis C., Schedl P. Chromatin fine structure of the histone gene complex of Drosophila melanogaster. Nucleic Acids Res. 1983 Jan 25;11(2):421–439. doi: 10.1093/nar/11.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]