Abstract
Leuciscine fishes represent an important component of freshwater ichthyofauna endemic to northern Mediterranean areas. This lineage shows high intra-specific morphological variability and exhibits high levels of hybridization, two characteristics that contribute to systematic uncertainties, misclassification of taxa and, potentially, the mismanagement of biodiversity. This study focused on brook chub, Squalius lucumonis, an endemic taxon of Central Italy. The taxonomic status of this species has long been questioned, and a hybrid origin from sympatric leusciscines (S. squalus x Rutilus rubilio, or S. squalus x Telestes muticellus) has been hypothesised. A phenotypic (evaluating shape and meristic counts) and genetic (using mitochondrial and nuclear markers) investigation of these four taxa was conducted to test species delimitation in sympatric areas and to evaluate the taxonomic status of S. lucumonis. One hundred and forty-five individuals of all four taxa were collected within streams of the lowest portion of the Tiber River basin and analysed; this region encompasses a large portion of the S. lucumonis distribution. The different morphological and genetic approaches were individually examined, compared, and then combined in a quantitative model to both investigate the limits of each approach and to identify cases of misclassification. The results obtained confirm the cladogenetic non-hybrid origin of S. lucumonis, highlight the need for immediate conservation actions and emphasise the value of an integrated approach in the study of leuciscines evolution.
Introduction
Numerous cases of hybridization and introgression events have been found by molecular studies in many organisms. The occurrence of such events makes the biological and other (e.g the phylogenetic) species concepts difficult to be applied univocally [1]. Most of the reported cases of hybridization in animals involve fishes, mainly freshwater species [1]. Among these, cyprinids are known to exhibit high rates of hybridization at both the intra- and intergeneric level. This is especially evident for leuciscine species [2]–[9]; approximately 62 different intra- and intergeneric hybrids were described for this group in the wild [10]. Leuciscine fishes were traditionally classified as a subfamily (Leuciscinae) within the Cyprinidae family [11], [12]. However, based on molecular data, Chen & Mayden [13] proposed that these fishes, along with nine other monophyletic groups of cyprinid, be classified each as a family within the superfamily Cyprinoidea. Thus, according to these authors, Leuciscinae should be considered as Leuciscidae.
In Europe, Cyprinoidea are dominant [14] and comprise a large number of endemic species in northern Mediterranean regions [15], including Italy [16]. Many leuciscine fishes are listed in the International Union for Conservation of Nature (IUCN) Red List [17], represent many of the threatened freshwater fishes of Europe [18] and are listed in Annex II of the European Union Habitats Directive 92/43/EEC (http://ec.europa.eu/environment/nature/legislation/habitatsdirective) and in Appendix III of Bern Convention [19], [20]. Moreover, many leuciscine species distributed within the Mediterranean region show high intraspecific morphological variability [21]. The described aspects of Leuciscidae biology make it difficult to resolve the phylogenetic and taxonomic status of several populations and species in the Mediterranean area. Thus efforts should be made to validate the taxonomy of this family and to identify diagnostic characters for species identification.
Over the last decade, multidisciplinary approaches have proven useful in cases of unclear taxonomic status that are very common when species tend to hybridize [22]–[24]. Resolving taxonomic uncertainties is particularly important for the identification of conservation units and relevant management strategies [25]. Combined morphological and genetic analysis have been successfully applied to resolve taxonomy [7], [26]–[29] and to interpret ecomorphology [9] within this family.
In the present study, morphological and molecular data were used mainly to investigate an Italian leuciscine, the brook chub Squalius lucumonis. This species is endemic to the Tuscano-Latium district of Central Italy. Its distribution appears to be restricted to the Tyrrhenian drainage basins of Ombrone-Serchio, Arno and Tiber [30]–[32], with approximately 12 main populations estimated [30]. Its first description by Bianco [33] was based exclusively on morphologic and meristic (countable) traits derived from the analysis of a few individuals caught within the Tiber and Ombrone River basins. The taxonomic status of S. lucumonis was questioned by Gandolfi et al. [34] and Zerunian [35], who argued that the brook chub might represent a hybrid between S. squalus (previously included in the S. cephalus taxon) and another sympatric species of Leuciscinae. Based on meristic characters, these authors proposed S. squalus x T. muticellus (previously included within the T. souffia complex) or S. squalus x R. rubilio as the potential parental species crosses. Manaresi et al. [36] rejected the hypothesis of a hybrid origin based on the absence of heterozygosity at diagnostic allozyme loci and at total protein bands between the presumed parental species. Furthermore, they found alternative alleles at one locus and the presence of two diagnostic bands (65 kDa and 26 kDa) in total protein analysis between S. lucumonis and S. squalus, and proposed that the two taxa may have diverged recently from a common ancestor.
Finally, morphological brook chub-like individuals were recorded in an area outside the natural range of S. lucumonis; genetic analysis identified these individuals as hybrids between S. squalus and R. rubilio [37]. This paper seeks to evaluate the morphological variation and taxonomic status of S. lucumonis in relation to three other sympatric Italian leuciscines: S. squalus, also distributed along the Dalmatian coasts, T. muticellus, also present along the south French-Italian border and in Switzerland, and the Italian endemic R. rubilio [30]. The latter two species are listed as endangered in Annex II of the European Habitat Directive [38]. To this purpose, both meristic traits (for distinguishing taxa by means of classification trees) and external morphology (investigated using geometric morphometrics) were analysed. In addition, morphological identification was verified on a subsample of specimens based on two nuclear regions, the Cyfun P (Cyprinid formerly unknown nuclear Polymorphism), a non-coding region of unknown function [39], and the Recombination Activating Gene 1 (RAG-1) and on the mitochondrial cytochrome b (cyt b) gene.
These different datasets were later combined in a quantitative statistical model of trait covariation.
Materials and Methods
Sampling
A total of 145 specimens of S. lucumonis, S. squalus, T. muticellus and R. rubilio were collected from 9 sampling sites within the lower section of the Tiber River basin (Table 1) using electrofishing procedures. Specimens were anesthetised with a 0.035% MS 222 (Tricaine Methanesulfonate) solution. The left lateral view of each specimen was photographed in the field for shape and meristic count analyses. A small portion of the pelvic fin was removed and fixed in 90% ethanol for genetic analysis. The procedures used for fish sampling were carried out in agreement with relevant legislation (CEN EN 14011/2003 - Water quality - Sampling of fish with electricity), avoided animal sacrifice and allowed for the live release of sampled specimens after data collection. Fish sampling were authorized by the Dipartimento Istituzionale e Territorio of the Regione Lazio (Prot. n. 526425). Specimens were initially identified according to external morphology using Bianco & Ketmaier [30] as reference for S. lucumonis and Gandolfi et al. [34] for the remaining three taxa. Taxonomic classification was further conducted in accordance with Kottelat & Freyhof [31]. Meristic counts and shape analyses through geometric morphometrics (GM) were carried out for the entire sample, whereas genetic analyses (based on nuclear and mitochondrial markers) were performed on a sub-sample.
Table 1. Details on the sampling sites in Tiber River basin and on number of individuals collected. Sl stands for Squalius lucumonis, Ss for Squalius squalus, Tm for Telestes muticellus, Rr for Rutilus rubilio, respectively.
| GPS Position – WGS1984 | LocationCode | N | |||
| Sl | Ss | Tm | Rr | ||
| 42°09′ 10.71″ N –12° 38′ 47.33″ E | Pc | 10 | 10 | − | 4 |
| 42° 09′ 58.00″ N –12° 43′ 27.00″ E | Sp | 10 | 9 | 6 | 2 |
| 42° 09′ 07.40″ N –12° 41′ 28.70″ E | Fc | 3 | − | − | − |
| 42° 00′ 32.20″ N –12° 28′ 33.70″ E | Pp | 5 | − | − | − |
| 42° 08′ 00.00″ N –12° 39′ 54.10″ E | Ro | 6 | − | − | − |
| 42° 05′ 00.70″ N –12° 41′ 48.90″ E | Ff | 6 | − | − | − |
| 42° 10′ 22.80″ N –12° 32′ 41.20″ E | Ra | 10 | − | 9 | 14 |
| 41° 55′ 54.00″ N –12° 43′ 55.10″ E | Fp | 5 | 6 | 2 | 2 |
| 41° 55′ 19.71″ N –12° 46′ 03.54″ E | Sv | 10 | 1 | 10 | 5 |
| Total | 65 | 26 | 27 | 27 | |
Analysis of Meristic Counts
Meristic characters are countable structures occurring in series (e.g. myomeres, vertebrae, fin rays). Six meristic characters were inspected in this study (Table S1), namely the number of scales of the lateral line (NSLL), above the lateral line (NSALL), under the lateral line (NSULL), and the number of rays of the dorsal fin (NRDF), left pectoral fin (NRPF), and anal fin (NRAF). A classification and regression tree (CRT) [40] was trained to build a species identification key based on meristic characters. This method has only recently been recognised as a useful tool for classification analysis by ecologists [41]. In the present study, all CRT procedures were performed using the software SPSS Statistics v17.0 (SPSS, Inc., 2009, Chicago, IL, www.spss.com). The six meristic counts were used as ordinal predictors of species, which was the dependent variable.
Shape Analysis
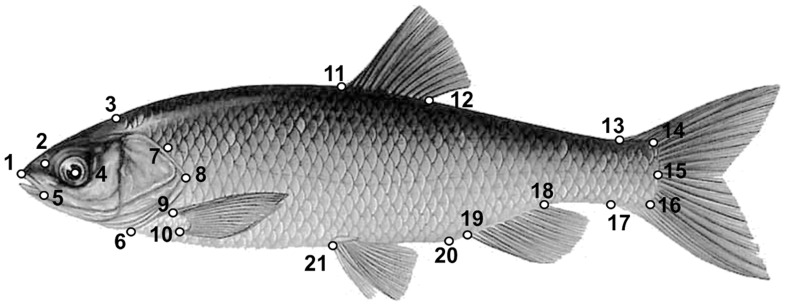
Each fish was measured (standard length, SL), and twenty-one landmarks (Fig. 1) were recorded on each digital image using the software TPSDIG [42].
Figure 1. Landmarks used for Geometric morphometrics analysis of external shape.
(1) Snout tip; (2) nostril; (3) beginning of scales coverage on the dorsal outline; (4) centre of the eye; (5) posterior extremity of the premaxillar; (6) insertion of the operculum on the ventral lateral profile; (7) beginning of the lateral line; (8) posterior extremity of the operculum; (9) superior and (10) inferior insertions of the pectoral fin; (11) anterior and (12) posterior insertions of the dorsal fin; (13) superior and (17) inferior insertions of the caudal fin; (14) superior and (16) inferior insertion of the caudal peduncle: (15) posterior body extremity; (18) posterior and (19) anterior insertion of the anal fin; (20) anus; (21) anterior insertion of the pelvic fin.
Configurations were scaled to unit centroid size, translated to a common centroid, and rotated to minimise the least squared residuals between corresponding landmarks [43]. Residuals from the registration were analysed with the thin-plate spline (TPS) interpolating function [44]. A canonical variate analysis (CVA) was performed on the W’ matrix of partial warps [45], [46] using MorphoJ [47]. CVA is often used to identify the shape features that best distinguish multiple groups of specimens by determining the linear combinations of the original variables that display the greatest variance between groups relative to the variance within groups. Group membership is assumed to be known a priori. The emphasis is on the degree of separation between groups and on the probability of correct and incorrect classification of each observation. The robustness of assignment was assessed through a leave-one-out resampling cross-validation procedure. Splines relative to the extreme positions of the canonical axes were obtained using TPSREGR [48]. Differences between shapes were visualised using deformation grids [44], which represent the most robust visualisation tool for synthesising the entire pattern of shape variation. In addition, the CVA procedure in MorphoJ carries out a leave-one-out cross-validation method (number of permutation runs = 1000) to assess the reliability of classifications.
Genetic Analyses
The DNA of 28 specimens (Table S2) (primarily from S. lucumonis and randomly chosen within each species) was extracted from ethanol-preserved fin tissue following the salt extraction protocol of Aljanabi & Martinez [49]. Amplifications of the three selected markers were performed using primers and protocols available in the literature [50]–[52] and reported in Table S3.
The Cyfun P band length (310–440 base pair, bp) was used for a rough species attribution and to verify the occurrence of nuclear hybrid genomes. This region is indeed known to show intergeneric diagnostic length and/or sequence polymorphism in leuciscine fishes [39]. Amplicons were also purified with SureClean (Bioline) and sequenced with an automated DNA sequencer (Macrogen Inc.). NCBI’s BLAST software was used for similarity searching of obtained sequences, that were then deposited in the GenBank database (Accession Numbers: JQ286163–JQ286167, JQ286169), aligned with ClustalX [53] and subsequently manually adjusted. Sequences allowed a more accurate species attribution.
The nuclear RAG1 (840 bp) and the mitochondrial cyt b (1131 bp) amplicons were processed as previously described (Accession Numbers: JQ286150–JQ286162 and JQ799135–JQ799137 for cyt b, and KC478779–KC478783 for RAG1) and used in subsequent phylogenetic analyses. Cyfun P sequences were excluded from these analyses as the presence of the numerous indels causes ambiguity in the alignment. In phylogenetic reconstructions,sequences of the four species retrieved from GenBank were also added (Table S4); for cyt b, only sequences longer than 1100 bp were considered. Pseudorasbora parva was used as outgroup, since sequences of both markers, from the same individual, were available. Phylogenetic analyses were performed independently on each gene and then on the combined dataset, using both maximum-likelihood (ML) and Bayesian inference (BI) analysis. Modeltest 3.7 [54] and Mrmodeltest 2.3 [55] were used to select the evolutionary model that best fits the data set for each data set for the ML and the BI analyses, respectively. The Akaike’s information criterion (AIC) [56] was used.
ML tree reconstructions were conducted in PhyML v2.4.4 [57], using heuristic search with the NNI swap algorithm and 1000 bootstrap replicates. Bayesian analyses were implemented in MrBayes v3.1.2 [58] performing two independent runs of four Markov chains each for 1,000,000 generations. The average standard deviation of split frequencies was close to 0.003 when the runs were finished. Trees were sampled every 1,000 generations, burn-in trees (25%) were discarded and a 50% majority-rule consensus tree was estimated.
Species tree reconstruction was performed using the coalescent-based Bayesian species tree inference method with the ∗BEAST (Bayesian Inference of Species Trees from Multilocus Data) protocol [59] implemented in the software BEAST v1.7.4 [60]. The input file was properly formatted with the BEAUti utility included in the software package, using the same partition scheme of the concatenated analysis. Two runs, each of 50×106 generations (samplefreq = 5000 and 10% burnin), were conducted. Tracer v1.5 [61] was used to check for convergence and normal distribution. The treefiles were combined in Logcombiner v1.7.4 and maximum clade credibility trees were created with TreeAnnotator v1.7.4, both programs are implemented in BEAST v1.7.4. Finally, a species tree was inferred and viewed using FigTree v1.4.0 [62].
We used the posterior predictive checking approach of Joly et al. [63], implemented in JML v1.0.1 [64] to test whether the incongruence between nuclear and mitochondrial genotypes was the result of incomplete lineage sorting or hybridization. We used both the RAG1 and cyt b datasets and the parameters yielded by Modeltest in JML running. The software calculates minimum pairwise sequence distances between the simulated sequences of individuals including all possible comparisons, and tests for each dataset whether the observed minimum distance between sequences of two species is smaller than those expected under the simulated model that does not account for hybridization. Gene trees and sequences were simulated separately for the mitochondrial (mt) and the nuclear (n) datasets using the relative substitution rates adopted in BEAST. Appropriate heredity scalars were selected for simulations of nuclear (2) and mitochondrial (0.5) markers, respectively. A significance level of 0.1 was specified in all runs.
Genetic distances obtained for the 28 RAG1 and cyt b sequences belonging to the four species (Table S5) were used as input for a principal coordinate analysis (PCOA) performed with the PAST program [65]. The scores of the specimens on these axes were used as variables in the morphology and genetics comparative analyses.
Integration between Morphology and Genetics: Partial Least-square Analysis
A partial least-square (PLS) analysis [42], [66] was conducted to explore the pattern of covariation between shape and genetic distance for the four species. The PLS analysis is a multivariate correlation technique that identifies pairs of linear combinations between two sets of input variables (here, shape and the two axes of the PCOA performed on the matrix of genetic distances). The PLS analysis produces ordered pairs of ‘latent’ vectors, one per each input dataset, that account for the maximum amount of information covariation between the two original sets of variables [66]–[68]. The amount of covariance explained by a pair of latent vectors (represented by the correlation coefficient ‘R’) and its ‘strength’ (estimated by permutations, TPSPLS [69]) provide a statistical assessment of the association between the two datasets. R can serve as a measure of the integration of shape and sets of other variables [43], [66]. This PLS approach has been successfully used to compare shape between different body regions [70] or to relate shape to other aspects of fish ecology, such as diet [71], [72]. However, this study represents the first attempt to use PLS analysis to relate shape and genetic data. For this analysis, we used as input (1) the landmarks data and (2) the scores on the PCOA axes for each specimen, which summarise the pattern arising from genetic analysis.
Results
Analysis of Meristic Counts
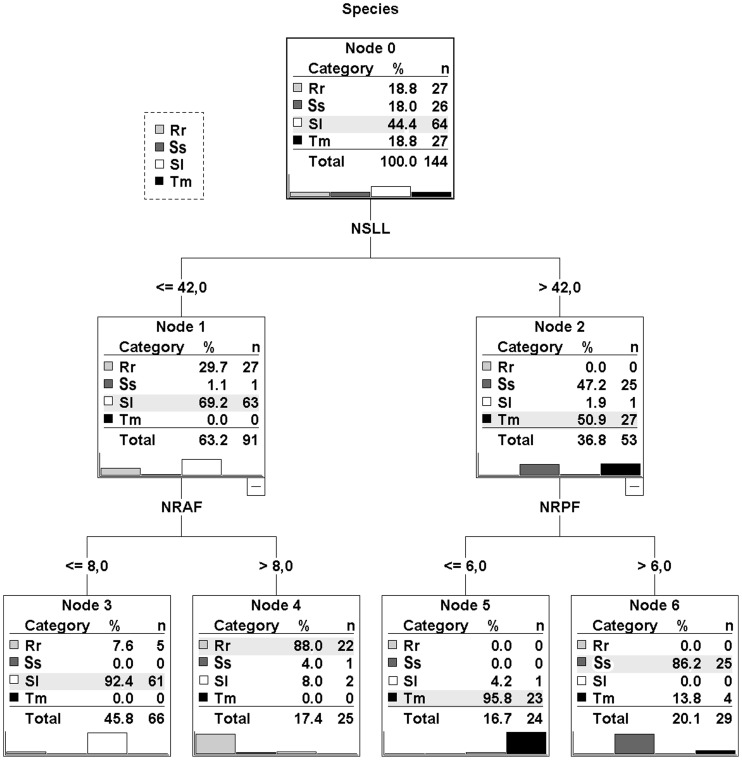
The CRT analysis produced a tree with four terminal nodes separating all four species (Fig. 2). The combination of NSLL, NRAF and NRPF characters allowed the discrimination among species The cross-validation procedure reported an overall correct classification percentage of 91%, ranging within species from 81.5% (R. rubilio) to 96.2% (S. squalus).
Figure 2. Classification tree obtained from the CRT analysis.
NSLL: number of scales of the lateral line; NRAF : number of rays of the anal fin; NRPF : number of rays of the left pectoral fin. Rr: R. rubilio; Ss: S. squalus; Sl: S. lucumonis; Tm: T. muticellus.
When misclassified specimens are excluded from analysis, some subtle differences can be detected in the ranges of the values of meristic characters for the four species (Table S1).
Shape Analysis
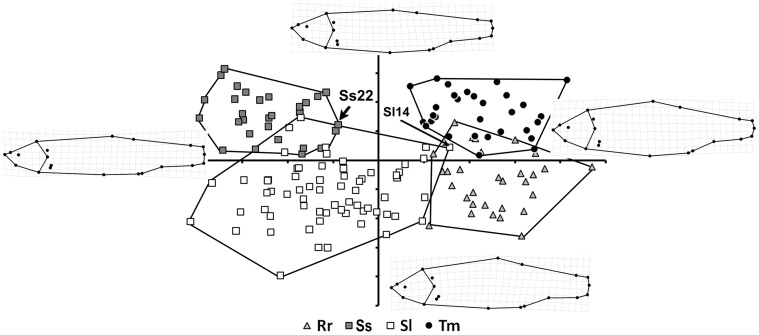
The results of the CVA were highly statistically significant for comparisons between the four species: the distribution along the two discriminant axes shows that intra-specific shape differences are much lower than the inter-specific ones (Procrustes distance = 0.038, Mahalanobis distance = 14.7161, p<0.01). Thus, the plot in Fig. 3 tends to segregate each taxon into a different quadrant, generating four, largely non-overlapping, groups of specimens. The magnitude of intra-group variation is largely consistent and comparable among three of the four species, whereas it is twice as large in S. lucumonis. The shape changes underpinning each axis can be examined by looking at the splines at the extremes of the axes. CVA1 describes a progressive increase in the lateral profile, which passes from a streamlined condition to a more discoidal one. This change mainly involves the head (see landmarks 2, 3 and 6) and trunk (see landmarks 11, 12, and 21) regions, whereas the tail region remains largely unchanged. The pattern depicted by this axis discriminates the two Squalius species from the R. rubilio and T. muticellus pair. In contrast, CVA2 describes a substantial change in the dorsal profile, which becomes more flattened in the transition from negative coordinates to positive ones along this axis. This shape change is captured by landmarks 3, 11, and 12. The dorsal profile of the head is also involved in this modification, such that the relative position of the eye (landmark 4) moves to a higher position. In addition, CVA2 describes a shortening of the height of the caudal peduncle (landmarks 13 and 17). All shape changes captured by this axis are crucial to distinguish between S. squalus and S. lucumonis and between R. rubilio and T. muticellus.
Figure 3. CVA output of the overall comparisons of the four taxa; the pattern described by the first two discriminant axes is shown.
Convex hulls are used to delimitate groupings; splines illustrate shape changes along each axis. Rr: R. rubilio; Ss: S. squalus; Sl: S. lucumonis; Tm: T. muticellus. See text for arrowed individuals.
The results of the cross-validation test are shown in Table 2. The model is efficient overall in identifying species (93.4% of cases). The highest rate of misclassification occurred in comparisons of R. rubilio with T. muticellus, whereas the lowest one occurred for the comparison between R. rubilio and S. squalus. In addition, the number of misclassified specimens differed from that arising from the observation of CVA pattern (Fig. 3). This difference reflects the fact that only the first two discriminant axes are visualised in Fig. 3, whereas the cross-validation test was carried out using all of the shape data.
Table 2. Results of the cross-validation test performed on shape data. Sl: S. lucumonis, Ss: S. squalus, Tm: T. muticellus and Rr: R. rubilio.
| Groups | N | Allocated to | Percent missclassified | |||
| Sl | Ss | |||||
| Sl | 65 | 62 | 3 | 4.6 | ||
| Ss | 26 | 4 | 22 | 15.3 | ||
| Sl | Tm | |||||
| Sl | 65 | 63 | 2 | 3.1 | ||
| Tm | 27 | 1 | 26 | 3.7 | ||
| Sl | Rr | |||||
| Sl | 65 | 62 | 3 | 4.6 | ||
| Rr | 27 | 2 | 25 | 7.4 | ||
| Ss | Tm | |||||
| Ss | 26 | 23 | 3 | 11.5 | ||
| Tm | 27 | 3 | 24 | 11.1 | ||
| Ss | Rr | |||||
| Ss | 26 | 26 | 0 | 0 | ||
| Rr | 27 | 0 | 27 | 0 | ||
| Tm | Rr | |||||
| Tm | 27 | 25 | 2 | 7.4 | ||
| Rr | 27 | 4 | 23 | 14.8 | ||
In the pairwise comparisons (Fig. S1), the landmarks that best captured shape differences between taxa are those of the dorsal (11–12), pelvic (21) and anal (18–19) fins and the anus (20). However, the landmarks for the pectoral fin (9–10), caudal fin (13–17), and eye position (4) have a non-negligible role in some comparisons. Looking at Figs. 3 and S1, it seems that S. lucumonis differs from the other taxa in the position of insertion of the dorsal fin (which is shifted in the caudal direction if compared to T. muticellus) and by its high lateral profile relative to R. rubilio and S. squalus.
Genetic Analyses
The presence of a single Cyfun P band was detected for each individual after amplicon size analysis on agarose gel. As expected [39], Cyfun P showed a larger variability at intergeneric (Squalius, Rutilus and Telestes) than at intrageneric (Squalius) level. Bands obtained ranged from approximately 310 bp in T. muticellus to 440 bp in R. rubilio. S. lucumonis and S. squalus showed similar intermediate band length (approximately 410 bp). Six different haplotypes were identified among the 28 analysed individuals, two typical of S. lucumonis (99.7% similarity), one of S. squalus, one of T. muticellus and two of R. rubilio (98.5% similarity). In spite of band length results, the alignment of the two Squalius species allowed to distinguish them by three diagnostic substitutions (two transversions and one transition) and by the presence of a 17 bp deletion at the 3′ end in S. lucumonis. Two individuals, one morphologically identified as S. squalus (Ss22) and one as S. lucumonis (Sl14), showed a S. lucumonis-like and a R. rubilio-like nuclear sequence, respectively. Sequence BLAST provided 99–100% similarity with GenBank specimens assigned to the same species, with the exception of S. lucumonis, whose sequences are not available in the database. In these case 99% similarity was obtained with the Balkan S. tenellus, S. microlepis and S. illyricus. When our sequences were aligned with those of other Squalius species present in Genbank (S. aphipsi, S. cephalus, S. illyricus, S. microlepis, S. squalus, S. tenellus, S. zrmanjae), the S. lucumonis 17 bp indel was shared only by the three above mentioned Balkan species.
Out of the 840 bp sequenced for RAG1 gene, 24 variable sites (2.8%), corresponding to 5 haplotypes, were identified. Sequence similarity within species ranged from 98.2% in S. squalus, to 100% in T. muticellus and R. rubilio. Higher percentages were detected for S. lucumonis and S. squalus, (99.9% and 100%, respectively), when the specimen Sl14 and Ss22 were excluded from the analysis. Indeed, RAG1 confirmed CyfunP results on these individuals (see above). Sequence similarity between species ranged from 98.1% (between S. lucumonis and S. squalus) to 98.8% (between R. rubilio and T. muticellus). BLAST search in GenBank did not provide informative data, as 99% similarity was obtained with many different leuciscine species, due to the general small substitution rate of nuclear genes and to the tendency of the program to round up the percentage of identity of the match to the nearest whole number. However when BLAST alignments were considered, the smaller number of substitutions for each species (1 for T. muticellus, 3 for S. lucumonis, 2–4 for S. squalus) was obtained with conspecific sequences, when available. The RAG1 phylogenetic trees obtained by the ML and BI analyses showed similar topologies (Fig. S2). The four species clustered in well-separated, well-supported clades, each of which included the conspecific sequences retrieved from GenBank.
Out of the 1131 bp sequenced for cyt b gene 248 variable sites (21.9%), corresponding to 16 haplotypes, were identified. Sequence similarity within species ranged from 87.1% (S. lucumonis) to 99.6% (S. squalus and R. rubilio). A higher percentage (99.1%) was detected for S. lucumonis when the specimen Sl14 was excluded from analysis. Indeed, this individual, according to nuclear marker data, showed a R. rubilio-like cyt b sequence. Sequence similarity between species ranged from 85.5% (between R. rubilio and T. muticellus) to 92.9% (between S. lucumonis and S. squalus). BLAST search of our sequences in GenBank, showed the highest percentage of similarity (99%) with specimens assigned to the same species. S. squalus sequences, in addition to exhibiting similarities to haplotypes of samples deposited with either this species name [73], [74] or the former valid one [2], [75], also showed 99% similarity with other species (S. lucumonis and S. zrmanjae) as already reported by Durand et al. [2].
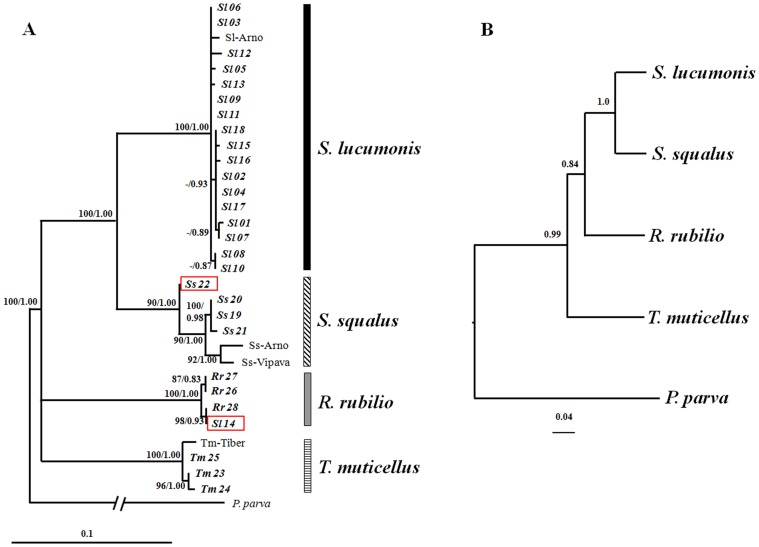
The phylogenetic trees obtained using separately nuclear (Fig. S2) and mitochondrial (Fig. S3) markers support the presence of four clades, each including conspecific sequences retrieved from GenBank. However their topologies show some incongruences in the branching of the four lineages. Two of the cyt b sequences retrieved from GenBank (AJ252818 and AJ252819) corresponding to specimens classified (based on morphology) as S. lucumonis but having S. squalus-like cyt b (Tib 1 and Tib 2) haplotypes [2] clustered with those of S. squalus. The topology of combined data trees (Fig. 4a) was identical to that obtained by cytochrome b, due to the higher number of variable sites of this gene compared to RAG1. The species tree (Fig. 4b) confirms the results obtained in gene genealogies supporting the distinction of four groups that correspond to the four species.
Figure 4. Species trees estimated with concatenation and coalescent approaches.
Panel A: ML and BI trees of concatenated dataset (RAG1 and cyt b sequences); bootstrap values (>70%) and Bayesian posterior probabilities (>0.7) are reported. Sl-Arno, Ss-Arno, Ss-Vipava, Tm-Tiber idicate specimens whose cyt b and RAG1 sequences were available in GenBank (see Table S4). Specimen Ss22 showing a S. lucumonis-like nuclear sequence, and Sl14 showing both mitochondrial and nuclear R. rubilio-like sequences, are boxed. Panel B: Phylogeny estimated with *BEAST; posterior probabilities shown for all nodes.
The analysis performed in JML showed consistent results for the nDNA and for the mtDNA markers, as minimum genetic distances observed between S. lucumonis and S. squalus were all non-significant smaller than simulated values. This indicate that incomplete lineage sorting alone can explain our data.
Integrating Morphology and Genetics: Partial Least-square Analysis
The complete suite of morphological (CRT and GM) and molecular (nuclear and mitochondrial) analyses was carried out for the 28 individuals (Table S2). In 21 specimens (75%), there was complete concordance of data. In the remaining 25% of cases, however, species assignment was equivocal, with disagreement between and within morphological (CART and Shape) and/or molecular (mitochondrial and nuclear) data. For example, the specimen Sl14 was assigned to S. lucumonis according to morphological data and to R. rubilio according to molecular data. In analyses of morphological data, five individuals (Table S2) were differently assigned by the CRT and Shape analyses. In two cases (Sl16 and Sl02) molecular data confirmed the CRT assignment, and in three cases (Rr26, Sl03, Tm23) molecular data confirmed the shape assignment. Nuclear and mitochondrial markers provided completely concordant data, with the exception of the already mentioned Ss22 individual that was initially classified as S. squalus according to meristic and morphometric traits.
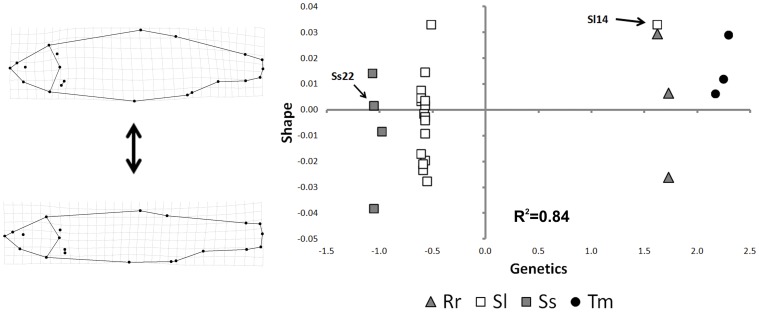
The PLS analysis detected a significant (p<0.05) monotonic relationship between shape and genetic distance for the four taxa (Fig. 5). When approximated by a linear relationship, it returned an R2 of 0.84. The two species of genus Squalius are close to each other in the negative half plane of latent vector 1, whereas R. rubilio and T. muticellus are located in the positive half plane. While the four species evidenced comparable levels of morphological and genetic variability, the taxa are mainly separated along the latent vector 1 (genetic information). This suggests that the degree of genetic distance between taxa is larger than those of morphological distance. Specimen Sl14, misclassified by the shape analysis, was the only specimen falling on the positive semi-plane of this axis.
Figure 5. Scatterplot representing the pattern obtained by partial least square (PLS) analysis of shape (GM) and genetic (PCOA axes) data.
Splines show shapes relative to the extremes of the y-axes. Specimen Ss22 and Sl14 are indicated by arrows.
Discussion
Our results, although based on specimens collected only in the Tiber River, and thus not representative of the entire distribution area, indicate that S. lucumonis, S. squalus, T. muticellus and R. rubilio are four valid and differentiated taxa and unambiguously confirm the status of S. lucumonis as a valid species.
The analyses of meristic counts and external shape reveal that the four species are characterised by significantly different, non-overlapping external morphologies. The specimens located in the regions of CVA overlap (Fig. 3), some of which were misclassified by the cross-validation procedure (Table 2), reflect a limitation in the efficiency of the model. The classification tree built on the basis of meristic characters separated the four taxa primarily by their NSLL and NRPF values; a third character (NRAF) appears to be important for distinguishing S. lucumonis from R. rubilio. Moreover, the two morphological analyses, each independently, separated the four taxa. In the CVA, a higher-order distinction was established between the genus Squalius (located at negative scores of axis CV1) and the two other species (R. rubulio and T. muticellus) (located at positive scores), suggesting some degree of intra-genus similarity. In contrast, the CRT analysis most strongly discriminated between pairs that did not reflect taxonomic relationships. However, the two critical meristic count characters identified by CRT analysis appear to be related to the shape changes identified by geometric morphometric analysis, as NSLL is a function of the absolute length of the body profile (being larger for a streamlined shape and shorter for a discoidal one) and both NRPF and NRAF are functions of the variation in the position of landmarks describing the insertions of these fins. For example, NRAF variation corresponds to one of the major external shape changes detected by geometric morphometrics in the pairwise comparison between R. rubilio and S. lucumonis (Fig. S1). Overall, the results of the morphological analyses reinforced one another, and the analyses provided different quantitative tools for distinguish among them.
Nuclear marker did not identify the presence of hybrid nuclear genomes either by amplicon band length analysis on agarose gel (Cyfun P) or by sequencing (Cyfun P and RAG1). However, the additional evaluation of the cyt b sequences allowed the identification of one individual (Ss22) with mixed genetic features, i.e., with a S. squalus cyt b haplotype and S. lucumonis nDNA. Consequently this individual shows an incongruent clustering in the two gene trees. Inconsistencies between gene trees, could be caused either by introgressive hybridization (ancient or recent) or incomplete lineage sorting [76], that can both generate very similar phylogenetic patterns [63]. Based on cyt b analysis, Durand et al. [2] already suggested incomplete lineage sorting between S. lucumonis and S. squalus. Indeed these authors identified three cytochrome b haplotypes from 9 brook chub individuals from Tiber Rivers, arranged into two different clades. One haplotype, shared by 6 specimens, formed the S. lucumonis clade, while the remaining two haplotypes (Tib1 and Tib2, 3 specimens) were included within the formerly “Adriatic lineage” of S. cephalus, now S. squalus. Neverthless, also introgressive hybridisation has been commonly reported in Squalius, often involving S. cephalus lineage [77]–[79]. Distinguishing between these two alternatives is not easy, and different methods were developed at this scope [63], [80], [81], [82]. The tests performed in JML suggests that the observed pattern is due to incomplete lineage sorting, a phenomenon that occurs when an ancestral species undergoes several speciation events in a short period of time. In this case the ancestral gene polymorphism is not fully resolved into two monophyletic lineages. However as “the difference in minimum distances between species simulated under incomplete lineage sorting and hybridization scenarios is not as distinct when short sequences are considered” [63], further and focused investigations are necessary to definitively solve this critical issue in every aspect.
Finally, the application of PLS analysis reconciles the results of the morphological (GM) and genetic analyses. The PLS analysis demonstrates that the information provided by these two different approaches is largely consistent and that their combined use can resolve cases of misclassification.
We detected a monotonic, progressive trend from R. rutilus and T. muticellus (the species characterised by a more discoidal body shape and with high scores on Shape axis) to the Squalius species (characterised by a more streamlined body shape and with low scores on Shape axis) (Fig. 5). S. lucumonis, however, spanned to the larger positive values on Shape axis, showing the largest shape variation among the four taxa. In this context, the misclassification of specimen Sl14 by the shape analysis can be explained by the scores at the extreme of the range that comprise S. lucumonis specimens along the second axis. Future studies could investigate if this pattern is related to ecomorphological factors (e.g., an adaptation to diverse ecological lotic systems, such as brooks, creeks, or small streams).
Conclusions
The morphological and genetic analyses reported here indicate that an integrated approach can be useful in resolving taxonomic relationships and in conservation management. This is particularly true for fish groups such as the Leuciscidae, which are characterised by large morphological intraspecific variation, interspecific overlap of morphological characters and frequent hybridisation events. Gilles et al. [7] reported that, in some Leuciscidae genera, the inferred number of species can vary depending on whether morphological or molecular criteria are adopted and that a comprehensive interpretation is possible only if a combination of approaches is applied. Our data provide evidence of morphological variation in the four taxa examined, with approximately 5 of 28 individuals ascribed to different taxa on the basis of CRT and shape characters. In contrast, if individuals with “hybrid” genotypes are present, they are undetectable in the population. Therefore, not only are morphological and genetic markers necessary for the correct assignment of individuals, both nuclear and mitochondrial datasets are also required for reliable genetic analyses. Our approach contributes to a broader knowledge of Italian leuciscines and provides robust elements for revisiting national and regional conservation strategies.
The presence in our dataset of an individual with mixed genetic traits (mtDNA of one Squalius species and nDNA of the other) poses a question on a strict application of the phylogenetic species concept to the two, otherwise monophyletic, groups (i.e. S. lucumonis and S. squalus). However Chambers [83], considering that speciation is a gradual process and that does not always exist a single perfectly-fitting species concept, suggests a synthetic model that allows the classification of pair of taxa applying a two step decision matrix. In the context of this synthetic model no further doubt on the status of S. lucumonis, at least in the Tiber River, is reasonable. This evidence, in light of the recent reassessment of this taxon into the “Endangered” IUCN category, highlights the need for immediate conservation actions. These actions require the inclusion of this species in the lists of fish species of conservational interest (sensu Habitat Directive), including the lists of the Latium Region [84], where a large number of populations are located. In addition conservation measures also need to address habitat restoration and specific legislation to protect residual and fragmented populations of this endemic species.
Supporting Information
Pairwise comparisons between the four taxa in which shape differences are stressed by a factor of 3.0X and visualised as vector arrows that start from the consensus configuration and end at the corresponding position of the taxon mean shape.
(TIF)
ML and BI trees based on RAG1 gene sequences using the TVMef+I G model of nucleotide substitution. Bootstrap values (>70%) and Bayesian posterior probabilities (>0.7) are reported. Specimen Ss22 and Sl14, showing a S. lucumonis-like and a R. rubilio-like nuclear sequence, respectively, are boxed.
(TIF)
ML and BI trees based on cyt b gene sequences using the GTR+I+G model of nucleotide substitution. Bootstrap values (>70%) and Bayesian posterior probabilities (>0.7) are reported. Specimen Ss22 showing a S. lucumonis-like nuclear sequence, and Sl14 showing both mitochondrial and nuclear R. rubilio-like sequences, are boxed.
(TIF)
Meristic characters (MC) inspected and ranges of values observed. In parenthesis values computed exclusively on specimens correctly classified by CRT.*marks meristic characters used for CRT.
(DOC)
Species assignment for each of the 28 individuals identified with both morphological (CART and Shape) and molecular (nuclear: Cyfun P and RAG1; mitochondrial: cyt b ) approaches. Sl stands for Squalius lucumonis, Ss for Squalius squalus, Tm for Telestes muticellus, Rr for Rutilus rubilio, respectively. In bold, individuals for which the species assignment was not univocal; in brackets Genbank accession numbers.
(DOC)
Primers and protocols used for amplifications.
(DOC)
Pairwise genetic distances used in PCOA analysis.
(DOC)
Available sequences for RAG 1, cyt b (GenBank Accession Number) and combined based phylogenetic analyses.
(DOC)
Acknowledgments
The authors acknowledge Riccardo Caprioli, Stefano Larsen, Fabio Campagna, Massimiliano Scalici, Mario Libianchi and Daniele Ciuffa for assistance with sampling activity; Domitilla Pulcini and Paolo Franchini for their technical help during data analysis.
Funding Statement
These authors have no support or funding to report.
References
- 1. Turner GF (1999) What is a fish species? Rev Fish Biol Fish 9: 281–297. [Google Scholar]
- 2. Durand JD, Ünlü E, Doadrio I, Pipoyan S, Templeton AR (2000) Origin, radiation, dispersion and allopatric hybridization in the chub, Leuciscus cephalus . Proc R Soc Lond B Biol Sci 267: 1987–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scribner KT, Page KS, Bartron ML (2001) Hybridization in freshwater fishes: a review of case studies and cytonuclear methods of biological inference. Rev Fish Biol Fish 10: 293–323. [Google Scholar]
- 4. Costedoat C, Pech N, Chappaz R, Salducci D, Lim P, et al. (2004) Study of introgressive hybridization between Chondrostoma t. toxostoma and Chondrostoma n. nasus (Teleostei, Cyprinidae) using multiple approaches. Cybium 28: 51–61. [Google Scholar]
- 5. Freyhof J, Lieckfeldt D, Bogutskaya NG, Pitra C, Ludwig A (2005) Molecules and morphology: evidence for introgression of mitochondrial DNA in Dalmatian cyprinids. Mol Phylogenet Evol 37: 347–54. [DOI] [PubMed] [Google Scholar]
- 6. Sousa-Santos C, Collares-Pereira MJ, Almada V (2007) Reading the history of a hybrid fish complex from its molecular record. Mol Phylogenet Evol 45: 981–96. [DOI] [PubMed] [Google Scholar]
- 7. Gilles A, Costedoat C, Barascud B, Voisin A, Bananrescu P, et al. (2010) Speciation pattern of Telestes souffia complex (Teleostei, Cyprinidae) in Europe using morphological and molecular markers. Zool Scr 39: 225–242. [Google Scholar]
- 8. Hayden B, Pulcini D, Kelly-Quinn M, O’Grady M, Caffrey J, et al. (2010) Hybridisation between two cyprinid fishes in a novel habitat: genetics, morphology and life-history traits. BMC Evol Biol 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toscano BJ, Pulcini D, Hayden B, Russo T, Kelly-Quinn M, et al. (2010) An ecomorphological frame work for the coexistence of two cyprinid fish and their hybrids in a novel environment. Biol J Linn Soc Lond 99: 768–783. [Google Scholar]
- 10. Yakovlev VN, Slyn’ko YV, Grechanov IG, Krysanov EY (2000) Distant hybridization in fish. J Ichthyol 40: 298–311. [Google Scholar]
- 11.Nelson JS (2006) Fishes of the world, 4th edn. New York: John Wiley & Sons.
- 12.Eschmeyer WN (2012) Catalog of Fishes electronic version (15 March 2012). http://research.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
- 13. Chen WJ, Mayden RL (2009) Molecular systematics of the Cyprinoidea (Teleostei: Cypriniformes), the world’s largest clade of freshwater fishes: further evidence from six nuclear genes. Mol Phylogenet Evol 52: 544–549. [DOI] [PubMed] [Google Scholar]
- 14. Reyjol Y, Hugueny B, Pont D, Bianco PG, Beier U, et al. (2007) Patterns in species richness and endemism of European freshwater fish. Glob Ecol Biogeogr 16: 65–75. [Google Scholar]
- 15.Crivelli AJ (1996) The freshwater fish endemic to the northern Mediterranean region, an action plan for their conservation. Arles: Station Biologique de la Tour du Valat.
- 16. Bianco PG (1995) Mediterranean endemic freshwater fishes of Italy. Biol Conserv 72: 159–170. [Google Scholar]
- 17.Crivelli AJ (2006) Squalius lucumonis IUCN 2011 IUCN Red List of Threatened Species Version 2011.2. www.iucnredlist.org. Downloaded on 11 November 2011.
- 18.Lelek A (1987) Threatened Fishes of Europe, vol.9. In: The Freshwater Fihses of Europe. Wiebelsheim: AULA-Verlag.
- 19.EC (1982) Council Decision 82/72/EEC of 3 December 1981 concerning the conclusion of the Convention on the conservation of European wildlife and natural habitats (Bern Convention). Official Journal L of 10.02.1982.
- 20.EC (1998) Council Decision of 21 December 1998 concerning the approval, on behalf of the Community, of amendments to Appendices II and III to the Berne Convention on the conservation of European wildlife and natural habitats adopted at the 17th meeting of the Convention's Standing Committee. Official Journal L 358 of 31.12.1998.
- 21. Manaresi S, Matovani B, Zaccanti F (2001) Egg to adult identification of 13 freshwater fishes from Italy: a biochemical-genetic key. Aquat Sci 63: 182–190. [Google Scholar]
- 22. Verspoor E, Hammar J (1991) Introgressive hybridization in fishes: the biochemical evidence. J Fish Biol 39: 309–334. [Google Scholar]
- 23. Gilles A, Lecointre G, Faure E, Chappaz R, Brun G (1998) Mitochondrial phylogeny of the European cyprinids: implication for their systematics, reticulate evolution, and colonization time. Mol Phylogenet Evol 10: 132–143. [DOI] [PubMed] [Google Scholar]
- 24. Coscia I, Rountree V, King JJ, Roche WK, Mariani S (2010) A highly permeable species boundary between two anadromous fish. J Fish Biol 77: 137–1149. [DOI] [PubMed] [Google Scholar]
- 25. Fraser DJ, Bernatchez L (2001) Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10: 2741–2752. [PubMed] [Google Scholar]
- 26. Salducci MD, Martin J, Pech N, Chappaz R, Costedoat C, et al. (2004) Deciphering the evolutionary biology of freshwater fish using multiple approaches - insights for the biological conservation of the vairone (Leuciscus souffia souffia). Conserv Genet 5: 63–78. [Google Scholar]
- 27. Freyhof J, Lieckfeldt D, Bogutskaya NG, Pitra C, Ludwig A (2007) Phylogenetic position of the Dalmatian genus Phoxinellus and description of the newly proposed genus Delminichthys (Teleostei: Cyprinidae). Mol Phylogenet Evol 38: 416–425. [DOI] [PubMed] [Google Scholar]
- 28. Costedoat C, Chappaz R, Barascud B, Guillard O, Gilles A (2006) Heterogeneous colonization pattern of european cyprinids, as highlighted by the dace complex (Teleostei: Cyprinidae). Mol Phylogenet Evol 41: 127–148. [DOI] [PubMed] [Google Scholar]
- 29. Doadrio I, Carmona JA (2006) Phylogenetic overview of the genus Squalius (Actinopterygii, Cyprinidae) in the Iberian peninsula, with description of two new species. Cybium 30: 199–214. [Google Scholar]
- 30. Bianco PG, Ketmaier V (2003) Threatened fishes of the world: Leuciscus lucumonis Bianco, 1983 (Cyprinidae). Environ Biol Fishes 68: 370. [Google Scholar]
- 31.Kottelat M, Freyhof J (2007) Handbook of European freshwater fishes. Cornol: Publication Kottelat.
- 32. Giannetto D, Franchi E, Pompei L, Lorenzoni M, Poercellotti S, et al. (2012) Proposed empirical standard weight equation for brook chub Squalius lucumonis . N Am J Fish Manage 32: 428–435. [Google Scholar]
- 33. Bianco PG (1983) Leuciscus lucumonis n. sp. from Italy (Pisces: Cyprinidae). Senckenb Biol 64: 81–87. [Google Scholar]
- 34.Gandolfi G, Zerunian S, Torricelli PM, Marconato A (1991) I Pesci delle Acque Interne Italiane. Roma: Ministero dell'Ambiente, Unione Zoologica Italiana.
- 35.Zerunian S (2002) Condannati all’estinzione? Biodiversità, biologia, minacce e strategie di conservazione dei Pesci d’acqua dolce indigeni in Italia. Bologna: Ed. Agricole.
- 36. Manaresi S, Matovani B, Zaccanti F (1997) Isozymic and muscle protein comparison of three taxa of Leuciscus from Northern Italy. Ital J Zool 64: 215–220. [Google Scholar]
- 37. Maldini M, Vaghi M, Nonnis Marzano F, Gandolfi G, Prigioni C, et al. (2006) Ibridazione intergenerica tra i ciprinidi Rutilus rubilio (Bonaparte 1873) e Leuciscus cephalus (L. 1758) valutata mediante l’analisi di aplotipi mitocondriali. Quaderni ETP 34: 63–67. [Google Scholar]
- 38. EC (1992) Council Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora. Official Journal L: 2006. [Google Scholar]
- 39. Lieckfeldt D, Hett AK, Ludwig A, Freyhof J (2006) Detection, characterization and utility of a new highly variable nuclear marker region in several species of cyprinid fishes (Cyprinidae). Eur J Wildl Res 52: 63–65. [Google Scholar]
- 40.Breiman L, Friedman JH, Olshen RA, Stone CJ (1984) Classification and regression trees. New York: Chapman and Hall.
- 41. Vayssières MP, Plant RE, Allen-diaz BH (2000) Classification trees: an alternative non-parametric approach for predicting species distribution. J Veg Sci 11: 679–694. [Google Scholar]
- 42.Rohlf FJ (2006) TPS dig Stony Brook, NY: Department of Ecology and Evolution, State University of New York. Available: http://life.bio.sunysb.edu/morph/.
- 43. Rohlf FJ, Slice DE (1990) Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Biol 39: 40–59. [Google Scholar]
- 44.Bookstein FL (1991) Morphometric tools for landmark data: geometry and biology. Cambridge: Cambridge University Press.
- 45.Zelditch ML, Swiderski DL, Sheets HD, Fink WL (2004) Geometric morphometrics for biologists: a primer. London: Elsevier Academic Press.
- 46. Klingenberg CP, Monteiro LR (2005) Distances and directions in multidimensional shape spaces: implications for morphometric applications. Syst Biol 54: 678–688. [DOI] [PubMed] [Google Scholar]
- 47.Klingenberg CP (2007) MorphoJ. UK: University of Manchester.
- 48.Rohlf FJ (2000) TpsRegr, Version 1.24. Stony Brook, NY: Department of Ecology and Evolution, State University of New York.
- 49. Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR- based techniques. Nucleic Acids Res 25: 4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quenouille B, Bermingham E, Planes S (2004) Molecular systematics of the damselfishes (Teleostei: Pomacentridae): Bayesian phylogenetic analyses of mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 31: 66–88. [DOI] [PubMed] [Google Scholar]
- 51. Milana V, Sola L, Congiu L, Rossi AR (2008) Mitochondrial DNA in Atherina (Teleostei, Atheriniformes): differential distribution of an intergenic spacer in lagoon and marine forms of Atherina boyeri . J Fish Biol 73: 1216–227. [Google Scholar]
- 52. Minegishi Y, Aoyama J, Inoue JG, Miya M, Nishida M, et al. (2005) Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Mol Phylogenet Evol 34: 134–146. [DOI] [PubMed] [Google Scholar]
- 53. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 55.Nylander JAA (2008) MrModeltest v2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- 56. Akaike H (1974) A new look at statistical model identification. IEEE Trans Automatic Control 19: 716–723. [Google Scholar]
- 57. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 58. Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 59. Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Drummond AJ, Suchard MA, Dong Xie, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambaut A, Drummond AJ (2009) Tracer v1.5. Available from http://beast.bio.ed.ac.uk/Tracer.
- 62.Rambaut A (2008) FigTree v1.4.0. Available from http://tree.bio.ed.ac.uk/software/figtree.
- 63. Joly S, McLenachan PA, Lockhart PJ (2009) A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am Nat 174: e54–e70. [DOI] [PubMed] [Google Scholar]
- 64. Joly S (2012) JML: testing hybridization from species trees. Mol Ecol Res 12: 179–184. [DOI] [PubMed] [Google Scholar]
- 65.Hammer Ø, Harper DAT, Ryan PD (2001) Past: paleontological statistics software package for education and data analysis. Palaeontol Electronica 4: 9 p.
- 66. Bookstein FL, Gunz P, Mitteroecker P, Prossinger H, Schaefer K, et al. (2003) Cranial integration in Homo: singular warps analysis of the midsagittal plane in ontogeny and evolution. J Hum Evol 44: 167–187. [DOI] [PubMed] [Google Scholar]
- 67. Fadda C, Corti M (2001) Three-dimensional geometric morphometrics of Arvicanthis : implications for systematics and taxonomy. J Zoolog Syst Evol Res 39: 235–245. [Google Scholar]
- 68. Rohlf FJ, Corti M (2000) The use of two-block partial least-squares to study covariation in shape. Syst Biol 49: 740–753. [DOI] [PubMed] [Google Scholar]
- 69.Rohlf FJ (2006) TPSPLS Ver. 1.18. Department of Ecology and Evolution, State University of New York: Stony Book.
- 70. Russo T, Pulcini D, Bruner E, Cataudella S (2009) Shape and size variation: growth and development of the dusky grouper (Epinephelus marginatus, Lowe, 1834). J Morph 270: 83–96. [DOI] [PubMed] [Google Scholar]
- 71. Kassam DD, Adams DC, Hori M, Yamaoka K (2003) Morphometric analysis on ecomorphologically equivalent cichlid species from Lakes Malawi and Tanganyika. J Zool 260: 153–157. [Google Scholar]
- 72. Russo T, Pulcini D, O’Leary A, Cataudella S, Mariani S (2008) Relationship between body shape and trophic niche segregation in two closely related sympatric fishes. J Fish Biol 73: 809–828. [Google Scholar]
- 73. Perea S, Böhme M, Zupančič P, Freyhof J, Šanda R, et al. (2010) Phylogenetic relationships and biogeographical patterns in Circum-Mediterranean subfamily Leuciscinae (Teleostei, Cyprinidae) inferred from both mitochondrial and nuclear data. BMC Evol Biol 10: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dubut V, Fouquet A, Voisin A, Costedoat C, Chappaz R, Gilles A (2012) From late Miocene to Holocene: processes of differentiation within the Telestes genus (Actinopterygii: Cyprinidae). PLoS ONE 7, E34423. [DOI] [PMC free article] [PubMed]
- 75. Sanjur OI, Carmona JA, Doadrio I (2003) Evolutionary and biogeographical patterns within Iberian populations of the genus Squalius inferred from molecular data. Mol Phylogenet Evol 29: 20–30. [DOI] [PubMed] [Google Scholar]
- 76. Maddison W (1997) Gene trees in species trees. Syst Biol 46: 523–536. [Google Scholar]
- 77. Wheeler A, Easton K (1978) Hybrids of chub and roach (Leuciscus cephalus and Rutilus rutilus) in English rivers. J Fish Biol 13: 467–473. [Google Scholar]
- 78. Bianco PG (1988) Leuciscus cephalus (Linnaeus), with records of fingerling adult males, Leuciscus pleurobipunctatus (Stephanidis) and their hybrids from western Grecee. J Fish Biol 32: 1–16. [Google Scholar]
- 79. Ünver B, Tatlidil H, Erk’akan F (2008) Biometrical features of intergeneric hybrid between Leuciscus cephalus(L.) and Chalcalburnus chalcoides (G.) (Osteichthyes-Cyprinidae) distributed in Lake Tödürge (Sivas-Turkey). Turk J Fish Aquat Sci 8: 207–213. [Google Scholar]
- 80. Holland B, Benthin S, Lockhart P, Moulton V, Huber K (2008) Using supernetworks to distinguish hybridization from lineage-sorting. BMC Evol Biol 8: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bloomquist EW, Suchard MA (2010) Unifying vertical and vonvertical evolution: a stochastic ARG-based Framework. Syst Biol 59: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yu Y, Cuong T, Degnan JH, Nakhleh L (2011) Coalescent histories on phylogenetic networks and detection of hybridization despite incomplete lineage sorting. Syst Biol 60: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chambers G (2012) The species problem: seeking new solutions for philosophers and biologists. Biol Philos 27: 755–765. [Google Scholar]
- 84.Zerunian S (2008) Pesci d’acqua dolce. In: Habitat e specie di interesse comunitario nel Lazio, Calvario E, Sebasti S, Copiz R, Salomone F, Brunelli M, Tallone G, Blasi C, eds.(Edizioni ARP), 225–226.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pairwise comparisons between the four taxa in which shape differences are stressed by a factor of 3.0X and visualised as vector arrows that start from the consensus configuration and end at the corresponding position of the taxon mean shape.
(TIF)
ML and BI trees based on RAG1 gene sequences using the TVMef+I G model of nucleotide substitution. Bootstrap values (>70%) and Bayesian posterior probabilities (>0.7) are reported. Specimen Ss22 and Sl14, showing a S. lucumonis-like and a R. rubilio-like nuclear sequence, respectively, are boxed.
(TIF)
ML and BI trees based on cyt b gene sequences using the GTR+I+G model of nucleotide substitution. Bootstrap values (>70%) and Bayesian posterior probabilities (>0.7) are reported. Specimen Ss22 showing a S. lucumonis-like nuclear sequence, and Sl14 showing both mitochondrial and nuclear R. rubilio-like sequences, are boxed.
(TIF)
Meristic characters (MC) inspected and ranges of values observed. In parenthesis values computed exclusively on specimens correctly classified by CRT.*marks meristic characters used for CRT.
(DOC)
Species assignment for each of the 28 individuals identified with both morphological (CART and Shape) and molecular (nuclear: Cyfun P and RAG1; mitochondrial: cyt b ) approaches. Sl stands for Squalius lucumonis, Ss for Squalius squalus, Tm for Telestes muticellus, Rr for Rutilus rubilio, respectively. In bold, individuals for which the species assignment was not univocal; in brackets Genbank accession numbers.
(DOC)
Primers and protocols used for amplifications.
(DOC)
Pairwise genetic distances used in PCOA analysis.
(DOC)
Available sequences for RAG 1, cyt b (GenBank Accession Number) and combined based phylogenetic analyses.
(DOC)