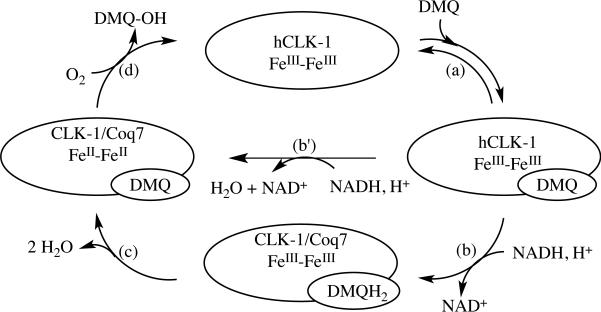
Abstract
The mitochondrial membrane-bound enzyme Clock-1 (CLK-1) extends the average longevity of mice and C. elegans, as demonstrated for Δclk-1 constructs for both organisms. Such an apparent impact on aging and the presence of a carboxylate-bridged diiron center in the enzyme inspired the present work. We expressed a soluble human CLK-1 (hCLK-1) fusion protein with an N-terminal immunoglobulin binding domain of protein G (GB1). Inclusion of the solubility tag allowed for thorough characterization of the carboxylate-bridged diiron active site of the resulting GB1-hCLK-1 by spectroscopic and kinetic methods. Both UV-vis and Mössbauer experiments provide unambiguous evidence that GB1-hCLK-1 functions as a 5-demethoxyubiquinone-hydroxylase (DMQ-hydroxylase), utilizing its carboxylate-bridged diiron center. The binding of DMQn (n = 0 or 2) to GB1-hCLK-1 mediates reduction of the diiron center by NADH and initiates O2 activation for subsequent DMQ hydroxylation. Deployment of DMQ to mediate reduction of the diiron center in GB1-hCLK-1 improves substrate specificity and diminishes consumption of NADH that is uncoupled from substrate oxidation. Both Vmax and kcat/KM for DMQ hydroxylation increase when DMQ0 is replaced by DMQ2 as substrate, which demonstrates that an isoprenoid side chain enhances enzymatic hydroxylation and improves catalytic efficiency.
Although the average human lifespan has increased steadily over the past two centuries, factors governing the aging process with its concomitant frailty and disease remain uncertain.1 An attempt to establish a model for studying the aging process led to the discovery of Clock-1 (CLK-1), an aging-associated enzyme.2 CLK-1 is conserved in yeast, C. elegans, and mammals including rats, mice, and humans.3 An increased lifespan, up to 30%, occurs in Δclk-1 C. elegans and mice.2,4 Long-lived clk-1+/- mice containing lower CLK-1 levels display decreased activity of the mitochondrial electron transport chain, reduced ATP synthesis, and increased mitochondrial oxidative stress.4 Impaired mitochondrial electron transport is accompanied by accumulation of 5-demethoxyubiquinone (DMQ), an immediate precursor of ubiquinone (UQ), as exhibited in mouse embryonic stem cells containing a knockout of clk-1.5 During respiration, UQ mediates delivery of electrons from Complex I or II to Complex III within the inner mitochondrial membrane of eukaryotes via interconversion of oxidized quinone and reduced hydroquinone.6
Based on the accumulation of 5-demethoxyubiquinone (DMQ9) in Δclk-1 mutants of C. elegans and mice, where the subscript indicates the length of the isoprenoid side chain (Chart 1), CLK-1 was proposed to function as a DMQ hydroxylase involved in the penultimate step of UQ biosynthesis.5,7 DMQ is converted to UQ by CLK-1 hydroxylation and subsequent O-methylation by Coq3, an O-methyltransferase.8 A structural model of CLK-1 from P. aeruginosa using bacterioferritin as a template revealed a four-helix bundle and, in addition, suggested a diiron active site within a conserved EXn1EXXHXn2EXn3EXXH binding motif.9-10 This motif is shared by the hydroxylase components in soluble methane monooxygenase (sMMO), toluene monooxygenase (ToMO), phenol hydroxylase (PH), and ribonucleotide reductase, supporting the hypothesis that CLK-1 is a member of the carboxylate-bridged diiron protein family (Supplementary Fig. S1).11-12 In addition, the structural model of human CLK-1 (hCLK-1) contains two conserved tyrosine residues having Fe···OTyr distances of 4.0 Å (Supplementary Fig. S1), reminiscent of the single conserved tyrosine responsible for radical initiation in ribonucleotide reductase.11,13 Thus far, the function of the tyrosine residues in CLK-1 remains unexamined. A docking model of rat CLK-1 with its substrate, DMQ10, was also reported.10 A previously proposed structural model of rat CLK-1 suggested several key structural features involving interactions between the substrate and the protein.10 Hydrophobic interactions occurring between the isoprenoid side chain of DMQ10 and a hydrophobic pocket within rat CLK-1 were proposed. In addition, hydrogen bonding between the carbonyl/methoxy group of DMQ10 and the protein motif Glu22/His110/Tyr111 were postulated for the DMQ10 adduct of CLK-1 (Supplementary Fig. S1).
Chart 1.
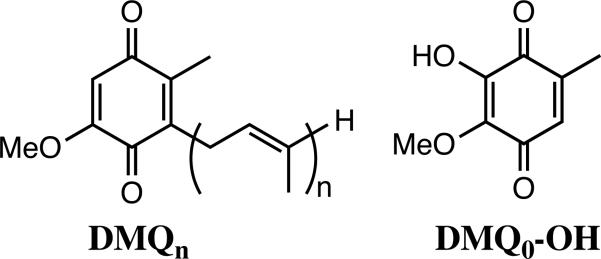
In the present study we report a robust expression system for, and substantially improved characterization of, CLK-1 as a follow-up of our preliminary work on this system.14 The solubility of the hCLK-1 membrane-bound enzyme was significantly improved through construction of an N-terminal immunoglobulin binding domain of protein G (GB1) fusion protein. The fusion protein designed and investigated here could be expressed in a highly efficient manner in E. coli.15 GB1-hCLK-1 uses its carboxylate-bridged diiron center to catalyze the hydroxylation of DMQ0 (Chart 1). Reduction of the diiron center by NADH occurs via a quinone-mediated electron transfer process, without the need for an additional reductase protein. As demonstrated here, DMQ mediates reduction of diiron center for subsequent O2 activation and DMQ hydroxylation in hCLK-1.
Material and Methods
Distilled water was purified with a Milli-Q filtering system. 2-Methoxy-5-methyl-1,4-benzoquinone (DMQ0), 2-methoxy-3-hydroxyl-5-methyl-1,4-benzoquinone (DMQ0-OH) and DMQ2 were synthesized based on published procedures.16-17 Other reagents were purchased from Sigma Aldrich and used as received.
Cloning and Plasmid Construction
The pET30a(+)-GBFusion vector was purchased from Dana-Farber/Harvard Cancer Center DNA resource core in Harvard medical school. pOTB7 containing human clk-1 (hclk-1) gene was obtained from ATCC and used as the starting vector (ATCC number: MGC-671). A PCR was run to amplify the human hclk-1 gene and introduce BamHI and EcoRI restriction sites into the 5’ and 3’ ends of the product using primers 5’-hclk-1 (5’-TCAGGAGGATCCATGACTTTAGACAATATCAGT-3’) and 3’-hclk-1 (5’-CACACTGAATTCTTATAATCTTTCTGATAAATA-3’). The gene product was digested with BamHI and EcoRI for 2.5 h at 37 °C and purified by extraction from a 1.5 % agarose gel (Qiagen). The digested product was then ligated into pET30a(+)-GBFusion vector that had also been treated with the same enzymes using 1 μL of T4 DNA ligase (New England Biolabs) and incubated at 16 °C for 16 hr. A 3 μL portion of the ligation reaction solution was transformed into E. coli DH5α cells (Invitrogen). The constructed plasmids were examined by agarose gel electrophoresis and then sequenced by the MIT Biopolymers facility.
Expression and Purification of GB1-hCLK-1
E. coli ArcticExpress(DE3)RP cells transformed with pET30a(+)-GBFusion-hclk-1 were cultured in 6 L of LB medium containing 50 μg/mL kanamycin at 37 °C until OD600 reached 0.4. Protein expression was induced by addition of IPTG to a final concentration of 100 μM. To maximize iron incorporation in recombinant GB1-hCLK-1, 100 μM (NH4)2Fe(SO4)·6H2O was added to the culture every hour in the first three hours. Growth was continued for 16 h at 25 °C. Cells were collected by centrifugation at 4 °C, and the cell paste was frozen in liquid nitrogen and stored at -80 °C. The cell paste (~25 g) was sonicated on ice using a Branson sonifier for 10 min (3 sec on, 20 sec off) at 48 % output in 200 mL of 20 mM Tris, 100 μM (NH4)2Fe(SO4)2·6H2O, 8 mM sodium thioglycolate, 2 mM cysteine, and 10% glycerol (buffer A, pH 7.0) containing 5 μL DNase I, 5 mM MgCl2 and 50 μM PMSF. Insoluble material was removed by centrifugation at 16,000 g for 90 min, and the supernatant was filtered through a 0.22 μm membrane and loaded onto a DEAE Sepharose FF column (500 mL, 5 cm diameter × 25 cm length) equilibrated in buffer A. The column was washed with 500 mL of buffer A and then eluted in 2500 mL by running a linear gradient from 20 mM NaCl to 500 mM NaCl at a rate of 3.0 mL/min. Fractions containing GB1-hCLK-1 eluted at ~ 250 mM NaCl were identified by SDS-PAGE. These fractions were pooled and concentrated to ~12 mL using a 3K MWCO Amicon filter (Millipore, Inc.). The resulting protein was loaded onto a Superdex S75 column (320 mL, 2.6 cm diameter × 60 cm length) equilibrated in 20 mM Tris, 10 % glycerol (buffer B, pH 7.0). Proteins were eluted by running buffer B over the column at a flow rate of 1 mL/min. These fractions were pooled and loaded onto a Mono Q column (20 mL, 1.6 cm diameter × 10 cm length) equilibrated with buffer B. The column was washed with 40 mL of buffer B and then eluted in 400 mL by running a linear gradient from 20 mM NaCl to 500 mM NaCl at a rate of 0.5 mL/min. Purified GB1-hCLK-1 confirmed by SDS-PAGE was concentrated and stored at -80 °C until further use.
Determination of Protein Concentration, Iron Content, and Oligomeric State of GB1-hCLK-1
The concentration of GB1-hCLK-1 was determined by a Bradford protein assay using BSA as the standard. Iron content was measured by inductively coupled plasma/optical emission spectroscopy with calibration using an Fe standard (BDH, ARISTAR® PLUS). GB1-hCLK-1 was digested by 1 mL of 2% HNO3 (BDH, ARISTAR® PLUS) and the precipitated protein was removed by centrifugation (13,000 rpm, 10 min). The supernatant was further diluted with 2% HNO3 and analyzed by ICP-OES at the MIT Center for Materials Science and Engineering. The oligomeric state of GB1-hCLK-1 was determined with the use of a HiLoad™ 16/60 Superdex™ 200 prep grade (GE Healthcare) size exclusion column. The elution time of GB1-hCLK-1 (74 min) falls between those of BSA (66 kDa, 60 min) and carbonic anhydrase (29 kDa, 80 min), suggesting that GB1-CLK-1 is dimeric in 20 mM Tris, 10% glycerol, pH 7.0 buffer. All concentrations of GB1-hCLK-1 in this work are computed on the basis of the monomer unit molecular weight.
Hydroxylase Activity GB1-hCLK-1
GB1-hCLK-1 hydroxylase activity was studied by product characterization using GC-MS. An aliquot (0.5 mL) of reaction solution containing 5 μM GB1-hCLK-1, 1 mM NADH, and 1 mM DMQ0 was quenched with 150 μL 0.7 M TCA after 30 min. The precipitated protein was removed by centrifugation. A 200 μL volume of chloroform was used to extract organic compounds from the supernatant, and the organic layer was analyzed by GC using an ZB-5MSi column (Phenomenex) attached to an Agilent 6890N gas chromatography system. The following temperature sequence was applied to analyze the products: 60 °C for 1 min, 15 °C per min to 100 °C, hold at 100 °C for 10 min, 20 °C per min to 260 °C. DMQ eluted at 17.10 min and the DMQ-OH at 17.39 min.
Steady State Activity of GB1-hCLK-1
The steady state activity of GB1-hCLK-1 was characterized by an NADH consumption assay using an HP 8452 diode array spectrophotometer. For DMQ0 (or DMQ2) concentration-dependence assays, 200 μM of NADH was added to 5 μM of GB1-hCLK-1 and DMQ0 (or DMQ2) at a specified concentration and the change in absorbance at 340 nm was monitored. All reactions were monitored continuously at 340 nm (ε340 = 6,220 M-1cm-1). Reaction mixtures were held at a constant temperature of 25.0 °C using a circulating water bath. Data were analyzed by fitting the initial time points to the linear function.
Redox Titration of GB1-hCLK-1
A redox titration was performed to determine the number of equivalents of electrons necessary to fully reduce GB1-hCLK-1. A 100-μM solution of GB1-hCLK-1 was made anaerobic with 12-15 cycles of vacuum gas exchange with O2 free N2. This solution was then titrated with 5 μL aliquots of 3.3 mM Na2S2O4 or 5 μL aliquots of 5 mM NADH in the presence of 100 μM DMQ0. The decrease in absorbance at 340 nm was monitored 10 min after addition of dithionite (or NADH) by UV/vis spectroscopy.
Redox Potential Determination for GB1-hCLK-1
The equilibrium midpoint potentials of GB1-hCLK-1 were determined by a series of reductive titrations based on a reported procedure using methylene blue as a redox indicator dye.18-19 GB1-hCLK-1 (ca. 20 μM) and dye (ca. 20 μM methylene blue) in a total volume 0.5 mL (100 mM Tris, pH 7.0) were made anaerobic with 12-15 cycles of vacuum gas exchange using O2-free N2 and titrated by a solution of sodium dithionite (ca. 500 μM). Multiple scans were taken after each dithionite addition until the system had reached equilibrium (usually 5-10 min, but as long as 40 min). The final spectrum was saved for data analysis. This process was repeated until the protein and dye were fully reduced. The solution potential and GB1-hCLK-1 midpoint potentials were computed for each titration point using modified Nernst equations (eq 1).18-19
| (1) |
Kinetic Studies of Substrate-Mediated Reduction of Oxidized GB1-hCLK-1
Reduction of oxidized GB1-hCLK-1 was monitored by using a HiTech DX2 stopped-flow UV-visible spectrophotometer. All reported concentrations are those after mixing. Reaction of oxidized GB1-hCLK-1 (4 μM) and NADH (60 μM) in the presence of DMQ2 (60 μM) was followed at 370 nm. Drive syringes and flow lines of the stopped-flow instrument were made anaerobic by flushing with at least 10 mL of an anaerobic solution of 4 mM sodium dithionite in 100 mM Tris, 10% glycerol, pH 7 buffer. Excess dithionite was removed by purging the syringes and flow lines with anaerobic buffer. Prior to conducting the stopped-flow experiments, a solution of 8 μM of GB1-hCLK-1 was incubated anaerobically with 120 μM of DMQ2 for 30 min. The solution was transferred to a drive syringe and loaded into the anaerobic stopped flow instrument. This solution was mixed with an equal volume of 120 μM of NADH in 100 mM Tris, 10% glycerol, pH 7 buffer. The temperature was maintained at 25 °C with a circulating water bath. The temperature of the reaction cell was verified by means of a thermocouple. Data were collected using a photomultiplier tube (PMT) with a tungsten lamp. Extinction coefficients at 370 nm for all reactants are as follows: oxidized GB1-hCLK-1, 4000 M-1cm-1; reduced GB1-hCLK-1, 1800 M-1cm-1; NADH, 2800 M-1cm-1; oxidized DMQ2, 1900 M-1cm-1; and reduced DMQ2, 1500 M-1cm-1. These values were used to fit time-dependent absorbance changes at 370 nm to one of two models shown in Scheme 1.20
Scheme 1.
Preparation of apo-GB1-hCLK-1
The expression and purification procedures were altered to obtain apo-GB1-hCLK-1. The (NH4)2Fe(SO4)·6H2O was omitted during expression and 100 μM (NH4)2Fe(SO4)·6H2O was replaced by 1 mM DTT in buffers A and B during the purification. No iron was detected in purified GB1-hCLK-1 with this procedure.
Mössbauer Spectroscopy
A 95.5% 57Fe-enriched sample of GB1-hCLK-1 was prepared as reported previously.14 The iron content for 57Fe-enriched GB1-hCLK-1 was 2.0 ± 0.3. Oxidized GB1-hCLK-1 containing DMQ0 was prepared by mixing the protein with 1.6 equivalents of the quinone and freezing the solution in a Mössbauer cup. Zero-field Mössbauer spectra were recorded at 78 K by using a conventional constant acceleration spectrometer equipped with a temperature controller maintaining temperatures within ± 0.1 K and a 57Co radiation source in a Rh matrix. Isomer shifts are referred to α-Fe metal at room temperature. Data were collected for 48 h at 78 K using protein samples containing approximately 40 μg of 57Fe. Data were fitted with a sum of Lorentzian quadrupole doublets by using a least-squares routine with WMOSS.
EPR Measurements
In order to confirm the absence of adventitious iron in the as-isolated GB1-hCLK-1 and to characterize FeII-FeIII mixed-valent or semiquinone species, EPR spectroscopy was employed. A 1 mM solution of GB1-hCLK-1 (or GB1-hCLK-1/DMQn, n = 0 or 2) in 20 mM Tris, 10 % glycerol, pH 7.0 buffer was mixed with 250 or 500 μM of sodium dithionite (or NADH) under anaerobic conditions. Reaction of 500 μM oxidized GB1-hCLK-1 and 500 μM NADH in the presence of 500 μM DMQ2 was rapidly freeze-quenched after mixing for 0.5, 1.5, and 5 s by spraying into a cold isopentane bath at -140 °C in order to characterize the transient formation of semiquinone or FeII-FeIII mixed-valent species. X-band EPR spectra were recorded on a Bruker ESP 300 spectrometer equipped with an Oxford EPR 900 liquid helium cryostat. Data were recorded under the following conditions: temperature, 4.2 or 77 K; microwave frequency, 9.38-9.40 GHz; microwave power, 2 mW; modulation frequency, 100 kHz; and modulation amplitude, 5 G. EPR quantitation was performed by double integration under a non-saturating condition by using 325, 160, and 80 μM of FeIII(EDTA) as a standard.
Results and Discussion
Expression of Human CLK-1 as a GB1 Fusion Protein
Spectroscopic and enzymatic studies of the membrane-bound protein human CLK-1 have been hampered by its limited solubility. To remedy this situation, we used the immunoglobulin binding domain of protein G (GB1) to create an N-terminal fusion of human CLK-1 (GB1-hCLK-1). The fusion protein improved both the expression level and the solubility of the enzyme.15 A pET30a(+)-GBFusion vector containing the human clk-1 gene was constructed having a small (6,367 Da) acidic (pI = 4.5) GB1 tag that is unlikely to interact with hCLK-1 (pI = 5.7) or alter its structure or ability to assembly the diiron center. Another benefit of having the GB1 fusion at the N-terminus is that the hydrophilic GB1 moiety is synthesized prior to the target gene, which often aids in proper folding of the protein of interest. GB1-hCLK-1 was expressed and purified in E. coli in high yield, ~13 mg per liter of soluble fusion protein. SDS-PAGE analysis of purified GB1-hCLK-1 exhibited a single band corresponding to a protein mass ~26 kDa, which is consistent with the calculated molecular weight of 26,507 Da, as well as the value of 26,505 Da determined by MALDI-TOF (Supplementary Fig. S2). The solubility of the hCLK-1 membrane-bound enzyme was significantly improved, to 3 mM (in 20 mM Tris, 10% glycerol, pH 7.0), through construction of the N-terminal GB1 fusion protein. Based on size exclusion chromatography, GB1-hCLK-1 is dimeric in solution.
Characterization of the Diiron Center
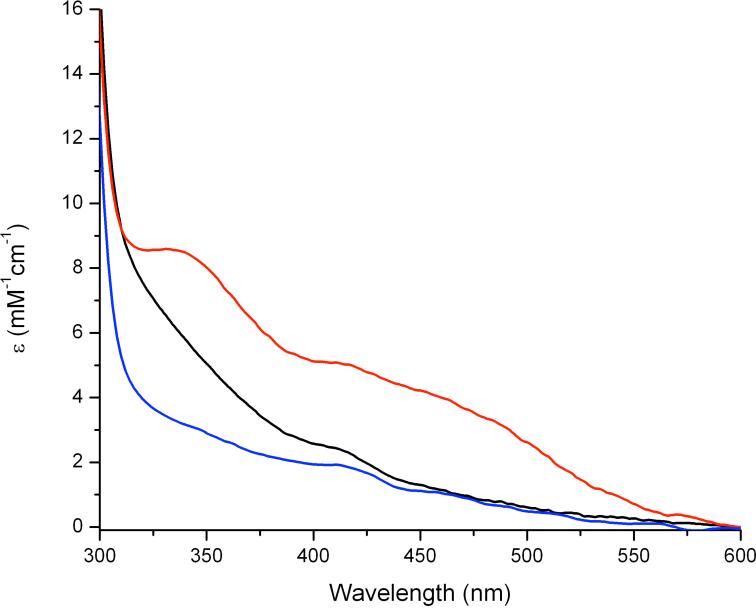
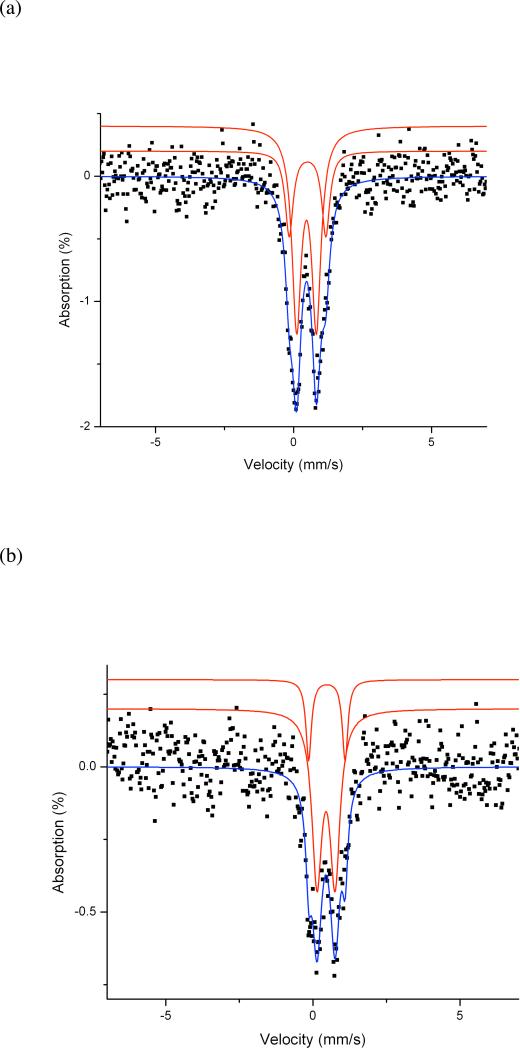
Compared to that of apo-GB1-hCLK-1, the UV-vis spectrum of the expressed GB1-hCLK-1 protein displays an absorption band at 340 nm with ε ~ 5300 M-1cm-1 (Fig. 1). This optical feature is similar to that observed in mouse CLK-1 containing an N-terminal maltose-binding protein (MBP-mCLK-1),14 which is typical of an oxo-bridged diiron(III) center.21 Following addition of NaN3, additional absorption bands at 342 and 450 nm appear (Fig. 1). The emergence of these optical features is consistent with other azide-adducts of carboxylate-bridged (μ-oxo)diiron(III) proteins, including stearoyl-acyl carrier protein Δ9 desaturase (Δ9D) and hemerythrin.22-23 The stoichiometry of Fe-to-GB1-hCLK-1 was determined to be 2.0(± 0.2):1, consistent with the present of a single diiron center. In order to further characterize this unit, we obtained the Mössbauer spectrum of a frozen solution of the protein that was 95.5% 57Fe-enriched at 78 K and zero magnetic field. As shown in Fig. 2a, there are two signals with isomer shifts typical of carboxylate-bridged high-spin diiron(III) units (Table 1).24-26 The simulation revealed two doublets best described as having isomer shifts (δ) of 0.47 ± 0.02 mm/s, with a corresponding quadrupole doublet (ΔEQ) of 0.72 ± 0.02 mm/s (73%), and δ of 0.50 ± 0.02 mm/s, with ΔEQ = 1.33 ± 0.02 mm/s (27%). Extensive studies of diiron model complexes suggest an assignment of these features to μ-hydroxo- and μ-oxo-diiron(III) centers, respectively.21,27 Together, these distinctive UV-vis and Mössbauer characteristics provide strong evidence that the isolated CLK-1 enzyme contains a non-heme, carboxylate-bridged diiron(III) center in a mixture of oxo- and hydroxo-bridged forms, similar to that observed in MBP-mCLK-1.14
Figure 1.
UV-vis spectra of expressed GB1-hCLK-1 (black), after addition of NaN3 (red), and apo-GB1-hCLK-1 (blue).
Figure 2.
DMQ0 binding perturbs the oxidized diiron center in GB1-hCLK-1 as revealed by Mössbauer spectroscopy. (a) Mössbauer spectrum of 1 mM oxidized GB1-hCLK-1 without DMQ0. (b) Mössbauer spectrum of 1 mM oxidized GB1-hCLK-1 in the presence of 1.6 mM DMQ0. The red lines are simulated spectra of the components, which combine as indicated by the blue line overlaid on the experimental spectrum.
Table 1.
| protein | isomer shift (δ) (mm/s) | quadruple splitting (ΔEQ) (mm/s) | ref |
|---|---|---|---|
| Human | 0.47 | 0.72 | this work |
| CLK-1 no DMQ0 | 0.50 | 1.33 | |
| Human | 0.45 | 0.61 | this work |
| CLK-1 with DMQ0 | 0.48 | 1.24 | |
| Human | 0.55 | 1.16 | 18b |
| DOHH | 0.58 | 0.88 | |
| Mouse | 0.47 | 1.32 | 18c |
| MIOX | 0.49 | 0.63 |
Abbreviation: DOHH, deoxyhypusine hydroxylase; MIOX, myo-inositol oxygenase.
Standard deviation for isomer shift and quadruple splitting is 0.02 mm/s.
Analysis of the Mössbauer signal of the GB1-tagged hCLK-1 differs from that of MBP-mCLK-1 in a number of ways.14 The ΔEQ values of 0.72 (73%) and 1.33 (27%) mm/s for GB1-hCLK-1, and of 0.62 (59%) and 1.59 (41%) mm/s for MBP-mCLK-1, suggest that the protonation states of the solvent-derived oxygen ligands may differ slightly for these two protein constructs. The differences may be a result of the protein source, mouse vs. human, but may also arise from perturbations introduced by the relative large MBP tag (42 kDa, pI = 5.7). In addition, there was adventitious iron in the samples of MBP-mCLK-1, which had to be accounted for in the Mössbauer spectral fits. In contrast, there is no adventitious iron in GB1-hCLK-1, a conclusion that is supported by Mössbauer spectroscopy and the lack of a characteristic g = 4.3 signal in the EPR spectrum of oxidized GB1-hCLK-1 (Supplementary Fig. S3). Moreover, the absence of adventitious Fe demonstrates the value of GB1 as a solubility-enhancement tag for the enzymatic study.
Hydroxylation of DMQ derivatives
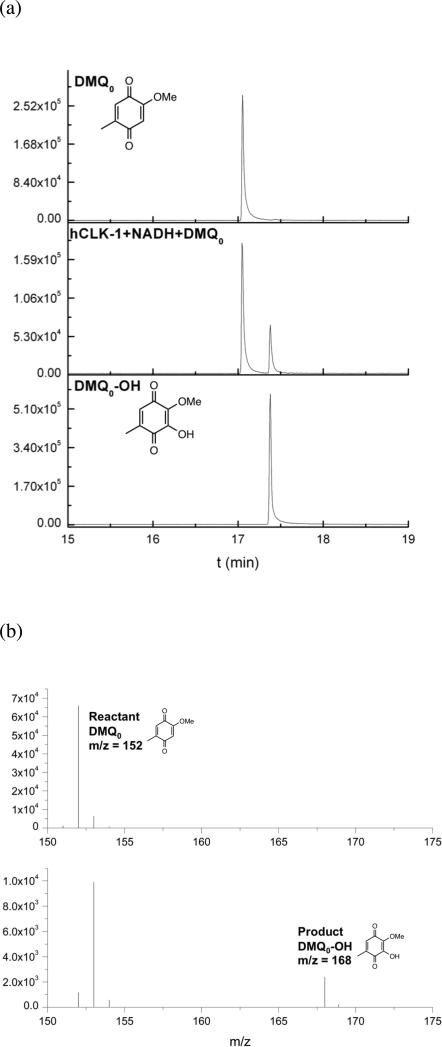
CLK-1 has been proposed as a DMQ hydroxylase because of the accumulation of DMQ9 in Δclk-1 mutants of C. elegans and mice.7 We therefore prepared 2-methoxy-5-methyl-1,4-benzoquinone (DMQ0) to evaluate its potential to serve as a substrate for hydroxylation by CLK-1 (Chart 1). The product formed in the reaction of 5 μM GB1-hCLK-1, 1 mM DMQ0, and 1 mM NADH, characterized by GC-MS, indicated that GB1-hCLK-1 catalyzes the hydroxylation of DMQ0 to generate DMQ0-OH (Fig. 3). Based on the time course of DMQ0-OH formation using GB1-hCLK-1, an apparent kcat of 2.3 min-1 was calculated (Supplementary Fig. S4).
Figure 3.
GB1-hCLK-1 catalyzes the hydroxylation of DMQ0. The product generated in the reaction of 5 μM GB1-hCLK-1, 1 mM DMQ0 and 1 mM NADH is identified by GC-MS. (a) GC-MS chromatograph of DMQ0 (top), reaction product (middle), and DMQ0-OH (bottom). (b) MS spectra of DMQ0 (top) and reaction product, DMQ0-OH (bottom).
The steady state activity of GB1-hCLK-1 was investigated by an NADH consumption assay. In contrast to the negligible consumption of NADH in the reaction of 5 μM GB1-hCLK-1 with 200 μM NADH (0.2 ± 0.1 μM/min), the NADH consumption rate was 12.7 ± 0.6 μM/min in the presence of 1 mM DMQ0 (Table 2). This enhanced NADH consumption suggests that continuous and catalytic reduction of the diiron center by NADH upon addition of DMQ0 occurs to activate dioxygen for DMQ0 hydroxylation. There was no DMQ0 hydroxylation or NADH consumption when apo-GB1-hCLK-1 was examined, further corroborating the requirement for a carboxylate-bridged diiron center to activate O2 and oxidize DMQ0.
Table 2.
NADH consumption rates, Vmax, KM and kcat/KM for GB1-hCLK-1 in the absence and presence of DMQ0 or DMQ2.
| Substrate | NADH Consumption (μM/min)a | Vmax (μM/min) | KM (μM) | kcat/KM (mM-1min-1) |
|---|---|---|---|---|
| none | 0.2 ± 0.1 | - | - | - |
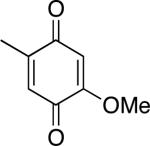
|
12.7 ± 0.6 | 12.8e | 68.1e | 37.6e |
| 12.6f | 204.0f | 12.4f | ||
| DMQ0 | ||||
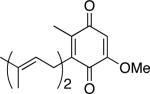
|
37.1 ± 4.2b | |||
| 36.0 ± 4.8c | 39.2 | 33.8 | 232.0 | |
| DMQ2 | 34.9 ± 3.5d |
5 μM of GB1-hCLK-1, 200 μM of NADH, and 1mM of DMQ0 or 400 μM of DMQ2 were used.
pre-mix GB1-hCLK-1 and DMQ2 followed by addition of NADH.
pre-mix GB1-hCLK-1 and NADH followed by addition of DMQ2.
pre-mix DMQ2 and NADH followed by addition of GB1-hCLK-1.
Reaction was performed in 20 mM Tris, pH 7.0.
Reaction was performed in duteriated Tris buffer.
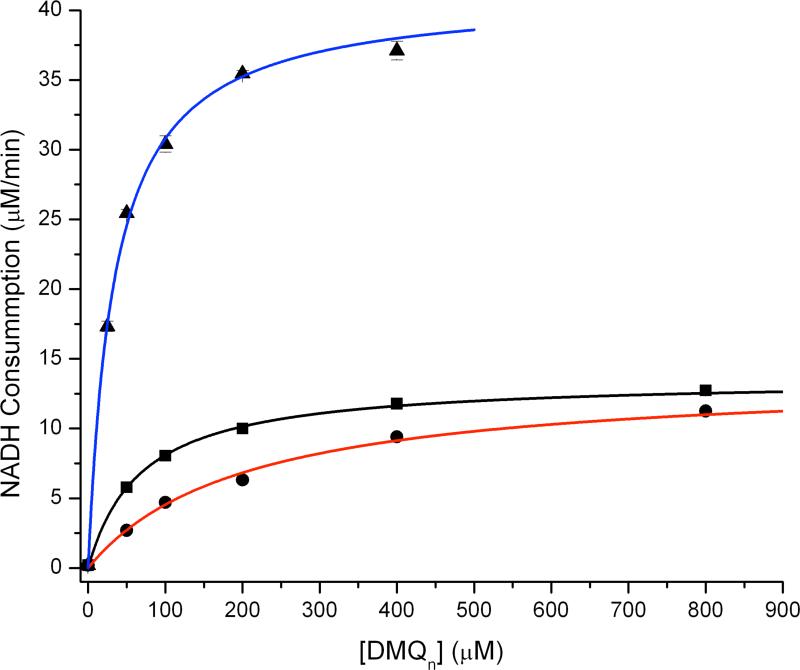
As shown in Fig. 4, a steady state activity analysis of NADH consumption using GB1-hCLK-1 in the presence of DMQ0 displayed typical Michaelis-Menten kinetics, with KM = 68.1 μM, Vmax = 12.8 μM/min, and kcat/KM = 37.6 min-1mM-1. The absence of a kinetic solvent isotope effect (KSIE) for kcat measured in H2O vs. D2O (kH/kD = 1.0) suggests that protonation/deprotonation is not involved in the rate-determining step during the catalytic cycle. Rather, an intermolecular isotope effect of 3 on KM(DK = DKM/HKM), was determined.28
Figure 4.
Steady state activity of GB1-hCLK-1 based on an NADH consumption assay. DMQn was used as substrate (n = 0, black and red; or n = 2, blue). To check for a possible kinetic solvent isotopic effect, NADH consumption in the presence of GB1-hCLK-1 and DMQ0 was carried out in H2O (black) and D2O (red). The NADH consumption rate was fit with Michaelis-Menten kinetics, as indicated by the solid lines.
The binding of DMQ0 to GB1-hCLK-1 was characterized by Mössbauer spectroscopy, and similar isomer shift values were observed in the absence and presence of the substrate (Fig. 2b and Table 1). The two quadrupole splitting parameters changed from 0.72 and 1.33 mm/s to 0.61 and 1.24 mm/s upon addition of DMQ0 to GB1-hCLK-1. Unlike the significant perturbation of the diiron active site by NaN3 revealed by optical spectroscopy (Fig. 1), similar absorption spectra were obtained for oxidized GB1-hCLK-1 in the absence and presence of DMQ0 or DMQ0-OH. A comparison of substrate- and product-exposed GB1-hCLK-1 spectra with that of the NaN3 GB1-hCLK-1 adduct indicates comparatively minor perturbation of the diiron(III) active site (Supplementary Fig. S5). Unlike the formation of a broad optical absorption band centered at 580 nm observed for product (catecholate)-bound ToMOH,29 the minor perturbation of the optical spectrum excludes DMQ0 (or DMQ0-OH) coordination to the active site iron atoms. These results support the previous proposal that substrate binds to the Glu22/His110/Tyr111 triplet of amino acids of CLK-1 rather than directly to the diiron site (Supplementary Fig. S1).10
In order to assess the influence of an isoprenoid side chain on substrate binding and steady state activity, DMQ2 was prepared and evaluated as a substrate (Chart 1). The KM of 33.8 μM for DMQ2, by comparison to the value of 68.1 μM for DMQ0, reveals that inclusion of an isoprenoid side chain increases the binding affinity of a quinone substrate to GB1-CLK-1 (Fig. 4). Moreover, the increase in Vmax from 12.8 μM/min to 39.2 μM/min and kcat/KM from 37.6 min-1mM-1 to 232.0 min-1mM-1 upon replacing DMQ0 with DMQ2 demonstrates that the isoprenoid side chain not only enhances enzymatic hydroxylation activity but also improves catalytic efficiency (Table 2). A similar dependence of catalytic efficiency on the length of acyl-chain of substrate is observed in Δ9 desaturase.30 Although a significant enhancement in GB1-hCLK-1 catalytic efficiency was expected using the natural substrate, DMQ10, the decrease in solubility of DMQ substrates when the isoprenoid side chain was increased from DMQ0 (~3 mM) to DMQ2 (~500 μM) prohibited the use of DMQn substrates with longer isoprenoid side chains. The association of DMQ10 and CLK-1 with the mitochondrial membrane through hydrophobic interactions most likely provides an orientational preference for facile electron transfer and DMQ-hydroxylation.
Substrate-Mediated Electron Transfer
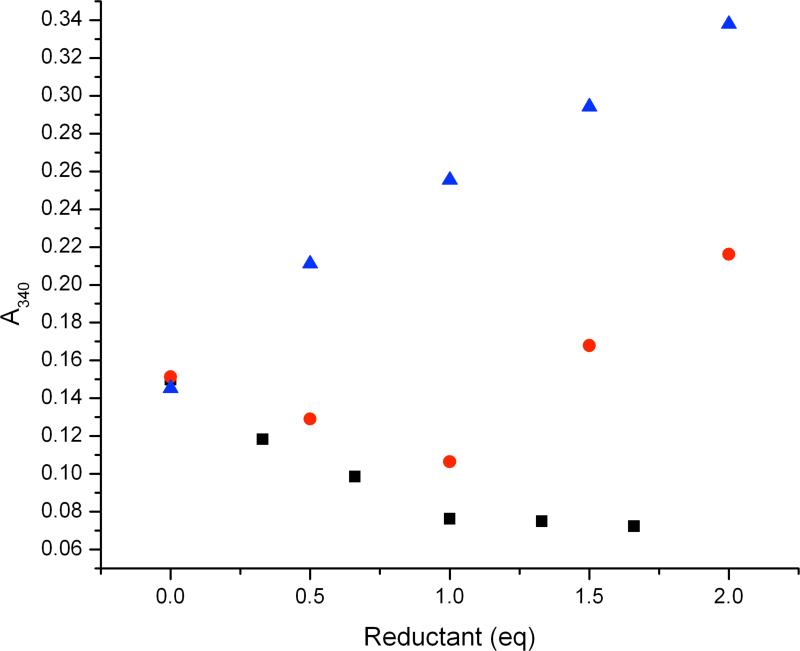
Because there is negligible NADH consumption by GB1-hCLK-1 in the absence of quinone, there appears to be substrate-gated electron transfer from NADH to the diiron center, leading to subsequent O2 activation and DMQ hydroxylation. Reductive titrations of GB1-hCLK-1 by sodium dithionite or NADH were therefore performed under anaerobic conditions in order to obtain more direct evidence for substrate-gated electron transfer. Titrations of GB1-hCLK-1 with sodium dithionite revealed that two electrons are required for complete reduction of the diiron center, as monitored by a decrease in the intensity of the putative oxo-to-FeIII charge transfer band at 340 nm (Fig. 5). This optical change further supports the conclusion that each GB1-hCLK-1 contains a single carboxylate-bridged diiron center. Addition of O2-saturated buffer to reduced GB1-hCLK-1 resulted in nearly quantitative reoxidation of diiron center (93% recovery, Supplementary Fig. S6). In contrast to the inability to reduce the diiron center in GB1-hCLK-1 by NADH in the absence of substrate, reduction of diiron center by NADH occurs in the presence of one equivalent of DMQ0 as indicated by the decrease of A340. These results support substrate-gated electron transfer (Fig. 5). The absence of a characteristic EPR signal for an FeII-FeIII mixed-valent or semiquinone during the reductive titration of oxidized GB1-hCLK-1 with dithionite or DMQ0/NADH excludes the formation of FeII-FeIII mixed-valent or semiquinone species in GB1-hCLK-1. The attempt to characterize the transient formation of such species is discussed below.
Figure 5.
Substrate-gated reduction of the diiron center in GB1-hCLK-1. The reduction of the diiron(III) center in resting state GB1-hCLK-1 was monitored by the decay of the optical band at 340 nm. Compared to the reduction of diiron center by dithionite (black) and the lack of NADH reactivity with GB1-hCLK-1 (blue), reduction of the diiron center by NADH in the presence of DMQ0 demonstrates substrate-gated electron transfer from NADH to diiron center in GB1-hCLK-1 (red).
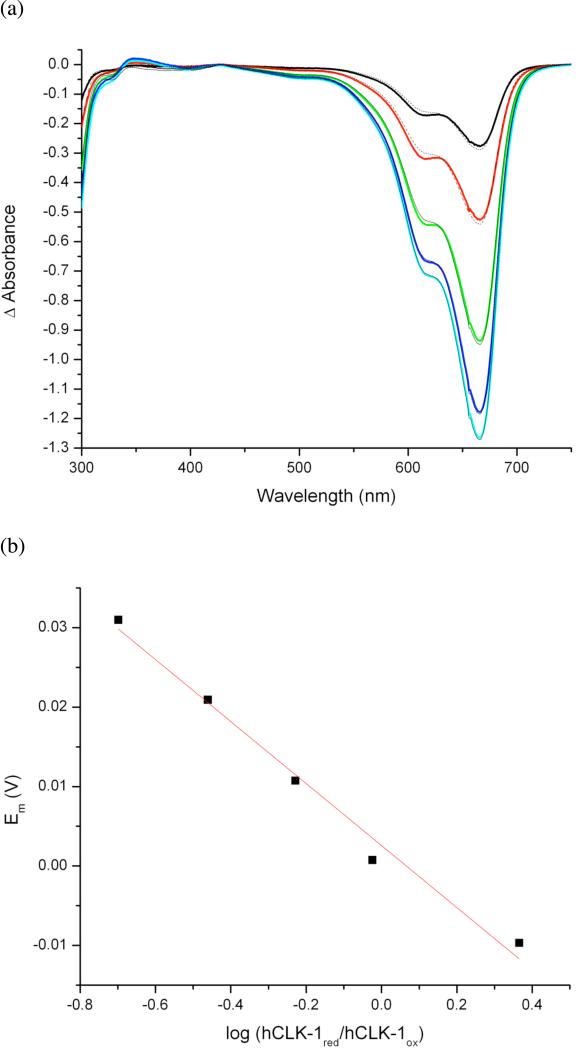
In Δ9D, a substrate-induced conformational change at the diiron center induced a positive shift in the reduction potential from –320 mV to –210 mV to facilitate its reduction by NADH (reduction potential -320 mV, vs NHE).31 In order to investigate whether DMQ0-binding similarly promotes NADH reduction of the diiron center in GB1-hCLK-1, the redox potential of the diiron center was determined by reductive titration using indicator dyes (Fig. 6). Unlike the thermodynamic gating of catalysis in Δ9D, the reduction potential of the diiron center in GB1-hCLK-1, 3 mV vs NHE, is sufficiently high such that NADH reduction is thermodynamically facile in the absence of substrate. This result suggests that a kinetic barrier prevents reduction of the diiron center and that binding of DMQ0 to GB1-hCLK-1 in some manner provides a pathway for electron transfer by NADH.
Figure 6.
Determination of the redox potential of the diiron center in GB1-hCLK-1. (a)Difference spectra for a reductive titration of 20 μM GB1-hCLK-1 and 20 μM methylene blue by 500 μM dithionite at pH 7.0 and 25 °C. Spectra correspond to the addition of 5, 10, 15, 20, and 25 μL of ca. 500 μM dithionite, respectively. Fits are shown as dashed lines. (b) Plot of the potential derived from log (dyered/dyeox) vs log (CLK-1red/CLK-1ox). The solid line indicates a fit to the modified Nernst equation (eq 1, see text).18-19 The y-intercept of 3 mV (vs NHE) represents the redox potential of GB1-hCLK-1.
We considered two possible mechanisms for this process, (i) quinone-mediated reduction by NADH (Schemes 1b and 1c) and (ii) quinone-gated conformational change of GB1-hCLK-1 for reduction by NADH (Scheme 1b’). In an attempt to distinguish between these two pathways, the reduction of oxidized GB1-hCLK-1 in the presence of DMQ2 by NADH under pre-steady-state conditions was investigated. Using this method, we observed that the decay in A370 did not fit well to a single exponential. Instead, the data fit well to a sum of two exponentials describing the substrate-mediated reduction process (). Fitting the data in this manner return rate constants k1’ = 1.15 ± 0.04 s-1 and k2 = 0.046 ± 0.002 s-1 (Figure S7).
| (2) |
In order to characterize the transient formation of semiquinone or FeII-FeIII mixed-valent species in the reduction of oxidized GB1-hCLK-1 by NADH, the reaction of oxidized GB1-hCLK-1 with NADH in the presence of DMQ2 was also rapid-freeze quenched after mixing for 0.5, 1.5, and 5 s. The absence of an EPR signal characteristic of a semiquinone or FeII-FeIII species supports quinone-mediated two-electron reduction of the diiron(III) center in GB1-hCLK-1 by NADH (Scheme 1b and 1c). Utilization of the intrinsic redox capability of a hydroquinone to mediate such two-electron transfer processes has been previously reported.6,32-33 Intermolecular electron transfer from Complex I or II to Complex III in the inner mitochondrial membrane of eukaryotes is similarly promoted, and hydroquinone-mediated intramolecular electron transfer occurs in the bacterial reaction center from Rb. Sphaeroides.6,32-33 The primary quinone, QA, is reduced by the excited bacterichlorophyll dimer (D) and then the reduced QA- further transfers an electron to the secondary quinone, QB (DQAQB → D+QA-QB → D+QAQB-).
NADH does not react with either hCLK-1ox or its quinone substrates in the absence of the enzyme. In the presence of hCLK-1ox and DMQox, however, the enzyme becomes reduced, implying the transient formation of DMQred and quinone-mediated electron transfer. We presume that this activity involves solvent-exchangeable hydrogen bonding interactions between the Glu22/His110/Tyr111 amino acid triad in GB1-hCLK-1 and quinones, which promotes their reduction by NADH. The enhanced reduction of quinones by NADH derivatives through hydrogen bonding interactions has precedence in the literature.34
Conclusion
We report the high-yield expression and purification of soluble human CLK-1 in E. coli as an N-terminal GB1 fusion. UV-vis and Mössbauer spectroscopic studies of this protein provide unambiguous evidence that human CLK-1 belongs to the carboxylate-bridged diiron protein family of proteins and activates O2 for DMQ hydroxylation at the diiron center. Human CLK-1 is the first membrane-bound diiron protein in humans that functions as a hydroxylase.
The binding of DMQn substrates in GB1-hCLK-1 gates and mediates reduction of the diiron center by NADH without an additional reductase protein component. Substrate-gated reduction of a diiron center for O2 activation and DMQ hydroxylation in human CLK-1 conveys specificity and prevents undesired consumption of NADH in mitochondria. The development of a highly efficient E. coli expression system to generate sufficient quantities of soluble human CLK-1 with GB1 fusion protein for further study should help to unravel the role of CLK-1, as well as its roles in the biosynthesis of ubiquinone and the aging process.
Supplementary Material
Acknowledgements
This work was supported by grant GM032134 from the National Institute of General Medical Sciences to SJL. T.T.L. received support from the Postdoctoral Research Abroad Program sponsored by National Science Council, R.O.C. and U.P.A. acknowledges the Alexander von Humboldt Foundation for fellowship support. We thank Drs. L. Do and Y. Li for preparing of DMQ0 and DMQ2 for these studies. We also thank Ms. A. Liang and Dr. W. Wang for helpful discussions and comments on the manuscript.
Abbreviations
- GB1
immunoglobulin binding domain of protein G
- DMQ
5-demethoxyubiquinone
- UQ
ubiquinone
- sMMO
soluble methane monooxygenase
- ToMO
toluene monooxygenase
- PH
phenol hydroxylase
- DMQ0
2-Methoxy-5-methyl-1,4-benzoquinone
- DMQ0-OH
2-methoxy-3-hydroxyl-5-methyl-1,4-benzoquinone
- PCR
polymerization chain reaction
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- PMSF
phenylmethanesulfonylfluoride
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- BSA
bovine serum albumin
- NADH
nicotinamide adenine dinucleotide
Footnotes
This work was supported by grant GM032134 from the National Institute of General Medical Sciences
Supporting Information
Additional details of the characterization of GB1-hCLK-1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kirkwood TB. A systematic look at an old problem. Nature. 2008;451:644–647. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- 2.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 3.Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- 4.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/- mice. J. Biol. Chem. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levavasseur F, Miyadera H, Sirois J, Tremblay ML, Kita K, Shoubridge E, Hekimi S. Ubiquinone is necessary for mouse embryonic development but is not essential for mitochondrial respiration. J. Biol. Chem. 2001;276:46160–46164. doi: 10.1074/jbc.M108980200. [DOI] [PubMed] [Google Scholar]
- 6.Ernster L, Forsmark-Andree P. Ubiquinol: an endogenous antioxidant in aerobic organisms. Clin. Investig. 1993;71:S60–65. doi: 10.1007/BF00226842. [DOI] [PubMed] [Google Scholar]
- 7.Miyadera H, Amino H, Hiraishi A, Taka H, Murayama K, Miyoshi H, Sakamoto K, Ishii N, Hekimi S, Kita K. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J. Biol. Chem. 2001;276:7713–7716. doi: 10.1074/jbc.C000889200. [DOI] [PubMed] [Google Scholar]
- 8.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Suppl):S62–71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stenmark P, Grunler J, Mattsson J, Sindelar PJ, Nordlund P, Berthold DA. A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane-bound hydroxylase involved in ubiquinone biosynthesis. J. Biol. Chem. 2001;276:33297–33300. doi: 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- 10.Rea S. CLK-1/Coq7p is a DMQ mono-oxygenase and a new member of the di-iron carboxylate protein family. FEBS Lett. 2001;509:389–394. doi: 10.1016/s0014-5793(01)03099-x. [DOI] [PubMed] [Google Scholar]
- 11.Nordlund P, Eklund H. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 12.Kurtz DM. Structural similarity and functional diversity in diiron-oxo proteins. J. Biol. Inorg. Chem. 1997;2:159–167. [Google Scholar]
- 13.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 14.Behan RK, Lippard SJ. The aging-associated enzyme CLK-1 is a member of the carboxylate-bridged diiron family of proteins. Biochemistry. 2010;49:9679–9681. doi: 10.1021/bi101475z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Wagner G. Overcoming the solubility limit with solubility-enhancement tags: successful applications in biomolecular NMR studies. J. Biomol. NMR. 2010;46:23–31. doi: 10.1007/s10858-009-9371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernini R, Mincione E, Barontini M, Provenzano G, Setti L. Obtaining 4-vinylphenols by decarboxylation of natural 4-hydroxycinnamic acids under microwave irradiation. Tetrahedron. 2007;63:9663–9667. [Google Scholar]
- 17.van der Klei A, de Jong RLP, Lugtenburg J, Tielens AGM. Synthesis and spectroscopic characterization of [1'-C-14]ubiquinone-2, [1'-C-14]-5-demethoxy-5-hydroxyubiquinone-2, and [1'-C-14]-5-demethoxyubiquinone-2. Eur. J. Org. Chem. 2002:3015–3023. [Google Scholar]
- 18.Blazyk JL, Lippard SJ. Expression and characterization of ferredoxin and flavin adenine dinucleotide binding domains of the reductase component of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Biochemistry. 2002;41:15780–15794. doi: 10.1021/bi026757f. [DOI] [PubMed] [Google Scholar]
- 19.Kopp DA, Gassner GT, Blazyk JL, Lippard SJ. Electron-transfer reactions of the reductase component of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Biochemistry. 2001;40:14932–14941. doi: 10.1021/bi015556t. [DOI] [PubMed] [Google Scholar]
- 20.Beauvais LG, Lippard SJ. Reactions of the peroxo intermediate of soluble methane monooxygenase hydroxylase with ethers. J. Am. Chem. Soc. 2005;127:7370–7378. doi: 10.1021/ja050865i. [DOI] [PubMed] [Google Scholar]
- 21.Kurtz DM. Oxo- and hydroxo-bridged diiron complexes: a chemical perspective on a biological unit. Chem. Rev. 1990;90:585–606. [Google Scholar]
- 22.Fox BG, Shanklin J, Somerville C, Munck E. Stearoyl-acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc. Natl. Acad. Sci. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richard C, Reem EIS. Spectroscopic studies of the binuclear ferrous active site of deoxyhemerythrin: coordination number and probable bridging ligands for the native and ligand-bound forms. J. Am. Chem. Soc. 1987;109:1216–1226. [Google Scholar]
- 24.Vu VV, Emerson JP, Martinho M, Kim YS, Munck E, Park MH, Que L., Jr. Human deoxyhypusine hydroxylase, an enzyme involved in regulating cell growth, activates O2 with a nonheme diiron center. Proc. Natl. Acad. Sci. 2009;106:14814–14819. doi: 10.1073/pnas.0904553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing G, Hoffart LM, Diao Y, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr. A coupled dinuclear iron cluster that is perturbed by substrate binding in myo-inositol oxygenase. Biochemistry. 2006;45:5393–5401. doi: 10.1021/bi0519607. [DOI] [PubMed] [Google Scholar]
- 26.Wallar BJ, Lipscomb JD. Dioxygen Activation by Enzymes Containing Binuclear Non-Heme Iron Clusters. Chem. Rev. 1996;96:2625–2658. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 27.Armstrong WH, Lippard SJ. Reversible Protonation of the Oxo Bridge in a Hemerythrin Model-Compound-Synthesis, Structure, and Properties of (μ-Hydroxo)mis(μ-acetato)bis[hydrotris(1-pyrazolyl)borato]diiron(III), [(HB(pz)3)Fe(OH)(O2CCH3)2Fe(HB(pz)3)]+. J. Am. Chem. Soc. 1984;106:4632–4633. [Google Scholar]
- 28.Bell LC, Guengerich FP. Oxidation kinetics of ethanol by human cytochrome P450 2E1. J. Biol. Chem. 1997;272:29643–29651. doi: 10.1074/jbc.272.47.29643. [DOI] [PubMed] [Google Scholar]
- 29.Murray LJ, Naik SG, Ortillo DO, Garcia-Serres R, Lee JK, Huynh BH, Lippard SJ. Characterization of the arene-oxidizing intermediate in ToMOH as a diiron(III) species. J. Am. Chem. Soc. 2007;129:14500–14510. doi: 10.1021/ja076121h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas JA, Fox BG. Role of hydrophobic partitioning in substrate selectivity and turnover of the Ricinus communis stearoyl acyl carrier protein Δ9 desaturase. Biochemistry. 1999;38:12833–12840. doi: 10.1021/bi991318a. [DOI] [PubMed] [Google Scholar]
- 31.Reipa V, Shanklin J, Vilker V. Substrate binding and the presence of ferredoxin affect the redox properties of the soluble plant Δ9-18:0-acyl carrier protein desaturase. Chem. Comm. 2004:2406–2407. doi: 10.1039/b409595b. [DOI] [PubMed] [Google Scholar]
- 32.Graige MS, Feher G, Okamura MY. Conformational gating of the electron transfer reaction QA-.QB -> QAQB-. in bacterial reaction centers of Rhodobacter sphaeroides determined by a driving force assay. Proc. Natl. Acad. Sci. 1998;95:11679–11684. doi: 10.1073/pnas.95.20.11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graige MS, Paddock ML, Bruce JM, Feher G, Okamura MY. Mechanism of Proton-Coupled Electron Transfer for Quinone (QB) Reduction in Reaction Centers of Rb. Sphaeroides. J. Am. Chem. Soc. 1996;118:9005–9016. [Google Scholar]
- 34.Yuasa J, Yamada S, Fukuzumi S. One-step versus stepwise mechanism in protonated amino acid-promoted electron-transfer reduction of a quinone by electron donors and two-electron reduction by a dihydronicotinamide adenine dinucleotide analogue. Interplay between electron transfer and hydrogen bonding. J. Am. Chem. Soc. 2008;130:5808–5820. doi: 10.1021/ja8001452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.