Abstract
A three-dimensional stereotaxic atlas of the human brainstem based on high resolution ex vivo diffusion tensor imaging (DTI) is introduced. The atlas consists of high resolution (125–255 μm isotropic) three-dimensional DT images of the formalin-fixed brainstem acquired at 11.7T. The DTI data revealed microscopic neuroanatomical details, allowing three-dimensional visualization and reconstruction of fiber pathways including the decussation of the pyramidal tract fibers, and interdigitating fascicles of the corticospinal and transverse pontine fibers. Additionally, strong grey-white matter contrasts in the apparent diffusion coefficient (ADC) maps enabled precise delineation of grey matter nuclei in the brainstem, including the cranial nerve and the inferior olivary nuclei. Comparison with myelin-stained histology shows that at the level of resolution achieved in this study, the structural details resolved with DTI contrasts in the brainstem were comparable to anatomical delineation obtained with histological sectioning. Major neural structures delineated from DTI contrasts in the brainstem are segmented and three-dimensionally reconstructed. Further, the ex vivo DTI data are nonlinearly mapped to a widely-used in vivo human brain atlas, to construct a high-resolution atlas of the brainstem in the Montreal Neurological Institute (MNI) stereotaxic coordinate space. The results demonstrate the feasibility of developing a 3D DTI based atlas for detailed characterization of brainstem neuroanatomy with high resolution and contrasts, which will be a useful resource for research and clinical applications.
Keywords: human brainstem, atlas, stereotaxic, diffusion tensor imaging
INTRODUCTION
The brainstem is the primary relay center for efferent and afferent connections between the cerebral cortex and the cerebellum and spinal cord (Carpenter, 1976). Prominent anatomic structures in the brainstem comprise densely-packed cranial nerve nuclei interspersed with ascending and descending white matter tracts, many of which, e.g. the corticospinal tract and the superior cerebellar peduncles, undergo decussations in this region. Fiber pathways in the brainstem are critically important for vital sensory and motor functions, and are implicated in the pathogenesis of a wide spectrum of neurologic disorders, including multiple sclerosis, Parkinson’s disease, and amyotrophic lateral sclerosis. The anatomy of the brainstem has been largely studied using two-dimensional histological sectioning and staining techniques (Afshar et al., 1978; Paxinos and Huang, 1995; Nolte, 2008). However, even in detailed atlases of myelin-stained sections, the orientations and exact three-dimensional anatomical locations of individual brainstem tracts, and their spatial positions relative to various nuclei and adjacent pathways, are sometimes difficult to comprehend. Because of the important neural structures concentrated in this relatively small region, lesions within the brainstem can lead to multiple neurologic deficits (Donaldson et al., 2006). Hence, three-dimensional imaging and detailed delineation of brainstem neuroanatomy are important to provide a complete understanding of its complex structure, and will also aid our ability to evaluate the impact of brainstem lesions and associated deficits.
Despite its highly organized structure, the brainstem appears relatively homogeneous in conventional relaxometry-based magnetic resonance images, e.g. T1- and T2-weighted images, which provide limited contrasts to visualize its neuroanatomy. In comparison, studies using diffusion tensor imaging (DTI) have shown the potential of this technique to identify key brainstem structures and tracts. DTI provides orientation-based tissue contrasts for better structural delineation, and has been routinely used for anatomical imaging of the brainstem (Virta et al., 1999; Stieltjes et al., 2001; Nagae-Poetscher et al., 2004; Salamon et al., 2005), and evaluation of brainstem pathology in clinical studies (Parvizi and Damasio, 2003; Chen et al., 2007; Lui et al., 2007; Helton et al., 2008). Typical in vivo resolutions for brainstem imaging in these studies range from 1.7–2.0 mm for two-dimensional DTI, with slice thickness ranging from 2.0–5.0 mm. Although the achievable resolution range with in vivo DTI enables the reconstruction of major brainstem tracts, it is too coarse to be used for delineation of microstructural details, and is far from comparable to histological staining methods. Axonal tracing techniques, such as silver impregnation, are therefore still used as the ‘gold standard’ to study pathways of the brainstem in postmortem specimens. Polarized light imaging, a relatively new technique, has been recently shown to reveal fiber orientations in sections of the postmortem brainstem with relatively high in-plane resolution (Axer et al., 2011). However, these techniques are currently limited to sectioning of the tissue specimen into multiple slabs of ~100–120 μm thickness.
Compared to in vivo imaging, ex vivo DTI of postmortem tissue specimens can allow data acquisition at significantly higher resolutions, which can reveal fiber architecture and details that are not resolvable in vivo. As indicated by previous studies of ex vivo tissue specimens (McNab et al., 2009; Augustinack et al., 2010; Miller et al., 2011), postmortem DTI contrasts can enable detailed anatomical characterization of brain structures. Previous studies have reported two-dimensional postmortem DTI of the brainstem, with through-plane resolutions of ~0.6 mm (Soria et al., 2011; Prats-Galino et al., 2012). Recently, an atlas of the postmortem brainstem based on DTI has also been introduced (Naidich et al., 2009). However, postmortem DTI studies of brainstem anatomy have been limited to two-dimensional imaging. The application of DTI for three-dimensional high resolution imaging of the brainstem is hindered by the long acquisition times required and the concomitant decrease in the signal-to-noise ratio with increasing resolutions. Additionally, the alteration of tissue properties with postmortem fixation is known to reduce the diffusion coefficient (Schmierer et al., 2008), which mandates the use of significantly high b-values to obtain sensitivity to diffusion for tissue contrasts. As such, there is currently no three-dimensional atlas of the brainstem with detailed anatomical delineation. A three-dimensional atlas of the brainstem is important for several applications. For instance, previous studies have used 3D interpolation of stacked two-dimensional histological sections in order to stereotaxically register patient MR images (Afshar and Dykes, 1982; Niemann et al., 1999). A detailed 3D MRI/DTI based atlas will enable direct stereotaxic registration of patient MR data, and will additionally lead to a better understanding of in vivo DTI contrasts in the brainstem.
Therefore, in the present study, we aimed to develop a three-dimensional human brainstem atlas based on high resolution ex vivo DTI. The use of fast acquisition techniques combined with high field strength on a preclinical scanner (11.7T) enabled three-dimensional DTI at high spatial resolution (125–255 μm isotropic) with strong tissue contrasts. The results revealed very fine neuroanatomical details in the brainstem, allowing delineation of tract features such as the decussation of the pyramidal fibers three-dimensionally. Comparison with histology shows that the level of structural details revealed by the high resolution DTI contrasts was comparable to anatomical delineation obtained with myelin-staining in the brainstem. Various neural structures are three-dimensionally reconstructed and the data registered to a widely-used in vivo MRI based brain atlas in the MNI stereotaxic coordinate space. The atlas developed in our study provides high-resolution visualization of brainstem neuroanatomy and reconstructed structures, and will be a useful resource for research and clinical applications.
MATERIAL AND METHODS
Specimen preparation
The brainstem specimen used in this study was obtained at autopsy from an adult woman with no history of neurological disease. The brainstem and adjacent cervical spinal cord were removed and immersion fixed in 10% neutral buffered formalin for one week. The specimen was then transferred to phosphate-buffered saline (PBS) with 2 mM gadopentetate dimeglumine (Magnevist, Berlex Imaging, Wayne, NJ, USA) for 48 h. For imaging, the specimen was placed in a polyethylene container that was filled with Fomblin (Fomblin Perfluoropolyether, Solvay Solexis, Thorofare, NJ, USA), to prevent dehydration and reduce susceptibility artifacts at tissue margins. Following imaging of the whole brainstem, the medulla oblongata and the cervical spinal cord at the level of the spinomedullary junction were extracted separately in order to fit smaller diameter coils for higher resolution imaging. During imaging, the temperature of the specimens was maintained constant at 37°C via the spectrometer’s temperature control system.
DTI acquisition
Imaging of the whole brainstem was performed on a horizontal bore 11.7-Tesla NMR spectrometer (Bruker Biospin, Billerica, MA, USA) with a triple-axis gradient system (maximum gradient strength of 740 mT/m), using an 8-channel volume coil. DTI data were acquired using a 12-segment diffusion-weighted echo planar imaging sequence, with echo time (TE)/repetition time (TR) = 27/500 ms, 1 signal average, partial Fourier acquisition factor of 1.4, 2 non-diffusion weighted (b0) images and 30 diffusion directions (Jones, 2004) (b-value=4000 s/mm2). The field of view (FOV) and matrix size were 4.3 × 4.3 × 6.5 cm3 and 168 × 168 × 256, respectively, for a native spatial resolution of 255 × 255 × 255 μm3 and total imaging time of 13.5 h. A T2-weighted image was acquired at the same resolution and FOV as the DTI data, using a 3D RARE sequence with TE/TR=31/2000 ms, 4 signal averages, rare-factor of 8, and imaging time 8.5 h.
DTI of the medulla and the spinomedullary junction was performed on a vertical bore 11.7-Tesla spectrometer (Bruker Biospin, Billerica, MA, USA), equipped with a Micro2.5 gradient system (maximum gradient strength of 1000 mT/m), using birdcage transmit/receive coils of 30- and 20-mm diameters, respectively. For the medulla, a diffusion-weighted multiple spin echo sequence was used with TE/TR=34, 45, 56/400 ms, 2 signal averages, 2 b0 images and 6 diffusion directions (b-value=2800 s/mm2), FOV 3.0 × 2.6 × 2.45 cm3, matrix size 176 × 152 × 144, resolution 170 × 170 × 170 μm3, and imaging time of 35 h. For the spinomedullary junction, a diffusion-weighted gradient and spin echo (GRASE) sequence with navigator phase correction was used (Aggarwal et al., 2010), with TE/TR=32/800 ms, 4 signal averages, 12 echoes per refocusing pulse, 2 b0 images and 16 diffusion directions (b-value=2100 s/mm2), FOV 1.6 × 1.25 × 1.72 cm3, matrix size 128 × 100 × 138, resolution 125 × 125 × 125 μm3, and scan time of 18.5 h. The spectral data were apodized by a trapezoidal function with symmetric 10% ramp widths, and zero-filled to twice the matrix size prior to Fourier transformation.
Data processing and tract reconstruction
Diffusion tensor fitting was done using the log-linear fitting function in DtiStudio (Jiang et al., 2006). The eigenvalues and associated eigenvectors (Basser et al., 1994) of the voxel-wise diffusion tensors were computed, from which maps of the apparent diffusion coefficient (ADC) and fractional anisotropy (FA) indices were derived. Isotropic diffusion-weighted (iDW) images were generated by averaging diffusion-weighted images along isotropically-distributed diffusion directions. The FA and primary eigenvector maps were combined to generate direction-encoded colormap (DEC) images, with the ratio between the red, green and blue components equal to the ratio of absolute values of x, y and z components of the primary eigenvector, and the intensity proportional to FA. Red, green and blue were assigned to anisotropy orientation along the medial-lateral, anterior-posterior, and superior-inferior axes, respectively (Pajevic and Pierpaoli, 1999).
For tractography, the fiber assignment by continuous tracking (FACT) algorithm (Mori et al., 1999) was used, with an FA threshold of 0.3 and inner product threshold of 0.8 (angles greater than 37° excluded from tracking). A brute force approach was used to track fibers originating from all voxels within the brainstem, and fibers that penetrated manually-defined regions of interest (ROIs) were assigned to specific tracts associated with each ROI. Anatomical definitions of tracts in corresponding axial brain sections in a histology-based atlas (Nolte and Angevine, 2000) were used to guide the placement of ROIs in the DTI images. The reconstructed fiber data were imported into the MRtrix package (Tournier et al., 2012) for three-dimensional visualization.
Coregistration to stereotaxic in vivo brain atlas
To incorporate the high-resolution neuroanatomical information of the brainstem in widely-used Montreal Neurological Institute (MNI) stereotaxic coordinates, the ex vivo DTI data of the brainstem were co-registered to an in vivo whole-brain MRI atlas (JHU brain atlas, www.mristudio.org). The JHU brain atlas contains single-subject and population-averaged (ICBM-DTI-81) images in the MNI coordinate system with T1-weighted, T2-weighted, and DTI-derived contrasts (Oishi et al., 2011). The registration was driven by DEC maps calculated from DTI, in which major structures can be readily identified in both in vivo and ex vivo coordinates. The JHU brain atlas was first cropped and resampled to match the FOV and resolution of the ex vivo brainstem data, followed by intensity-based rigid registration. To remove residual misalignment and correct for possible postmortem tissue distortion, the registered brainstem data were then nonlinearly deformed to the in vivo atlas via large deformation diffeomorphic metric mapping (Miller et al., 2002) based on placement of 40 manually-defined anatomic landmarks. Transformation of the tensor field was carried out as described previously by Alexander et al. (2001).
RESULTS
Anatomical details in the brainstem resolved with DTI contrasts
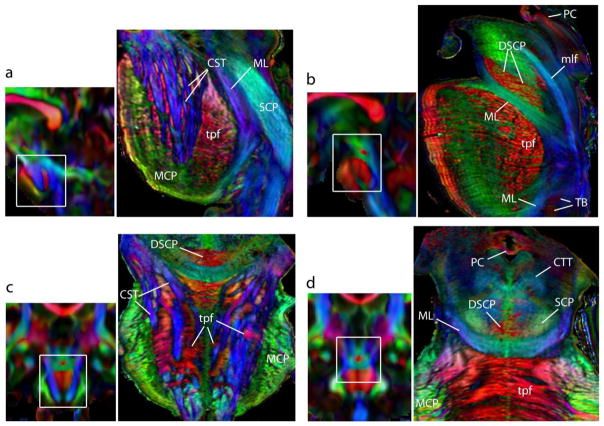
Fig. 1 shows 2D planar views generated from the 3D DEC maps of the brainstem acquired at 255 μm resolution. Corresponding T2-weighted axial sections are shown for anatomic comparison. Structural orientation based contrasts in the DEC maps enabled detailed anatomical delineation in the brainstem. At the medullary level, major longitudinal pathways including the corticospinal tract, medial lemniscus, and medial longitudinal fasciculus were identified by their predominantly inferior-superior orientations (blue in the DEC maps). At the level of the pons, the corticospinal tract splits into multiple fiber bundles, that descend through interdigitating transverse pontine fibers. The DEC maps provided striking contrasts to delineate the craniocaudally-oriented fiber bundles of the corticospinal tract (blue) and the interleaving mediolaterally-oriented transverse pontine fibers (red), based on their respective structural orientations (Fig. 1). The superior cerebellar peduncle was distinctly delineated (crescent-shaped in axial sections) in the pontine tegmentum, which appeared relatively homogeneous in T2-weighted contrasts. Fig. 2 further illustrates the level of structural detail resolved by the high-resolution DTI data at different sagittal and coronal levels in the brainstem. The decussation of the superior cerebellar peduncles was clearly demarcated in the DEC maps as red crossing fibers at the mid sagittal level (Fig. 2b–d). Crossing fibers of the posterior commissure and the trapezoid body can also be clearly seen in the DEC maps in Fig. 2, delineated by their mediolateral orientation (Fig. 2b,d).
Fig. 1.
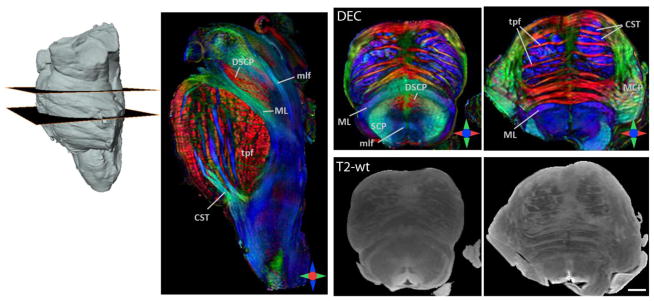
DTI of the ex vivo brainstem at 255 μm isotropic resolution. Sagittal and axial sections from the direction-encoded colormap (DEC) derived from DTI are shown. Corresponding T2-weighted (T2-wt) axial sections are shown for anatomic comparison. In the DEC maps, red, blue and green represent anisotropy along medial-lateral, superior-inferior, and anterior-posterior orientations, respectively. Abbreviations used are: CST: corticospinal tract, ML: medial lemniscus, mlf: medial longitudinal fasciculus, MCP: middle cerebellar peduncle, SCP: superior cerebellar peduncle, DSCP: decussation of the superior cerebellar peduncle, tpf: transverse pontine fibers. Scale bar = 4 mm.
Fig. 2.
Neuroanatomical details in the brainstem resolved with the high-resolution ex vivo DTI. Sagittal (a, b) and coronal (c, d) sections from DEC maps show the different structures delineated at each level. The anatomic locations of the sections are indicated in corresponding scout in vivo brain images. Red, green and blue represent anisotropy along medial-lateral, anterior-posterior, and superior-inferior orientations, respectively. Structural abbreviations are: CST: corticospinal tract, CTT: central tegmental tract, DSCP: decussation of the superior cerebellar peduncle, ML: medial lemniscus, mlf: medial longitudinal fasciculus, PC: posterior commissure, SCP: superior cerebellar peduncle, TB: trapezoid body, tpf: transverse pontine fibers.
Fig. 3 shows three-dimensional reconstruction of major white matter tracts and grey matter nuclei from the tensor data. Brainstem nuclei, including the cranial nerve and the inferior olivary nuclei, were three-dimensionally segmented based on their hyperintense appearance in ADC maps. The pontine nuclei, dispersed among the corticospinal and transverse pontine fibers in the basal pons, were delineated as hyperintense bands in the ADC map (inset in Fig. 3a). Tractography results from the tensor data enabled three-dimensional reconstruction of fiber fascicles of the corticospinal tract interleaving through the transversely-oriented pontine fibers (Fig. 3b,c). The transverse pontine fibers were traced coursing dorso-laterally and merging with the contralateral middle cerebellar peduncle (Fig. 3c,d). The shift in the dorso-ventral position of the medial lemniscus along the longitudinal axis of the brainstem can be seen in Fig. 3. The medial lemniscus coursed along the ventral side in the caudal medulla, and turned sharply towards the dorsal side of the brainstem as it entered the pons. These trajectories agree well with the 2D histological description of these tracts (Nieuwenhuys et al., 1983; Nolte, 2008).
Fig. 3.
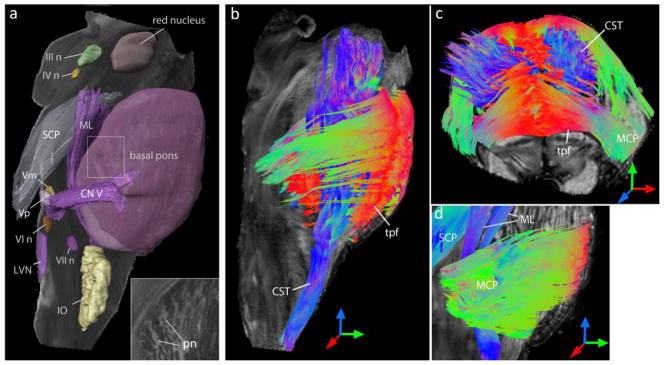
Three-dimensional reconstruction of brainstem nuclei and white matter tracts from the DTI data. a) Reconstructed cranial nerve and inferior olivary nuclei are overlaid on the ADC map for anatomic reference. The pontine nuclei in the basal pons are visible as hyperintense bands in the ADC map (inset). b–d) Three-dimensional rendering of fibers reconstructed from tractography are overlaid on fractional anisotropy images for anatomic reference, with color representing the orientations of the fibers. Red, green and blue indicate fiber orientations along the medial-lateral, anterior-posterior, and superior-inferior axes, respectively. Abbreviations are: III n: oculomotor nucleus, IV n: trochlear nucleus, Vp: primary sensory nucleus of trigeminal nerve, Vm: trigeminal motor nucleus, VI n: abducens nucleus, VII n: facial motor nucleus, CN V: trigeminal nerve, CST: corticospinal tract, IO: inferior olivary nucleus, LVN: lateral vestibular nucleus, MCP: middle cerebellar peduncle, ML: medial lemniscus, pn: pontine nuclei, SCP: superior cerebellar peduncle, tpf: transverse pontine fibers.
High-resolution DTI of the medulla oblongata and cervicomedullary junction
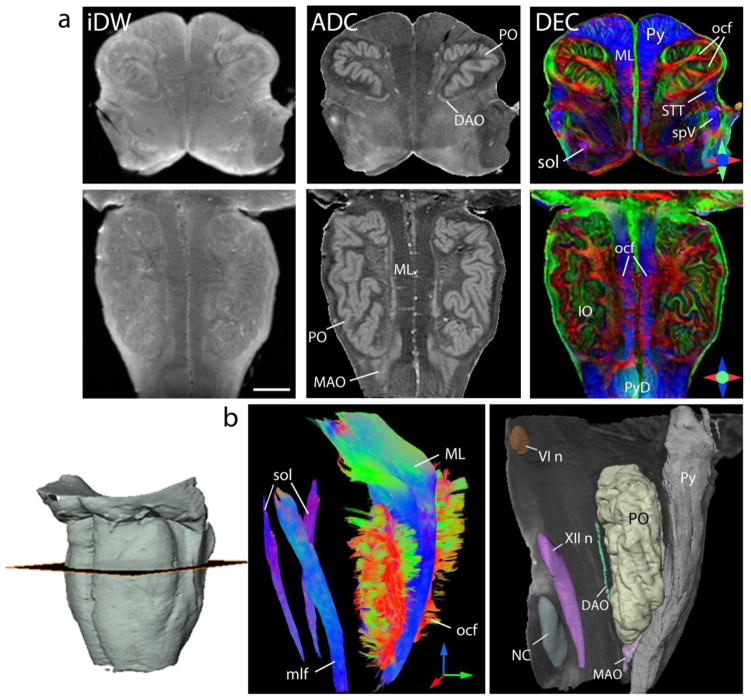
Fig. 4 shows the neuroanatomical details revealed by diffusion contrasts in the medulla at 170 μm resolution. ADC maps of the medulla exhibited strong grey-white matter contrast, with ADC in the grey matter (0.57 ± 0.024 μm2/ms) being significantly higher than that of white matter (0.39 ± 0.019 μm2/ms). This can be seen in Fig. 4, that shows prominent delineation of the inferior olivary nuclei (ADC maps in Fig. 4a). The high grey-white matter contrast enabled delineation of the smaller dorsal and medial accessory nuclei in the inferior olivary complex (Fig. 4a,b). DEC maps revealed several finer fiber tracts in the medulla, e.g. the solitary tract and internal arcuate fibers, which were not resolved at lower resolution. A number of fine mediolaterally-oriented fibers (red bands in the DEC maps) interleaved between the predominantly superior-inferior orientation of the medial lemniscus fibers could be resolved, and were identified as decussating olivocerebellar fibers (Fig. 4a). The relatively less-myelinated spinothalamic and spinal trigeminal pathways were also clearly delineated in the DEC maps. Fig. 4b shows three-dimensional reconstruction of structures in the medulla from the tensor data. Tractography-based reconstruction of the medial lemniscus fibers showed the sharp dorsolateral bend at the level of the rostral medulla. The solitary tract, with a diameter of approximately 1-mm, was traced coursing obliquely along the craniocaudal axis of the medulla (Fig. 4b). Placement of seed regions in the inferior olivary nuclei resulted in tracing of a large number of fine olivocerebellar fibers. The olivocerebellar fibers originate at the inferior olivary nuclei, and decussate in the raphe to join the contralateral inferior cerebellar peduncle (Nieuwenhuys et al., 1983). Although decussation of the olivocerebellar fibers could be identified in the DEC maps as loosely-arranged fascicles coursing mediolaterally through the raphe, 3D reconstruction of the fiber trajectories to the contralateral inferior cerebellar peduncle could not be performed owing to the very small diameters of the individual fascicles.
Fig. 4.
Anatomical details in the medulla oblongata resolved with ex vivo DTI at 170 μm isotropic resolution. a) Axial (top panel) and coronal (bottom panel) sections through the medulla show isotropic diffusion-weighted (iDW), apparent diffusion coefficient (ADC), and direction encoded colormap (DEC) contrasts derived from DTI. In the DEC maps, red, blue and green represent anisotropy along medial-lateral, superior-inferior, and anterior-posterior orientations, respectively. Scale bar = 4 mm. b) Three-dimensional reconstruction of fiber tracts and nuclei in the medulla from the tensor data. Tracts reconstructed using tractography are color-coded based on fiber orientations and shown in a sagittal view in the left panel. Red, green and blue denote fiber orientations along medial-lateral, anterior-posterior, and superior-inferior axes, respectively. The right panel shows 3D rendering of grey matter nuclei and the pyramidal tract overlaid on the ADC map for anatomical reference. VI n: abducens nucleus, XII n: hypoglossal nucleus, DAO: dorsal accessory olivary nucleus, IO: inferior olivary complex, MAO: medial accessory olivary nucleus, ML: medial lemniscus, mlf: medial longitudinal fasciculus, NC: nucleus cuneatus, ocf: olivocerebellar fibers, PO: primary olivary nucleus, Py: pyramids, PyD: pyramidal decussation, sol: solitary tract, spV: spinal trigeminal tract, STT: spinothalamic tract.
Fig. 5 shows DTI of the cervical spinal cord at the level of the spinomedullary junction at 125 μm resolution. A conspicuous feature of the spinomedullary junction is the decussation of the pyramids. In the DEC maps, the crossing fiber bundles of the pyramidal tracts could be clearly resolved, with the angle of decussation oriented obliquely to the axial plane (Fig. 5a,b). Because of their oblique orientations, the topographic organization of the decussating fibers cannot be well visualized with axial 2D histology. Tractography revealed the 3D topography of the pyramidal decussation, the pyramidal fibers crossed over in interdigitating bundles of unequal sizes (Fig. 5c,d), with upper crossed fibers migrating more dorsally than lower ones. Please note the fundamental differences with the manner in which the superior cerebellar peduncles decussate (Figs. 1–2), where the decussation takes place at a microscopic scale (much lower than the image resolution) and individual crossing fibers cannot be resolved. Tractography results showed a majority of the pyramidal fibers crossing over in successive bundles to the contralateral spinal lateral funiculus to form the crossed lateral corticospinal tract, and uncrossed pyramidal fibers were traced continuing into the ipsilateral anterior corticospinal tract adjacent to the anterior median fissure (Fig. 5e).
Fig. 5.
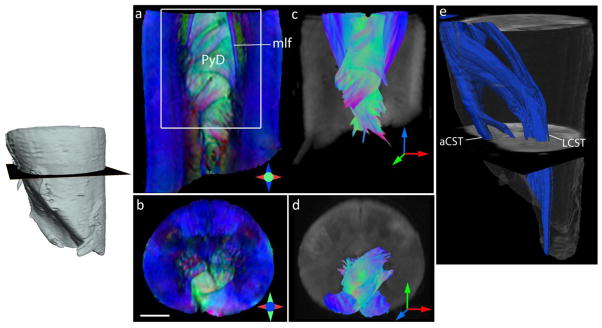
Ex vivo DTI of the cervical spinal cord at the level of the spinomedullary junction showing the decussation of the pyramidal tracts (PyD). a–b) Coronal and axial sections through the DEC maps show delineation of the PyD and medial longitudinal fasciculus (mlf). Scale bar = 2 mm. c–d) Fiber tractography shows interdigitating bundles from the left and right pyramidal (Py) tracts (reconstructed fibers are cropped to coronal and axial sections in c and d, respectively). Fibers are colored by orientation, with red, green and blue denoting fiber orientations along medial-lateral, anterior-posterior, and superior-inferior axes, respectively. e) Three-dimensional reconstruction of the left pyramidal tract (rendered in blue), showing crossed fibers forming the lateral corticospinal tract (LCST) and uncrossed pyramidal fibers continuing into the ipsilateral anterior corticospinal tract (aCST).
Comparison of anatomical details obtained from DTI and histological staining
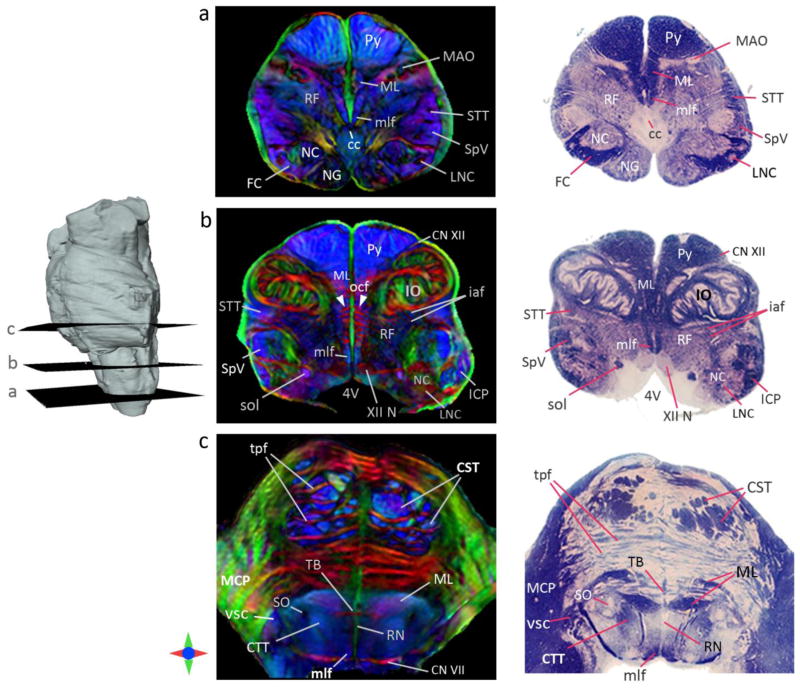
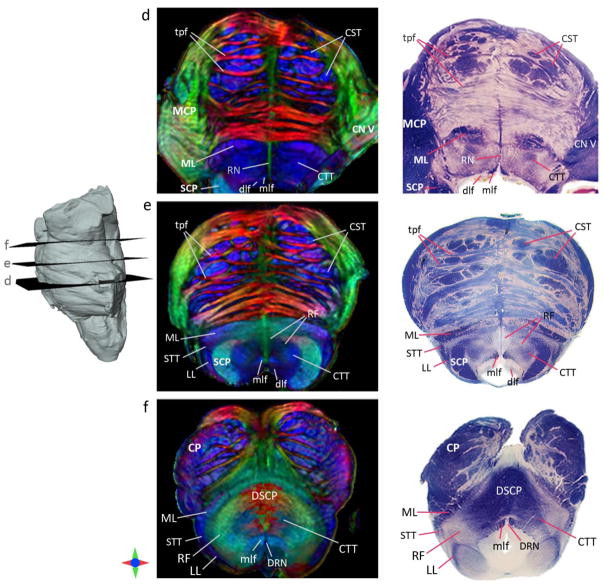
Fig. 6 compares 2D axial brainstem sections from the DEC maps with corresponding myelin-stained sections from Nolte’s human brain atlas (Nolte, 2008). As shown in Fig. 6, the level of anatomical detail resolved by the high-resolution DTI contrasts was comparable to structural delineation obtained with myelin-stained histology of the brainstem. Additionally, the DEC maps provided complementary contrasts to resolve the orientations of various white matter tracts. For instance, the solitary and spinal trigeminal tracts both extend longitudinally in the medulla, and in DTI contrasts the oblique orientation of the solitary tract along the mediolateral and superior-inferior axes (purple in the DEC maps) compared to the predominantly superior-inferior orientation of the spinal trigeminal tract (blue in the DEC maps) can be clearly appreciated, which was difficult to resolve with myelin staining (Fig. 6a). Additional structures that could be delineated from orientation-based DTI contrasts included the olivocerebellar fibers, visible as distinct mediolaterally-oriented fascicles traversing orthogonal to the medial lemniscus (white arrowheads in Fig. 6a), which were difficult to discern in corresponding myelin-stained histology sections.
Fig. 6.
Fig. 6a. Comparison of DTI contrasts in the brainstem with myelin-stained histology. The left panel shows DEC maps of axial sections at the level of the caudal medulla (a), rostral medulla (b), and caudal pons (c). Corresponding histology sections from Nolte’s human brain atlas (Nolte, 2008) are shown in the right panel (reproduced with permission). Structural abbreviations are: 4V: fourth ventricle, XII N: hypoglossal nucleus, cc: central canal, CN XII: hypoglossal nerve, CST: corticospinal tract, CTT: central tegmental tract, FC: fasciculus cuneatus, iaf: internal arcuate fibers, ICP: inferior cerebellar peduncle, IO: inferior olivary nucleus, LNC: lateral cuneatus nucleus, MAO: medial accessory olivary nucleus, MCP: middle cerebellar peduncle, ML: medial lemniscus, mlf: medial longitudinal fasciculus, NC: nucleus cuneatus, NG: nucleus gracilis, ocf: olivocerebellar fibers, Py: pyramid, RF: reticular formation, RN: raphe nuclei, SO: superior olivary nucleus, sol: solitary tract, SpV: spinal trigeminal tract, STT: spinothalamic tract, TB: trapezoid body, tpf: transverse pontine fibers, vsc: ventral spinocerebellar tract.
Fig. 6b. Comparison of DTI contrasts in the brainstem with myelin-stained histology. The left panel shows DEC maps of axial sections at the level of the midpons (d), rostral pons (e), and caudal midbrain (f). Corresponding histology sections from Nolte’s human brain atlas (Nolte, 2008) are shown in the right panel (reproduced with permission). Structural abbreviations are: CN V: trigeminal nerve, CP: cerebral peduncle, CST: corticospinal tract, CTT: central tegmental tract, dlf: dorsal longitudinal fasciculus, DRN: dorsal raphe nucleus, DSCP: decussation of the superior cerebellar peduncles, LL: lateral lemniscus, MCP: middle cerebellar peduncle, ML: medial lemniscus, mlf: medial longitudinal fasciculus, RF: reticular formation, RN: raphe nuclei, SCP: superior cerebellar peduncle, tpf: transverse pontine fibers.
Registration to in vivo brain atlas in MNI coordinates
To construct a high-resolution atlas of the brainstem in the MNI stereotaxic coordinate space, the ex vivo DTI data of the brainstem were co-registered to the JHU in vivo brain atlas. Fig. 7 illustrates the degree of registration accuracy, with a representative coronal section (y = −26 mm in MNI coordinates) from the in vivo atlas and the co-registered ex vivo brainstem data. The mean deformation necessary to warp the ex vivo data to the in vivo atlas was 0.65 ± 0.49 mm (mean ± standard deviation), with a maximum displacement of upto 2.28 mm. The determinant of the Jacobian matrix of the transformation provides the quantitative local volumetric differences between the atlas and the ex vivo data (values greater than 1 indicate local expansion and values less than 1 indicate local volumetric contraction of the ex vivo data). As shown in Fig. 8c, the Jacobian map revealed local tissue expansion in most regions within the brainstem under the transformation, representing the degree of local tissue shrinkage in the ex vivo specimen. Fig. 8 shows orthogonal planar views through the in vivo whole brain atlas (2.2-mm isotropic resolution) and the resulting high-resolution brainstem atlas (0.225-mm isotropic resolution) registered to the MNI coordinate space. The ex vivo atlas provides delineation of several brainstem structures that are difficult to observe with the low field in vivo DTI contrasts, for e.g. in the basal pons, where the in vivo atlas provides limited resolution to resolve orthogonal interdigitating fibers of the corticospinal tract and the transverse pontine fibers (Fig. 8).
Fig. 7.
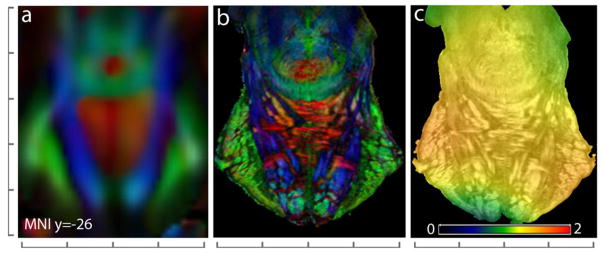
Co-registration of the high-resolution ex vivo brainstem DTI data to the JHU in vivo brain atlas (Oishi et al., 2011). A representative coronal section (y = −26 mm in MNI coordinates) through the in vivo atlas (a) and the corresponding co-registered section from the ex vivo data (b) show the degree of registration accuracy. The color-coded map of the Jacobian determinant overlaid on the anatomical image (c) represents the local volumetric scaling applied by the transformation to warp the ex vivo data to the in vivo atlas. In the Jacobian map, values greater than 1 represent local expansion, and values less than 1 represent local volumetric contraction of the ex vivo image under the deformation. Scales shown are in units of mm with 10 mm spacing.
Fig. 8.
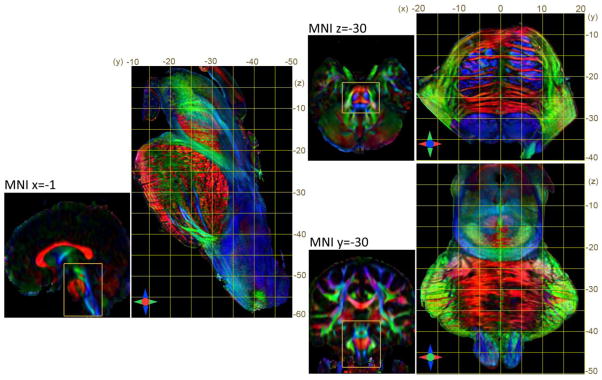
The high-resolution DTI atlas of the brainstem registered to the MNI stereotaxic coordinate space. Orthogonal planar views from the in vivo whole-brain atlas (2.2 mm resolution) and corresponding sections through the constructed brainstem atlas (0.225 mm resolution) are shown for comparison of anatomical details. Red, green and blue in the DEC maps represent anisotropy orientation along the medial-lateral, anterior-posterior, and superior-inferior axes, respectively. All scales are in units of mm.
DISCUSSION
Our study demonstrated DTI of the brainstem at a level of resolution that enabled precise delineation of white matter pathways, their orientations and detailed morphologic features, e.g., the ascending course of the medial lemniscus fibers and the decussation of the pyramidal fibers. The results showed the feasibility of tractography based three-dimensional reconstruction of interdigitating fibers of the corticospinal tract and transverse pontine fibers, which is difficult at lower resolutions due to the well-known limitation of the tensor model with respect to intravoxel ‘crossing’ of fibers. In addition to detailed white matter anatomy, DTI contrasts in our study allowed delineation of grey matter structures including the cranial nerve and inferior olivary nuclei. The dissociative effect of increasing b-values on grey and white matter has been demonstrated previously in the in vivo human brain (Yoshiura et al., 2001). Consistent with these findings, strong grey-white matter contrasts were observed in the ADC maps in our study for b-values of 2800–4000 s/mm2 (Fig. 4), which enabled precise delineation of grey matter nuclei. The strong grey as well as white matter delineation offers another advantage over brainstem atlases based on conventional histological techniques, where the anatomical contrasts available are dependent on the specific stains used.
Currently, both histology and MRI-based atlases of the brainstem are scarce (Afshar et al., 1978; Paxinos and Huang, 1995; Naidich et al., 2009). Histology-based atlases are limited to two-dimensional sections at limited axial levels, and the sectioning orientations are not consistent across different atlases. Three-dimensional MRI-based atlases of the human brain often cover the brainstem, but they have at most 1-mm resolution. Resolution and strong tissue contrast are both crucial factors for detailed MRI-based neuroanatomical delineation. In this respect, conventional relaxation-based MR contrasts have several limitations. At high field strengths, the difference in T1-relaxation times between different tissue types reduces. T2-weighted imaging tends to require long repetition intervals and thus long scan times, with structures in the brainstem displaying relatively limited T2-contrasts as shown in Fig. 1. Intermediate-weighted images have been previously shown to provide better grey matter-white matter contrasts in the brainstem (Naidich et al., 2009). As indicated in our study (Figs. 1, 2, and 4), the orientation-dependent contrasts derived from DTI are important to resolve fine structural details in the brainstem. Currently, the only existing DTI-based atlases of the brainstem to our knowledge (Naidich et al., 2009; Soria et al., 2011) are based on two-dimensional acquisition with through-plane resolutions of 0.5–0.6 mm. Our study demonstrates the feasibility of generating a three-dimensional DTI based atlas that provides detailed visualization of brainstem anatomy with high resolution and contrasts. The understanding of DTI contrasts at this resolution level also yields important insights for the interpretation of relatively low resolution clinical DTI contrasts. For instance, we usually look for structures with strong medial-lateral alignment (appearing red in DEC maps) for all decussating tracts. These include the anterior and posterior commissures, and decussations of the superior cerebellar peduncles and the corticospinal tract. As seen in Fig. 2, the decussation of the superior cerebellar peduncles takes place at a microscopic level where the axonal bundles from the left and right peduncles cannot be differentiated at 255-μm resolution, but as expected, the decussation has a strong medial-lateral orientation. Interestingly, the decussation of the corticospinal tract takes place at a much more macroscopic scale, such that the axonal bundles from the left and right tracts could be clearly resolved at 170-μm resolution. However, the angle of the decussation is significantly oblique along both the dorsal-ventral (green) and superior-inferior (blue) planes, which leads to a distinctive cyan color in the DEC maps (Fig. 5). The registration of the high-resolution DTI atlas to MNI stereotaxic coordinates also enables its application as a useful research resource. If patient data are registered to the MNI coordinate space, our stereotaxic atlas can be directly applied to the patient data in order to estimate the precise stereotaxic coordinates of various brainstem structures.
In addition to developing a stand-alone atlas of the brainstem, our study is the first step towards developing a multi-resolution atlas of the whole human brain. As shown in Fig. 8, high-resolution DTI is important to resolve microscopic structural details in the brain. However, imaging resolution is a function of specimen size, SNR, and imaging time, and whole human brain coverage with 100–300 μm resolution is currently unfeasible from both hardware and computational points of view. The lower magnetic field strength and maximum gradient amplitudes on existing clinical scanners necessary to accommodate whole brain samples pose significant hardware constraints on the achievable SNR with increasing resolution. Additionally, the exceedingly long acquisition time necessary for larger samples in order to maintain the same resolution level is practically unfeasible, due to issues of scanner instability over extended periods as well as specimen degradation. Based on these technical constraints, ultra-high resolution examination of human samples is likely to be limited to a portion of the brain. Several studies have previously reported segmented brain MRI (Vandersteen et al., 1994; Fatterpekar et al., 2002) and DTI (Shepherd et al., 2007; Augustinack et al., 2010; Soria et al., 2011). As shown in our study with multi-resolution DTI of smaller tissue segments, by using deformable registration techniques to link the multi-resolution data to global stereotaxic coordinates, it is possible to create an atlas with detailed three-dimensional views of brain anatomy. In our study, images from three DTI datasets (from the whole brainstem, and the isolated medulla and cervicomedullary junction) acquired with different resolutions (255, 170, and 125 μm, respectively) are registered to the 2.2-mm whole brain atlas using diffeomorphic image transformation, establishing the feasibility of generating such a multi-resolution atlas of the human brain. In the future, recent advances in MR hardware, e.g. the 300 mT/m human gradient system (Tisdall et al., 2012) may make it feasible to further push the achievable resolution for diffusion MRI of the brain.
It is also important to note the limitations of our study. Firstly, some tissue deformation is usually unavoidable during excision and fixation of postmortem specimens. For in vivo anatomical studies, it is common to use data from multiple subjects from which population-based atlases can be generated (Mori et al., 2008; Fonov et al., 2011). This procedure, however, is effective for reducing sample-specific bias when the individual images have high anatomic fidelity. Because imaging studies based on postmortem samples can be influenced by an unknown amount of postmortem tissue deformation, it is difficult to estimate inherent biological variability based on population-averaged anatomy. Therefore, the anatomical framework for postmortem studies needs to be provided by in vivo images. In our study, global deformation of the specimen was corrected for by applying deformable registration to the anatomic coordinates established by in vivo population-based studies. Further studies using registration to in vivo MRI data may be necessary to provide estimates of the degree of inter-subject variability. Secondly, the effects of fixatives on postmortem tissue properties are not precisely understood at present, and need to be taken into account while extrapolating the contrasts of the ex vivo atlas to in vivo DTI studies. Several studies have shown that ADC in brain tissue changes with formalin fixation, although the degree of anisotropy (FA) stays unaffected (Guilfoyle et al., 2003; Sun et al., 2005). Therefore, although anisotropy-based orientation contrasts can be directly compared with in vivo DTI data, quantitative comparison of diffusivity indices is not feasible and should be avoided. Lastly, the six-element tensor model and deterministic tractography approach have some known limitations (Mori and van Zijl, 2002). The use of more sophisticated diffusion sampling schemes, such as high angular resolution diffusion imaging (HARDI), and tractography algorithms (Behrens et al., 2003; Tuch et al., 2003) may possibly reveal further structural details in the postmortem brainstem. However, HARDI-type data acquisition requires a large number of diffusion-encoding directions, typically more than 60 orientations, requiring significantly longer scanning time. In addition, the higher b-values necessary compared to DTI can further reduce the achievable SNR at increasing resolutions. The gain in scan time/SNR can be traded for image resolution enhancement of DTI, which could also potentially reveal finer microstructures. Therefore, the relative advantages between DTI and HARDI need to be carefully evaluated.
In conclusion, the results of this study will significantly enhance existing 2D atlases of the human brainstem, and improve our understanding and interpretation of relatively low resolution in vivo DTI contrasts. The high-resolution DTI atlas will also serve as a useful neuroanatomical reference for future research and clinical imaging studies of the brainstem.
Acknowledgments
This study was supported by National Institutes of Health grants R01AG020012 and R01EB003543.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afshar F, Dykes E. A three-dimensional reconstruction of the human brain stem. J Neurosurg. 1982;57(4):491–495. doi: 10.3171/jns.1982.57.4.0491. [DOI] [PubMed] [Google Scholar]
- Afshar F, Watkins ES, et al. A Variability Study. New York: Raven Press; 1978. Stereotaxic Atlas of the Human Brainstem and Cerebellar Nuclei. [Google Scholar]
- Aggarwal M, Mori S, et al. Three-dimensional diffusion tensor microimaging for anatomical characterization of the mouse brain. Magn Reson Med. 2010;64(1):249–261. doi: 10.1002/mrm.22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DC, Pierpaoli C, et al. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Transactions on Medical Imaging. 2001;20(11):1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- Augustinack JC, Helmer K, et al. Direct visualization of the perforant pathway in the human brain with ex vivo diffusion tensor imaging. Front Hum Neurosci. 2010;4:42. doi: 10.3389/fnhum.2010.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axer H, Beck S, et al. Microstructural analysis of human white matter architecture using polarized light imaging: views from neuroanatomy. Front Neuroinform. 2011;5:28. doi: 10.3389/fninf.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, et al. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50(5):1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- Carpenter M. Human neuroanatomy. Baltimore: Williams & Wilkins; 1976. [Google Scholar]
- Chen X, Weigel D, et al. Diffusion tensor imaging and white matter tractography in patients with brainstem lesions. Acta Neurochir (Wien) 2007;149(11):1117–1131. doi: 10.1007/s00701-007-1282-2. [DOI] [PubMed] [Google Scholar]
- Donaldson SS, Laningham F, et al. Advances toward an understanding of brainstem gliomas. J Clin Oncol. 2006;24(8):1266–1272. doi: 10.1200/JCO.2005.04.6599. [DOI] [PubMed] [Google Scholar]
- Fatterpekar GM, Naidich TP, et al. Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 Tesla. AJNR Am J Neuroradiol. 2002;23(8):1313–1321. [PMC free article] [PubMed] [Google Scholar]
- Fonov V, Evans AC, et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle DN, Helpern JA, et al. Diffusion tensor imaging in fixed brain tissue at 7.0 T. NMR Biomed. 2003;16(2):77–81. doi: 10.1002/nbm.814. [DOI] [PubMed] [Google Scholar]
- Helton KJ, Weeks JK, et al. Diffusion tensor imaging of brainstem tumors: axonal degeneration of motor and sensory tracts. J Neurosurg Pediatr. 2008;1(4):270–276. doi: 10.3171/PED/2008/1/4/270. [DOI] [PubMed] [Google Scholar]
- Jiang H, van Zijl PCM, et al. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Computer Methods and Programs in Biomedicine. 2006;81(2):106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: A Monte Carlo study. Magn Reson Med. 2004;51:807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- Lui YW, Law M, et al. Brainstem corticospinal tract diffusion tensor imaging in patients with primary posterior fossa neoplasms stratified by tumor type: a study of association with motor weakness and outcome. Neurosurgery. 2007;61(6):1199–1207. doi: 10.1227/01.neu.0000306098.38141.81. [DOI] [PubMed] [Google Scholar]
- McNab JA, Jbabdi S, et al. High resolution diffusion-weighted imaging in fixed human brain using diffusion-weighted steady state free precession. Neuroimage. 2009;46(3):775–785. doi: 10.1016/j.neuroimage.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Miller KL, Stagg CJ, et al. Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. Neuroimage. 2011;57(1):167–181. doi: 10.1016/j.neuroimage.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Trouve A, et al. On the metrics and Euler-Lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, et al. Three dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, Oishi K, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15(7–8):468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Nagae-Poetscher LM, Jiang H, et al. High-resolution diffusion tensor imaging of the brain stem at 3 T. American Journal of Neuroradiology. 2004;25(8):1325–1330. [PMC free article] [PubMed] [Google Scholar]
- Naidich TP, Duvernoy HM, et al. Duvernoy’s Atlas of the Human Brain Stem and Cerebellum. Springer-Verlag; 2009. [Google Scholar]
- Niemann K, van den Boom R, et al. A brainstem stereotactic atlas in a three-dimensional magnetic resonance imaging navigation system: first experiences with atlas-to-patient registration. J Neurosurg. 1999;90(5):891–901. doi: 10.3171/jns.1999.90.5.0891. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R, Voogd J, et al. The Human Central Nervous System: A Synopsis and Atlas. Springer-Verlag; 1983. [Google Scholar]
- Nolte J. The Human Brain: An Introduction to its Functional Anatomy. Mosby; 2008. [Google Scholar]
- Nolte J, Angevine JB. The human brain: In photographs and Diagrams. St. Louis: Mosby; 2000. [Google Scholar]
- Oishi K, Faria AV, et al. MRI Atlas of Human White Matter. Academic Press; 2011. [Google Scholar]
- Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med. 1999;42:526–540. [PubMed] [Google Scholar]
- Parvizi J, Damasio AR. Neuroanatomical correlates of brainstem coma. Brain. 2003;126(Pt 7):1524–1536. doi: 10.1093/brain/awg166. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Huang X. Atlas of the Human Brainstem. Academic Press; 1995. [Google Scholar]
- Prats-Galino A, Soria G, et al. Functional anatomy of subcortical circuits issuing from or integrating at the human brainstem. Clin Neurophysiol. 2012;123(1):4–12. doi: 10.1016/j.clinph.2011.06.035. [DOI] [PubMed] [Google Scholar]
- Salamon N, Sicotte N, et al. Analysis of the brain-stem white-matter tracts with diffusion tensor imaging. Neuroradiology. 2005;47(12):895–902. doi: 10.1007/s00234-005-1439-8. [DOI] [PubMed] [Google Scholar]
- Schmierer K, Wheeler-Kingshott CA, et al. Quantitative magnetic resonance of postmortem multiple sclerosis brain before and after fixation. Magn Reson Med. 2008;59(2):268–277. doi: 10.1002/mrm.21487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Ozarslan E, et al. Diffusion tensor microscopy indicates the cytoarchitectural basis for diffusion anisotropy in the human hippocampus. AJNR Am J Neuroradiol. 2007;28(5):958–964. [PMC free article] [PubMed] [Google Scholar]
- Soria G, De Notaris M, et al. Improved assessment of ex vivo brainstem neuroanatomy with high-resolution MRI and DTI at 7 Tesla. Anat Rec (Hoboken) 2011;294(6):1035–1044. doi: 10.1002/ar.21383. [DOI] [PubMed] [Google Scholar]
- Stieltjes B, Kaufmann WE, et al. Diffusion tensor imaging and axonal tracking in the human brainstem. NeuroImage. 2001;14:723–735. doi: 10.1006/nimg.2001.0861. [DOI] [PubMed] [Google Scholar]
- Sun SW, Neil JJ, et al. Formalin fixation alters water diffusion coefficient magnitude but not anisotropy in infarcted brain. Magn Reson Med. 2005;53(6):1447–1451. doi: 10.1002/mrm.20488. [DOI] [PubMed] [Google Scholar]
- Tisdall MD, Witzel T, et al. Improving SNR in high b-value diffusion imaging using Gmax=300 MT/m human gradients. Proceedings of International Society for Magnetic Resonance in Imaging.2012. [Google Scholar]
- Tournier JD, Calamante F, et al. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22(1):53–66. [Google Scholar]
- Tuch DS, Reese TG, et al. Diffusion MRI of complex neural architecture. Neuron. 2003;40(5):885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- Vandersteen M, Beuls E, et al. High field magnetic resonance imaging of normal and pathologic human medulla oblongata. Anat Rec. 1994;238(2):277–286. doi: 10.1002/ar.1092380213. [DOI] [PubMed] [Google Scholar]
- Virta A, Barnett A, et al. Visualizing and characterizing white matter fiber structure and architecture in the human pyramidal tract using diffusion tensor MRI. Magn Reson Imaging. 1999;17:1121–1133. doi: 10.1016/s0730-725x(99)00048-x. [DOI] [PubMed] [Google Scholar]
- Yoshiura T, Wu O, et al. Highly diffusion-sensitized MRI of brain: dissociation of gray and white matter. Magn Reson Med. 2001;45(5):734–740. doi: 10.1002/mrm.1100. [DOI] [PubMed] [Google Scholar]