Abstract
Phospholipase D (PLD) has an important role in various biological functions including vesicular transport, endocytosis, exocytosis, cell migration, and mitosis. These cellular biological processes are deregulated in the development of various human tumors. In order to explore the relationship between the PLD1 gene and risk of non-small cell lung cancer (NSCLC), single nucleotide polymorphisms (SNP) in the PLD1 exon region were surveyed in 211 NSCLC patients and 205 normal controls. In this study, we identified six SNPs at exon 23 in the PLD1 gene. Among the six SNPs, the most notable was a heterozygous A to C transition at nucleotide 2698 (A2698C, p<0.001). In addition, the genotype frequencies of A2744C (AC+CC) and A2756C (AC+CC) were associated with gender (female, A2744C and A2756C: p=0.071) in NSCLC patients. Interestingly, although the SNP A2698C did not cause change in amino acid, correlation between odd ratio of NSCLC patients and the SNP A2698C was observed to be statistically significant.
Keywords: biomarker, DGGE, lung cancer, NSCLC, phospholipase D, single nucleotide polymorphism
INTRODUCTION
Phospholipase D (PLD) is a ubiquitous enzyme that catalyzes the hydrolysis of phosphatidylcholine (PC) to phosphatidic acid (PA) and choline (1). PLD has two isoforms, PLD1 and PLD2, which differ in their mechanisms of activation and subcellular localization (2). In numerous human cancers, both PLD expression and activity are aberrantly increased (3-5). Recently, several studies have shown that expression levels of PLD1 mRNA and protein as well as PLD activity were markedly increased in human breast cancer tissue (6) and human renal cancer (7). We have previously shown that PLD and molecules involved in PLD signaling can be valuable targets for therapeutic intervention in cancers (1). These observations strongly imply that PLD is implicated in tumorigenesis, but the molecular mechanism remains unknown.
Lung cancer is the leading cause of cancer mortality in Korea and worldwide (8-11). Non-small cell lung cancer (NSCLC) accounts for ~85% of all cases of lung cancer (12). Polymorphisms in genes coding for enzymes involved in the metabolic activation and detoxification of tobacco carcinogens in the repair of DNA damage have been associated with an increased risk of lung cancer in case-control studies (13-15). Molecular epidemiologic studies have reported relationships between lung cancer and polymorphisms in genetic susceptibility genes, including metabolic enzymes (cytochrome P450s, glutathione S-transferases) and DNA repair enzymes (hOGG1, XRCC1), with the goal of elucidating their relationships with lung cancer susceptibility (13, 15, 16). Furthermore, a possible association between cancer susceptibility and variation in genes involved in chromatin structure and histone methylation has been investigated (17-19), suggesting that genetic susceptibility plays an important role in lung carcinogenesis.
This study was undertaken to examine the possible relationships between six novel SNPs found in the PLD1 exon 23 region and the risk of NSCLC in Korean NSCLC patients. We observed one and two other SNPs are associated with susceptibility and gender, respectively. Particularly, SNP A2698C leads us to conclude that SNPs on PLD1 may contribute to genetic susceptibility to NSCLC.
METHODS
Patients
This study evaluated 211 patients diagnosed with NSCLC who underwent surgical resection at Samsung Medical Center between 2005 and 2008. For comparison, a total of 205 control subjects were individually matched with lung cancer patients for age (± 10 years) and gender (Table 1). The control subjects had no prior histories of cancer and were recruited from a pool of visitors to our institution who were participating in a cancer-screening program. Information on demographic characteristics, including gender, age, and smoking habits, was obtained from self-administered questionnaires (for controls) or personal interviews (for cases) administered by a trained personnel after written informed consent was obtained. This study was performed after approval by the institutional review board at Samsung Medical Center (Seoul, Korea).
Table 1.
Characteristics of non-small cell lung cancer patients and controls
| Variable | Controls | Non-small cell lung cancer cases (n=211) |
|---|---|---|
| Age (years) | 48.3 (± 10) | 59.8 (± 10) |
| Sex | ||
| Male | 124 (60.5%) | 139 (66.8%) |
| Female | 81 (39.5%) | 72 (33.2%) |
| Smoking status | ||
| Current | 87 (42.5%) | 102 (48.3%) |
| Never | 118 (57.5%) | 109 (51.7%) |
DNA extraction and PCR amplification
Whole blood samples (1 ml) from the 416 subjects were collected in EDTA tubes. Genomic DNA was extracted using a Puregene DNA Purification kit (Gentra, MN, USA). The 26 exons of the PLD1 gene were amplified by PCR using primers synthesized according to sequences derived from GenBank (NM_001130081) (Table 2). For a standard 30-cycle amplification was performed using the following conditions: 94°C for 1 min, 55°C for 1 min, 72°C for 1 min (for exons 1-14, 18, and 20) or 94°C for 1 min, 60°C for 1 min, 72°C for 1 min (for exons 15-17, 19, and 21-26).
Table 2.
List of Primers for Detection of SNPs in PLD1 gene
| Exon No. | Direction | Sequences | Exon No. | Direction | Sequences |
|---|---|---|---|---|---|
| 1 | Forward | 5’-[40GC]GCCCTTTGCTTTTACTCTGTC-3’ | 14 | Forward | 5’-[40GC]CATGTCTTATGCAGTGTCTTTT-3’ |
| Reverse | 5’-CGCTCAGATCATCCGTCTTTAC-3’ | Reverse | 5’-GATAAATTCTAGTCAAGGCCA-3’ | ||
| 2 | Forward | 5’-AGTGTATATCCCTTTCTCTGC-3’ | 15 | Forward | 5’-GATCAGCTTTGCTTTCCAGTTT-3’ |
| Reverse | 5’-GAGTCCATAAACGCTCTGAC[40GC]-3’ | Reverse | 5’-TAAGGGAGTTCTGCCACTTCA[40GC]-3’ | ||
| 3 | Forward | 5’-[40GC]ATGTATCACTGTAGGTACCAAG-3’ | 16 | Forward | 5’-ACTCACCTGAACCACAGTGT-3’ |
| Reverse | 5’-AAATGGTTACCTTCTAGTGGG-3’ | Reverse | 5’-AATATAACCAGCACCCCACCA[40GC]-3’ | ||
| 4 | Forward | 5’-[40GC]GGTGTTTGCATTCTGTGTGT-3’ | 17 | Forward | 5’-[40GC]AGACTTTGCCCCAACACTGAA-3’ |
| Reverse | 5’-CTTACTCTTCTACCAAGGAATA-3’ | Reverse | 5’-GATAAATCATGATAGCAACATCC-3’ | ||
| 5 | Forward | 5’-[40GC]GATCTCATCATTGTCACTACTG-3’ | 18 | Forward | 5’-CACAAAGTAGGGAGAATGAATC-3’ |
| Reverse | 5’-TGACTAGTACTTACTGTGGCAT-3’ | Reverse | 5’-AAGGGAAGGCAGTTTCTCACA[40GC]-3’ | ||
| 6 | Forward | 5’-TGAATTGTTTTGCTTGCAAAAG-3’ | 19 | Forward | 5’-[40GC]CTGATGTCCTCTCCATTGCTAA-3’ |
| Reverse | 5’-ATGGCATGCTGCTACGTTA[40GC]-3 | Reverse | 5’-AAGGGAAGTCTAGTAGGTGG-3’ | ||
| 7 | Forward | 5’-[40GC]TGTTGGGAGGCTGTACGAG-3’ | 20 | Forward | 5’-CAGTATTGTTCTTACGTATATTGC-3’ |
| Reverse | 5’-GTAAAACTAGCCCAAATACC-3’ | Reverse | 5’-AATACAAGAACATCTGCAGCGA[40GC]-3’ | ||
| 8 | Forward | 5’-[40GC]CTTACTACCTTCTTACAGATGG-3’ | 21 | Forward | 5’-[40GC]TGAACTGCTTGGCTGTCATCTA-3’ |
| Reverse | 5’-CCTTGAAAGATTATCAATTCGG-3’ | Reverse | 5’-ATGATGCATGACCGAAAGCTCA-3’ | ||
| 9 | Forward | 5’-[40GC]TGAGATAGAACAGAGTGACC-3’ | 22 | Forward | 5’-AGGATTAAACCTACAGATACTGC-3’ |
| Reverse | 5’-ATTAGATGCTATGACTGCCCTTG-3’ | Reverse | 5’-ATGATTACTGATACCTCACCTTC[40GC]-3’ | ||
| 10 | Forward | 5’-[40GC]TGGAGATCTAGGCAGTGG-3’ | 23 | Forward | 5’-[40GC]TGTACGTTTATTGAGCTTGGTCA-3’ |
| Reverse | 5’-GTGTAATTCTTGAACAGCACTA-3’ | Reverse | 5’-TAAGTCAACTGGCAAGGAATACA-3’ | ||
| 11 | Forward | 5’-CCTTTGCTTCCTATGACACA-3’ | 24 | Forward | 5’-[40GC]GTTGTTCAGCCTCACTGTTTCT-3’ |
| Reverse | 5’-GTGTAATTCTTGAACAGCACTA[40GC]-3’ | Reverse | 5’-TGGAAATGCATCAGAGAGACAC-3’ | ||
| 12 | Forward | 5’-[40GC]CGGTTTTCTCCTGTGACAG-3’ | 25 | Forward | 5’-CAGTTACTCAATGTGGAGGTCA-3’ |
| Reverse | 5’-ATCAAGATGAACCTGAATACC-3’ | Reverse | 5’-GAGGAGGGGAATACGTGAACT[40GC]-3’ | ||
| 13 | Forward | 5’-[40GC]TTTCAGGGAGAACACAGACC-3’ | 26 | Forward | 5’-TAAAGGCCATGTGCTCGCTT-3’ |
| Reverse | 5’-GGTTAAATATACTTACTGGGAGG-3’ | Reverse | 5’-TGGAAGTCTTTGAGCTGCCAA[40GC]-3’ | ||
Denaturing gradient gel electrophoresis (DGGE)
After performing of DGGE to screen whether the sample has SNP, PCR products were loaded on a 20 × 27 cm, 0.75 mm-thick polyacrylamide gel (acrylamide:bisacrylamide, 37.5:1) containing a linear denaturing gradient (100% UF = 7 M urea/40% deionized formamide). The percentage of polyacrylamide varied between experiments. A 9% polyacrylamide stacking gel was poured to create solid slots for efficient loading of the PCR products, preventing difficulties caused by high urea concentration. Electrophoresis was performed in 1 × TAE buffer (40 mM Tris-acetate, 20 mM sodium acetate, 1 mM EDTA, pH8.0) at 59°C. For all experiments performed in this study, fresh buffer was used and only a single experimental condition was changed per test. Time-travel parallel DGGE was performed according to an established protocol. Gels were stained with ethidium bromide and photographs were taken under a UV transilluminator.
Detection and genotyping of PLD1 polymorphisms
To identify novel polymorphisms in PLD1, DGGE was performed for screening of two or multi bands on 26 exons of PLD1 prior to sequencing. We found several SNPs on exons of PLD1 through direct sequencing using PCR product. Among these SNPs we focused on exon 23 region of PLD1 within a 210 kb segment of genomic DNA. 147 out of 211 NSCLC patients and 77 out of 205 controls were examined by PCR and then analyzed by direct sequencing. If there was no detection of multi bands on DGGE, we considered it as a wild type PLD1. Variants were identified by comparison with traces of the PLD1 sequence relative to the reference GenBank sequence (NM_001130081) and confirmed by re-amplification and re-sequencing. The genotypes of PLD1 SNPs were identified by direct sequencing; PCR primers for PLD1 SNPs at exon23 were as follows: forward: 5’-TGTACGTTTATTGAGCTTGGTCA-3’; reverse: 5’-TAAGTCAACTGGCAAGGAATACA-3’.
Immunohistochemistry staining
The tissues were fixed overnight in 4% paraformaldehyde at 4°C, deparaffinized, rehydrated and immersed in normal goat blocking serum. The rabbit anti-human PLD1 antibody was used as primary antibody. The secondary antibody was biotinylated goat anti-rabbit antibody and the detection kit was DAB (3, 3′-diaminobenzidine). The assay was repeated five times on each kind of tissues. The immunostaining slides were examined under light microscopy and the digital images were captured with a Leica DM5000B digital camera.
Statistical analysis
The relationships between clinical factors and PLD1 SNPs were analyzed using the χ2-test. The association between NSCLC patients and each individual SNP was estimated using unconditional logistic regression after adjustment for clinical characteristics. The probability of survival was estimated using the Kaplan-Meier method. Differences in survival were evaluated using the log-rank test. All p-values were two-sided, and p-values less than 0.05 were considered to be statistically significant. Statistical analyses were performed using SPSS software version 13.0 (SPSS Institute Inc., Cary, NC).
RESULTS
Identification of PLD1 polymorphisms
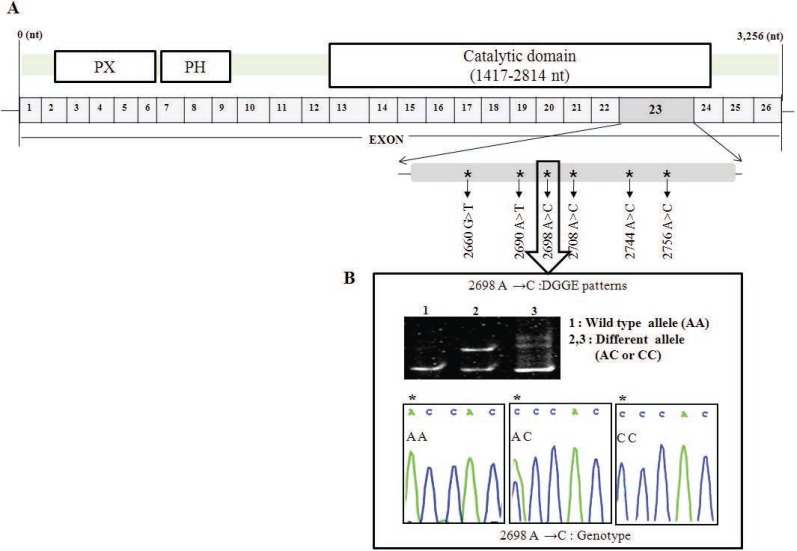
We included 211 NSCLC patients and 205 controls in this analysis (Table 1). To identify novel polymorphisms in PLD1, DGGE was performed for screening two or multi bands for every 26 exons of PLD1. For tissues that showed multi bands, direct sequencings were performed. After screening, we found several SNPs on PLD1. Among those SNPs we focused on exon 23 of PLD1, since it has 6 SNPs on a single exon and the frequency of these SNPs in NSCLC patient was high enough to deep search. If there was no detection of multi bands on DGGE, we considered it normal as a wild type of PLD1. We identified six genetic variants by direct DNA sequencing analysis and DGGE within the full 3.2 kb genome, including the exon 23 region (2612 bp ~ 2863 bp) of PLD1: 2660 G→T, 2690 A→T, 2698 A→C, 2708 A→C, 2744 A→C and 2756 A→C. These are novel SNPs within the 26 exons of the PLD1 gene (Figure 1). There were frequency differences in the distribution of variant alleles of the six SNPs between NSCLC patients and controls. Of the novel SNPs, the most notable SNP was a heterozygous A to C transition at nucleotide 2698 (exon 23, allele A/C: A2698C, p<0.001, OR=4.619) (Table 3).
Figure 1.
Locations of the six observed SNPs in PLD1 cDNA. (A) Distribution of single nucleotide polymorphism in the PLD1 gene. Substituted nucleotides are indicated by an arrow; (B) Top: DGGE pattern of A2698C showing a different allele (lanes 2 and 3). Bottom: DNA sequencing electropherograms of wild type (AA) and different alleles (AC and CC) of A2698C identified in PLD1 SNPs in this study. Polymorphic sites are indicated by asterisks (*).
Table 3.
Allelic frequencies of PLD1 SNPs in non-small cell lung cancer patients and controls
| Polymorphism | Non-small cell lung cancer (n) | Controls (n) | P value (n) (p<) | Adjusted OR |
|---|---|---|---|---|
| 2660 G>T | 0.94% (2/211) | 0% (0/205) | 0.246 | 0.615 |
| 2690 A>T | 1.44% (3/211) | 0% (0/205) | 0.195 | 0.784 |
| 2698 A>C | 65.9% (139/211) | 37.6% (77/205) | 0.001 | 4.619 |
| 2708 A>C | 0.47% (1/211) | 0% | 0.437 | 0.511 |
| 2744 A>C | 0.47% (1/211) | 0% | 0.437 | 0.511 |
| 2756 A>C | 0.47% (1/211) | 0% | 0.437 | 0.511 |
p-values and odd ratio (OR) for allelic frequencies were obtained from Pearson’s χ2-test.
Genotyping SNPs
The genotypic distributions of the six PLD1 SNPs in NSCLC and controls are summarized in Table 4. Even though the number of sample is not enough to define whether they are significantly associated with NSCLC, there were differences in genotype distributions G2660T (p=0.013), A2698C (p<0.001), A2744C (P=0.005), and A2756C (p=0.005) between NSCLC cases and controls. However, there was no significant difference in the genotype frequencies of A2690T (p=0.092) and A2708C (p=0.612) between NSCLC cases and controls.
Table 4.
Genotype distribution of the PLD1 polymorphisms in non-small cell lung cancer and controls
| SNP | Group | Genotypes of PLD1 polymorphisms |
|||
|---|---|---|---|---|---|
| W/S (%)a | W/V (%)a | V/V (%)a | P-value (p<) | ||
| 2660 G→T | Lung cancer | 192 (91.0) | 17 (8.1) | 2 (0.9) | 0.013 |
| control | 200 (97.6) | 5 (2.4) | 0 (0.0) | ||
| 2690 A→T | Lung cancer | 190 (90.0) | 19 (9.0) | 2 (0.9) | 0.092 |
| control | 195 (95.1) | 10 (4.9) | 0 (0.0) | ||
| 2698 A→C | Lung cancer | 50 (23.7) | 120 (56.9) | 41 (19.4) | 0.001 |
| control | 113 (55.1) | 69 (33.7) | 23 (11.2) | ||
| 2708 A→C | Lung cancer | 190 (90.0) | 20 (9.5) | 1 (0.5) | 0.612 |
| control | 186 (90.7) | 19 (9.3) | 0 (0.0) | ||
| 2744 A→C | Lung cancer | 184 (87.2) | 27 (12.8) | 0 (0.0) | 0.005 |
| control | 195 (95.1) | 10 (4.9) | 0 (0.0) | ||
| 2756 A→C | Lung cancer | 184 (87.2) | 27 (12.8) | 0 (0.0) | 0.005 |
| control | 195 (95.1) | 10 (4.9) | 0 (0.0) | ||
p-values for genotypes was obtained from Pearson’s χ2-test.
W means wild-type allele and V means variant allele of each SNP.
Association analysis between PLD1 SNPs and clinical characteristics in NSCLC patients
We investigated whether PLD1 SNPs are related to clinical characteristics using logistic regression analysis. Of the six PLD1 SNPs we studied, A2698C (AC+CC) was the only genotype that was highly associated with survival in NSCLC patients (carrying one variant allele vs. none, p=0.012) (Table 5). This result indicates that the A2698C PLD1 SNP may be related to risk of NSCLC. The genotypes A2744C and A2756C (AC+CC) were associated with gender in NSCLC patients, although the p value did not reach statistical significance (p=0.071).
Table 5.
Association analysis among PLD1 SNPs, smoking status, age and gender in non-small cell lune cancer patients
| SNP | Geno-type | Gender (%) |
Survival (%) |
Smoking (%) |
Age-years (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | P | Death | N/Da | P | Never | Ever | P | Age ≥60 | Age <60 | P | ||
| G2660T | GG | 125 (89.9) | 67 (93.0) | 0.578 | 88 (82.3) | 104 (91.2) | 0.967 | 94 (92.2) | 98 (89.9) | 0.648 | 106 (91.4) | 86 (90.5) | 0.642 |
| GT+TT | 14 (10.1) | 5 (7.0) | 9 (17.7) | 10 (8.8) | 8 (7.8) | 11 (10.1) | 10 (8.6) | 9 (9.5) | |||||
| A2690T | AA | 123 (88.5) | 67 (93.0) | 0.150 | 88 (82.3) | 102 (89.5) | 0.991 | 92 (90.2) | 98 (89.9) | 0.932 | 107 (92.2) | 83 (76.3) | 0.131 |
| AT+TT | 16 (11.5) | 5 (7.0) | 9 (17.7) | 12 (10.5) | 10 (9.8) | 11 (10.1) | 9 (7.8) | 12 (23.7) | |||||
| A2698C | AA | 34 (24.4) | 16 (22.2) | 0.536 | 14 (14.4) | 36 (31.6) | 0.012 | 21 (20.6) | 29 (26.6) | 0.481 | 29 (25.0) | 21 (22.1) | 0.369 |
| AC+CC | 105 (75.6) | 56 (77.8) | 83 (85.6) | 78 (68.4) | 81 (79.4) | 80 (73.4) | 87 (75.0) | 74 (77.9) | |||||
| A2708C | AA | 127 (91.4) | 63 (87.5) | 0.369 | 88 (90.7) | 102 (89.5) | 0.626 | 100 (91.7) | 90 (88.2) | 0.372 | 103 (88.8) | 87 (91.6) | 0.331 |
| AC+CC | 12 (8.6) | 9 (12.5) | 9 (9.3) | 12 (10.5) | 9 (8.3) | 12 (11.8) | 13 (11.2) | 8 (8.4) | |||||
| A2744C | AA | 117 (84.2) | 67 (93.0) | 0.071 | 83 (73.2) | 101 (88.6) | 0.661 | 94 (86.2) | 90 (88.2) | 0.662 | 105 (90.5) | 79 (83.2) | 0.236 |
| AC+CC | 22 (15.8) | 5 (7.0) | 14 (26.8) | 13 (11.4) | 15 (13.8) | 12 (11.8) | 11 (9.5) | 16 (16.8) | |||||
| A2756C | AA | 117 (84.2) | 67 (93.0) | 0.071 | 84 (86.6) | 100 (87.7) | 0.651 | 96 (88.1) | 88 (86.3) | 0.628 | 99 (85.3) | 85 (89.5) | 0.528 |
| AC+CC | 22 (15.8) | 5 (7.0) | 13 (13.4) | 14 (12.3) | 13 (11.9) | 14 (13.7) | 17 (14.7) | 10 (11.5) | |||||
p-values for gender, survival, smoking and age were obtained from logistic regression analysis.
Non Death.
Expression of PLD1 in NSCLC
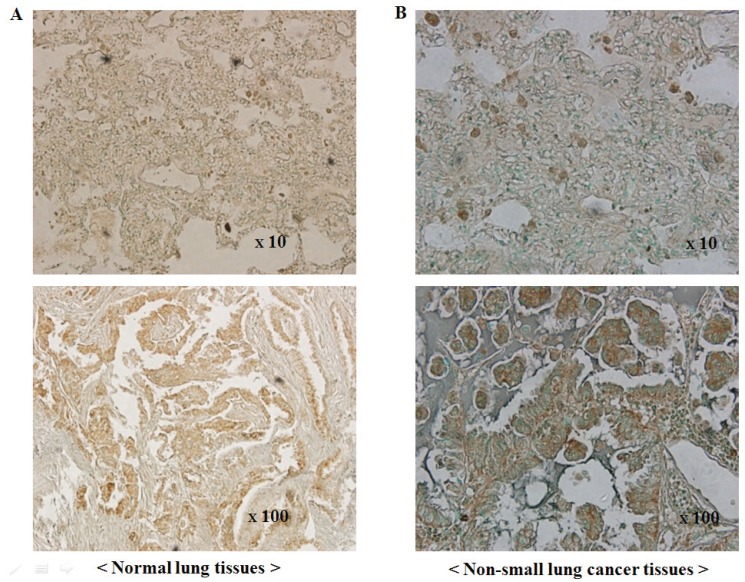
The expression of PLD1 protein was examined by immunohistochemical staining using surgical specimens from an independent set of 215 NSCLC patients (Figure 2). Expression was detected in 179 (83.3%) of 215 NSCLC tissues, while PLD1 was not readily detected in normal lung tissues. However, there was not a significant difference in overall survival between the PLD1-positive and the PLD1-negative group in NSCLC patient (data not shown).
Figure 2.
PLD1 immunohistochemistry in human non-small cell lung cancers and normal lung tissues. (A) Immunoreactivity for PLD1 in normal alveolar lung tissues. The acinar epithelia and intra-alveolar connective tissue exhibit a negative reaction and positive staining, respectively; (B) Immunoreactivity for PLD1 in non-small cell lung cancer tissues (NSCLC). NSCLC tissues exhibit a strong positive reaction (top: ×10, bottom: ×100).
DISCUSSION
Individual susceptibility to lung cancer has been shown to vary with the presence of single nucleotide polymorphisms (SNPs) in a number of critical genes (20-22). It is quite possible that DNA sequence variations in PLD1, a gene that has recently been studied for its association with the development of many cancers, may lead to alteration in the activity of PLD, which can cause individual differences in lung cancer susceptibility. Indeed, PLD is a critical regulator of cell proliferation, survival, and abnormalities in many cancer cells, such as PC 12 cells, v-src-transformed rat fibroblasts, and MDA-MB-231 breast cancer cells (6, 23-25). Recently, it has been reported that PLD and molecules involved in PLD signaling may be valuable targets in therapeutic interventions for cancers, given that a substantial portion of tumor cells apparently has elevated PLD activity (1). Furthermore, activation of PLD1 by bradykinin and sphingosine 1 is involved in the protein kinase C signaling pathway in A549 human lung adenocarcinoma cells (26, 27), suggesting a possible association between PLD1 and NSCLC. Recently, it has been reported that PLD polymorphisms are closely associated with cancer. For examples, the C1814T (Thr577Ile) polymorphism in the human PLD2 gene is associated with the prevalence of colorectal cancer (28), and a naturally-occurring variant of human PLD2 in which Gly901 in the COOH-terminal region is replaced by the charged amino acid Asp is catalytically inactive (29).
Interestingly, the six PLD1 SNPs in the present study were located in the catalytic domain of the PLD1 gene (Figure 1). We analyzed whether six PLD1 SNPs (G2660T, A2690C, A2698C, A2708C, A2744C, and A2756C) function as a biomarker which contributes to prediction or risk of NSCLC based on the results from direct sequencing and DGGE of the whole genomic region in 211 Korean NSCLC patients and 205 normal controls. Even though tobacco smoking has a well-established critical role in the development of lung cancer, association study of PLD1 SNP genotypes using smoking status and age in NSCLC patients did not show any significant correlation with smoking status (Table 5). However, the variant alleles A2744C (AC+CC) and A2756C (AC+CC) have correlation with gender, particularly female NSCLC group (p=0.072), although this relationship was not statistically significant. Among the six SNPs, the variant (AC+CC) A2698C SNP was the only genotype associated with an increased risk of developing NSCLC. Unfortunately, A2698C is a synonymous mutation (GTAval → GTCval); however, it is reported that altering nucleotide between the same amino acids does affect tRNA affinity during translation. According to Elf and Nilsson et al’s reported that GTAval increased tRNA affinity to mRNA more than GTCval (30), PLD expression level should be decreased in NSCLC patients. However, as shown in Figure 2 and Table 3, PLD expression level and PLD SNP occurrence in NSCLC patients are higher compared to normal. Indeed, these findings suggest that there is a possible mechanism for regulation of PLD expression or maintenance. To the best of our knowledge, this is the first study to demonstrate a statistically significant association between PLD1 SNPs and NSCLC risk. Furthermore, it is the first to show that one of the PLD SNPs, A2698C, causes an increase in NSCLC susceptibility. These results suggest that the presence of A2698C SNP in PLD1 may be involved in the development of NSCLC and could be one of important markers of genetic susceptibility to lung cancer. Precise mechanism by which susceptibility to NSCLC increases remains uncovered, yet.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MEST) (2010-0029503 and 2010-0026844).
CONFLICT OF INTEREST STATEMENT
The authors declare that no conflicting interests exist.
REFERENCES
- 1.Cho JH, Hong SK, Kim EY, Park SY, et al. Overexpression of phospholipase D suppresses taxotere-induced cell death in stomach cancer cells. Biochim. Biophys. Acta. 2008;1783(5):912–923. doi: 10.1016/j.bbamcr.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Park SY, Cho JH, Oh DY, Park JW, et al. House dust mite allergen Der f 2-induced phospholipase D1 activation is critical for the production of interleukin-13 through activating transcription factor-2 activation in human bronchial epithelial cells. J. Biol. Chem. 2009;284(30):20099–20110. doi: 10.1074/jbc.M109.010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchida N, Okamura S, Nagamachi Y, Yamashita S. Increased phospholipase D activity in human breast cancer. J. Cancer Res. Clin. Oncol. 1997;123(5):280–285. doi: 10.1007/BF01208639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999;19(1B):671–675. [PubMed] [Google Scholar]
- 5.Hsu YL, Hung JY, Ko YC, Hung CH, et al. Phospholipase D signaling pathway is involved in lung cancer-derived IL-8 increased osteoclastogenesis. Carcinogenesis. 2010;31(4):587–596. doi: 10.1093/carcin/bgq030. [DOI] [PubMed] [Google Scholar]
- 6.Noh DY, Ahn SJ, Lee RA, Park IA, et al. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161(2):207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, Ehara H, Akao Y, Shamoto M, et al. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem. Biophys. Res. Commun. 2000;278(1):140–143. doi: 10.1006/bbrc.2000.3719. [DOI] [PubMed] [Google Scholar]
- 8.Lei Z, Liu RY, Zhao J, Liu Z, et al. TGFBR1 haplotypes and risk of non-small-cell lung cancer. Cancer Res. 2009;69(17):7046–7052. doi: 10.1158/0008-5472.CAN-08-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spinola M, Leoni VP, Galvan A, Korsching E, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251(2):311–316. doi: 10.1016/j.canlet.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 10.Engels EA, Wu X, Gu J, Dong Q, et al. Systematic evaluation of genetic variants in the inflammation pathway and risk of lung cancer. Cancer Res. 2007;67(13):6520–6527. doi: 10.1158/0008-5472.CAN-07-0370. [DOI] [PubMed] [Google Scholar]
- 11.Park JY, Park JM, Jang JS, Choi JE, et al. Caspase 9 promoter polymorphisms and risk of primary lung cancer. Hum. Mol. Genet. 2006;15(12):1963–1971. doi: 10.1093/hmg/ddl119. [DOI] [PubMed] [Google Scholar]
- 12.Shields PG. Molecular epidemiology of smoking and lung cancer. Oncogene. 2002;21(45):6870–6876. doi: 10.1038/sj.onc.1205832. [DOI] [PubMed] [Google Scholar]
- 13.Larsen JE, Colosimo ML, Yang IA, Bowman R, et al. CYP1A1 Ile462Val and MPO G-463A interact to increase risk of adenocarcinoma but not squamous cell carcinoma of the lung. Carcinogenesis. 2006;27(3):525–532. doi: 10.1093/carcin/bgi227. [DOI] [PubMed] [Google Scholar]
- 14.Wenzlaff AS, Cote ML, Bock CH, Land SJ, et al. CYP1A1 and CYP1B1 polymorphisms and risk of lung cancer among never smokers: a population-based study. Carcinogenesis. 2005;26(12):2207–2212. doi: 10.1093/carcin/bgi191. [DOI] [PubMed] [Google Scholar]
- 15.Le Marchand L, Donlon T, Lum-Jones A, Seifried A, et al. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol. Biomarkers Prev. 2002;11(4):409–412. [PubMed] [Google Scholar]
- 16.Ito H, Matsuo K, Hamajima N, Mitsudomi T, et al. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25(8):1395–1401. doi: 10.1093/carcin/bgh153. [DOI] [PubMed] [Google Scholar]
- 17.Tsuge M, Hamamoto R, Silva FP, Ohnishi Y, et al. A variable number of tandem repeats polymorphism in an E2F-1 binding element in the 5’ flanking region of SMYD3 is a risk factor for human cancers. Nat. Genet. 2005;37(10):1104–1107. doi: 10.1038/ng1638. [DOI] [PubMed] [Google Scholar]
- 18.Cebrian A, Pharoah PD, Ahmed S, Ropero S, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27(8):1661–1669. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- 19.Yoon KA, Hwangbo B, Kim IJ, Park S, et al. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27(11):2217–2222. doi: 10.1093/carcin/bgl084. [DOI] [PubMed] [Google Scholar]
- 20.Cargill M, Altshuler D, Ireland J, Sklar P, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22(3):231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 21.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic. Acids. Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang DG, Fan JB, Siao CJ, Berno A, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280(5366):1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Rodrik V, Foster DA. Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene. 2005;24(4):672–679. doi: 10.1038/sj.onc.1208099. [DOI] [PubMed] [Google Scholar]
- 24.Kim KO, Lee KH, Kim YH, Park SK, et al. Anti-apoptotic role of phospholipase D isozymes in the glutamate-induced cell death. Exp. Mol. Med. 2003;35(1):38–45. doi: 10.1038/emm.2003.6. [DOI] [PubMed] [Google Scholar]
- 25.Park MA, Lee MJ, Lee SH, Jung DK, et al. Anti-apoptotic role of phospholipase D in spontaneous and delayed apoptosis of human neutrophils. FEBS Lett. 2002;519(1-3):45–49. doi: 10.1016/s0014-5793(02)02705-9. [DOI] [PubMed] [Google Scholar]
- 26.Meacci E, Vasta V, Moorman JP, Bobak DA, et al. Effect of Rho and ADP-ribosylation factor GTPases on phospholipase D activity in intact human adenocarcinoma A549 cells. J. Biol. Chem. 1999;274(26):18605–18612. doi: 10.1074/jbc.274.26.18605. [DOI] [PubMed] [Google Scholar]
- 27.Meacci E, Nuti F, Catarzi S, Vasta V, et al. Activation of phospholipase D by bradykinin and sphingosine 1-phosphate in A549 human lung adenocarcinoma cells via different GTP-binding proteins and protein kinase C delta signaling pathways. Biochemistry. 2003;42(2):284–292. doi: 10.1021/bi026350a. [DOI] [PubMed] [Google Scholar]
- 28.Yamada Y, Hamajima N, Kato T, Iwata H, et al. Association of a polymorphism of the phospholipase D2 gene with the prevalence of colorectal cancer. J. Mol. Med. 2003;81(2):126–131. doi: 10.1007/s00109-002-0411-x. [DOI] [PubMed] [Google Scholar]
- 29.Yamada Y, Banno Y, Yoshida H, Kikuchi R, et al. Catalytic inactivation of human phospholipase D2 by a naturally occurring Gly901Asp mutation. Arch. Med. Res. 2006;37(6):696–699. doi: 10.1016/j.arcmed.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300(5626):1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]