Summary
The importance and role of the cellular epigenome in cell fating and development has been studied for decades. The epigenome encompasses a range of attributes including DNA methylation, histone modifications, and chromatin remodelers; together these components define the cellular transcriptome, identity, and function. The cellular epigenome is dynamic in response to environmental signals, modifiable during normal cell differentiation and is heritable in daughter cells. This plasticity, however, poses a risk for misregulation and may underlie a number of hereditary disorders, development defects, and cancer. Although the first epigenetic change described in cancer was gene hypomethylation [1,2], we know that cancers display global hypomethylation, as well as, site-specific gene hypermethylation in addition to changes in chromatin modifications. Mechanisms explaining the sometimes paradoxical epigenetic changes observed in cancer, their contributions to tumor initiation and progression and how epigenetics relate to genetic events are poorly understood. In this review we will briefly discuss recent findings on the epigenomic states observed in colon cancer, in particular, how perturbations to the genome and epigenome together may contribute to initiation and progression of colon cancer.
Introduction
Intestinal Epithelial Regeneration poses a risk for colon cancer development
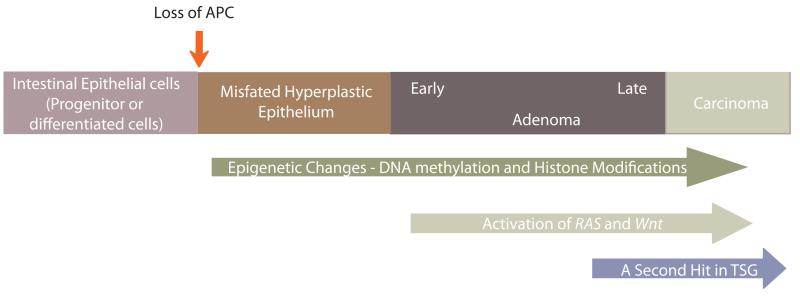
Epithelia is a continuous sheet of tightly linked cells that line the digestive tract, urogenital, and respiratory tract. These epithelial layers protect from the external environment and aid in nutrient/water absorption and glandular secretions. Most epithelial layers are constantly regenerated in order to maintain normal adult organ function. Within the intestine a cyclical regeneration process [3-6] is maintained by adult stem cell populations that reside within the intestinal crypt [7-12]. The stem cells from the crypt bottoms give rise to a rapidly dividing transit-amplifying (TA) population. Near the mouth of the crypt, TA cells exit mitosis and differentiate into all mature cell types of the intestinal epithelium including absorptive enterocytes and three secretary cells types; goblet, enteroendocrine and Paneth cells [13]. Eventually, differentiated epithelial cells undergo apoptosis and are shed into the intestinal lumen. The average life span of a cell in the intestinal epithelium is just 3-5 days [14], so the mechanisms that regulate stem cell maintenance, proliferation, differentiation, and apoptosis must be precisely tuned to ensure proper organ maintenance. An imbalance in the proliferation, differentiation, and apoptosis patterns within the intestinal crypts can lead to aberrant crypt foci [3-6], which are thought to later progress to an adenoma. The progression from an adenoma to carcinoma in colon cancer may take decades, supporting the notion that accumulated genetic and epigenetic changes underlie the multistep developmental process of colorectal cancer (Figure 1).
Figure 1.
Model Figure
Colon cancer development is a multistep process with known perturbations to the genome and/or epigenome of colonic epithelial or progenitor cells. These perturbations together contribute to initiation and progression of colon cancer. Although, this may be an over simplification of the process, the loss of APC represents a common starting point. Upon loss of APC, the attenuated retionic acid levels and the concomitant upregulation in the DNA demethylases may perturb the differentiated cell fate or may prevent proper colonic progenitor cell differentiation. The upregulation in the DNA demethylases may contribute to the global hypomethylation or facilitate the second hit mutation in a tumor suppressor gene, which provides cancer cells a growth advantage and metastatic potential.
The Molecular Genetics of Colon Adenoma Formation
Familial Adenomatous Polyposis (FAP) results from mutations in a single gene known as adenomatous polyposis coli, APC [15,16]. This syndrome is defined by the appearance of hundreds to thousands of adenomatous polyps in affected individuals. The APC gene was discovered by genetic linkage analysis in FAP families [17-19]. Mutations in the APC gene appear in aberrant crypt foci and early adenomas, suggesting inactivation of APC very early in adenoma formation [20-22]. Furthermore, mutations in APC have been observed in 70-80% of sporadic colon cancers [23,24]. In support of APC loss as an initiating event in adenoma, mice lacking functional APC develop numerous intestinal adenomas[25] [26]. Together the data from human and mice place APC as a gatekeeper of colonic epithelial cell proliferation and differentiation; whose loss may lead to an imbalance in cell turnover. Supporting this gatekeeper role, a number of studies have investigated whether germline or de novo somatic mutations in genes such as p53 and RAS, commonly mutated genes in colon cancer [21,27-29], can efficiently initiate neoplastic processes. These studies show that the loss of these genes alone does not appear to lead to colorectal neoplasia, instead they assist in progression from adenoma to carcinoma. Therefore, these studies suggest that the sequence of mutations and the accumulation of mutations will determine the propensity of neoplasia [30,31]. A perplexing question in cancer biology is how a single gene mutation can lead to polyp formation or a marked predisposition to colorectal cancer? A possible answer to this question originates from the recent data suggesting that the loss of APC in a cell may affect both intestinal cell fating and cell proliferation, therefore, possibly explaining the enhanced disease penetrance in FAP patients. These findings suggest a model wherein genetic lesions coordinate with epigenetic changes that cause improper cell fating and may help dictate the response to subsequent transforming events.
The role DNA methylation in cancer and cell fating
Although genetic mutations have been implicated in the initiation of many cancers, epigenetic and genetic alterations are likely to act synergistically in cancer development. One of the first epigenetic abnormalities discovered in a number of cancers was the loss of DNA methylation at CpG dinucleotides [2,32]. This loss of methylation was observed in very early stages of premalignant adenomas with no significant bulk changes in methylation from adenoma to carcinoma [33,34]. This hypomethylation was thought to have significant implications on gene activation, loss of heterozygosity, and global chromosomal stability [35-37]. At the time, however, there were no obvious mechanisms explaining DNA demethylation or its biological importance. In parallel, however, the field made rapid progress in understanding DNA hypermethylation and the enzymes that facilitate this process. Evidence for targeted and predictable hypermethylation changes came from analysis of patient samples that resulted in a characteristic pattern of methylation referred to as a CpG island methylator phenotype or CIMP+ [38]. Clear examples of gene hypermethylation leading to inactivation of important tumor suppressor genes was first observed at the mismatch repair enzyme MLH1 in colon cancers [39]. This list of hypermethylated gene promoters in colorectal cancer (CRC) has grown extensively and now includes key tumor suppressor such as retinoblastoma (RB) [40], P16 [41], RARB, and SFRP [42,43]. It is important to note that epigenetic alterations commonly observed in colon cancers such as candidate gene hypermethylation and genome-wide DNA hypomethylation are also evident in normal aged colonic tissue [44-46]. These observations raise the question whether epigenetic marks acquired during aging or in response to oncogene activation might play important roles in priming tumorigenesis and cancer progression. Reversing the methylation state at these hypermethylated promoters/loci, such as MLH1, can be achieved using drugs that inhibit DNA methyltransferase and remain an interesting approach for reprogramming tumor cell with aberrant methylation. [47,48].
Today genome-wide analysis of DNA methylation using expanded promoter arrays have expanded our view of how and where methylation changes happen. Furthermore these genome-wide array studies have revealed that most of the methylation changes observed in a variety of cancers occur in CpG island shores rather than promoters [49]. In addition, this analysis showed that cancer specific differentially methylated loci varied across normal and colon, lung, breast, thyroid and wilms tumor subtypes [50]. Interestingly separate, parallel studies have revealed that these variable cancer loci are often differentially methylated or misregulated when comparing embryonic stem cells (ES) and induced pluripotent stem cells (iPSCs) [51]. Thus suggesting that the mechanisms of differentiation and reprogramming that are employed in normal development maybe shared in reprogramming toward cancer.
In addition to the targeted changes at specific loci, factors such as nuclear organization, architecture, and genomic sequence may impart profound changes in genome wide methylation patterns. For example, a close examination of the genome using shotgun bisulfite sequencing in normal mucosa, three colorectal cancers, and two adenomatus polyps verified previous findings and revealed many new and interesting insights [52]. First, when investigating global DNA methylation changes by comparing the methylation of tumor to adjacent normal mucosa three distinct methylation profiles were found: (1) regions hypomethylated in both tumors and normals, (2) regions that are demethylated in tumors only (3) regions that acquired methylation in tumor. The methylation prone regions in tumors corresponded with CpG islands in and outside promoters, and were also highly enriched for marks of polycomb repressive complex 1 and 2 activity in hESCs and methylated in normal development [53-56]. Furthermore, the methylation prone loci were generally depleted of certain transcription factor sequences such as Sp1, YY1, and NRF1, which confer methylation protection in cancer [57,58]. Notably, the authors find that focal and large blocks of the genome are demethylated [50], and the regions of hypomethylation within tumors corresponded with an increase in gene expression in these tumors. Interestingly, these large blocks of hypomethylation overlapped with previously described partially methylated domains (PMDs) in IMR90 cells [59], demonstrating a shared attribute between immortalized cell lines and tumor cells. Furthermore, these large blocks of hypomethylation are defined by the nuclear lamina associated domains (LADs) [52]. These observations clearly indicate that changes in DNA methylation can occur throughout the genome and may require a variety of mechanisms all of which may serve to improperly fate cells. These changes could, therefore, be a major mechanism for initiating transformation or generating transformation competent cells.
Although the hypomethylation observed in adenomas and carcinomas [60-64] suggests altered cell plasticity and potential growth advantage, the mechanism through which this happens and the biological impacts are unclear. Many have speculated that this global change may be achieved passively (absence of maintenance methyltransferase activity) or actively (targeted enzymatic removal of mC mediated by the DNA demethylases) or a combination of both. Recent work, however, has shed light on this possibility by demonstrating that aberrant DNA methylation can occur soon after the loss of APC. For example, human FAP adenomas and apcmcr zebrafish have elevated transcript levels of the DNA demethylase machinery components (i.e. Mbd4, AID, Apobec2a, Gadd45a) [65-69]. The increase in DNA demethylase components corresponded with an increase in expression and reciprocal decrease in methylation at a number gene promoters implicated in intestinal cell fate specification and colorectal cancers such as aldh, and hoxd13 determined by MEDIP arrays. Knockdown of the DNA demethylase components restored DNA methylation in apcmor zebrafish. Furthermore, the upregulation of the DNA demethylase machinery was due to a previously described lack of retinoic acid [65-69]. Treatment of apcmcr zebrafish with retinoic acid, reduced the DNA demethylase components and restored intestinal cell fating; fating determined by an increase in IFABP expression and reduction in aldh1a2 levels. Furthermore, knockdown of the DNA demethylase components alone in apcmcr zebrafish also induced intestinal cell differentiation, suggesting that the upregulation of DNA demethylases maintained intestinal cells in a progenitor like state [65-69]. These data support a role for APC in controlling cell fate specification through its regulation of the DNA demethylases and place this demethylation as the initiating event that precedes disregulated cell proliferation. Consistent with this notion, Apc min mice carrying a genetic deletion for Apobec1 −/−, a cytidine deaminase, have reduced polyp formation [70].
Changes in Chromatin Packaging in Cancer Initiation and Progression
The histone code at or outside promoters affects DNA methylation dynamics in a stem/progenitor or differentiated cell. For example regions highly enriched for marks of the polycomb repressive complex 1 and 2 activity in hESCs commonly acquire DNA methylation in normal development/differentiation [53-56]. Interestingly, these same gene promoters acquire methylation in colon adenomas. Recently, the misregulation of post-translational histone modifications has become increasingly apparent in a number of human cancers, and is caused by the deregulation of factors that mediate the reading, writing, and removal. For example, in addition to global changes to DNA methylation and H3K27me, a generalized loss of H4K16 acetylation and H4K20 methylation is found in both lymphoma and colorectal cancer and correlated with transcriptional silencing [71]. Future studies are needed to explore the role of histone modifications, remodelers, and transcription factors in the facilitation in intestinal cell turnover and tumor initiation and progression. Achieving a comprehensive understanding of the roles of chromatin remodelers and modifiers in normal intestinal fating and how they are misregulated in cancer may be of therapeutic potential.
Uncontrolled cell proliferation and its role in Colon cancer initiation and progression
The current data now point to misregulation of epigenetics as a major factor in governing intestinal cell fating and colon tumor initiation. Changes in the intestinal cell epigenome may precede and/or enhance the activity of other oncogenes such as Wnt, RAS and p53, which are needed for neoplastic progression. Indeed, a number of studies suggest that loss of cell fating precedes disregulation of proliferation stimulated by signaling pathways such as Wnt/beta - catenin [23,75-82]. In support of this possibilitiy, a number of studies that have failed to correlate the loss of APC with activation of WNT signaling as determined by the presence of nuclear b-catenin, particularly in early adenomas [83-87]. An absence of nuclear b-catenin suggests a role for others factors, such as mehthylation changes as key to adenoma initiation upon loss of APC. Support for this model comes from work in human cells lines, human FAP adenomas, and APC morphant zebrafish [86] showing a need for loss of APC as well as RAS activation in promoting proliferation of undifferentiated intestinal cells. These findings are consistent with previous observations in mice, wherein, loss of APC and KRAS mutation causes an increase in adenoma size, number and invasiveness [88,89]. Further, this enhanced proliferation appears to expand the number of cells bearing putative stem cell markers within the tumor epithelium [90]. Taken together the data suggest that epigenetic changes work in concert with proliferative signals for initiation and progression of colon tumors. Future studies with help define whether specific epigenetic landscapes may be necessary and permit transformation by oncogenes such as RAS.
Concluding remarks
Emerging evidence suggests that both genetic alterations and epigenetic aberrations contribute to the initiation and progression of human cancers, including colon cancer. Loss of a major tumor suppressor, APC, appears to induces aberrant DNA methylation and that this misprogramming contributes to mis-fating of intestinal cells as a common mechanism to drive colon tumorigenesis. These changes in DNA methylation, along with changes in histone modifications, create a new landscape for the correct interpretation of cell signals that usually govern normal cell turnover. Mis-interpretation of these signals due an altered epigenetic state can lead to the misregulation of gene expression and selected growth advantage of tranformed cells. Therefore, further definition of mechanisms targeting epigenetic modifications in the normal intestinal epithelium and in precancerous, and cancerous lesions offers the promise of identifying opportunities for early cancer detection and intervention.
APC modulates the intestinal epigenomic landscape and cell fate through its regulation of RA/DNA demethylases.
Misregulation of the epithelial cell epigenome governs intestinal cell fate and colon tumor initiation.
Global intestinal epigenomic changes may enhance oncogene activity and may facilitate neoplastic progression
Acknowledgements
This work is supported by grants from the National Cancer Institute (CA073992 and CA96934) awarded to D.A.J., the National Institute of Child Health and Development (HD058506 to B.R.C. and D.A.J), the Howard Hughes Medical Institute (to B.R.C), the Huntsman Cancer Foundation, The Helen Hay Whitney awarded to S. H., and by the Cancer Center Support Grant (CA042014).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended Readings
Papers of particular interest, published within the period of the review, have been highlighted as:
*of special interest
** of outstanding interest
- 1.Holliday R, Jeggo PA. Mechanisms for changing gene expression and their possible relationship to carcinogenesis. Cancer Surv. 1985;4:557–581. [PubMed] [Google Scholar]
- 2*.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. The first study examining the methylation changes in primary human tumors and adjacent grossly uninvolved samples. Interestingly this report shows that a number of gene promoters in cancers and metastasis were hypomethylated.
- 3.Karam SM. Lineage commitment and maturation of epithelial cells in the gut. Front Biosci. 1999;4:D286–298. doi: 10.2741/karam. [DOI] [PubMed] [Google Scholar]
- 4.Wright NA. Epithelial stem cell repertoire in the gut: clues to the origin of cell lineages, proliferative units and cancer. Int J Exp Pathol. 2000;81:117–143. doi: 10.1046/j.1365-2613.2000.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth C, Potten CS. Gut instincts: thoughts on intestinal epithelial stem cells. J Clin Invest. 2000;105:1493–1499. doi: 10.1172/JCI10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clatworthy JP, Subramanian V. Stem cells and the regulation of proliferation, differentiation and patterning in the intestinal epithelium: emerging insights from gene expression patterns, transgenic and gene ablation studies. Mech Dev. 2001;101:3–9. doi: 10.1016/s0925-4773(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 7*.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. One of many reports identifying intestinal stem cell markers.
- 8*.Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012 doi: 10.1038/emboj.2012.166. One of many reports identifying intestinal stem cell markers.
- 9*.Snippert HJ, Es JH, van den Born M, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187–2194. e2181. doi: 10.1053/j.gastro.2009.03.002. One of many reports identifying intestinal stem cell markers.
- 10*.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. One of many reports identifying intestinal stem cell markers.
- 11.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. One of many reports identifying intestinal stem cell markers.
- 13.Ayabe T, Ashida T, Kohgo Y, Kono T. The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol. 2004;12:394–398. doi: 10.1016/j.tim.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Hooper CE. Cell turnover in epithelial populations. J Histochem Cytochem. 1956;4:531–540. doi: 10.1177/4.6.531. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura Y, Nishisho I, Kinzler KW, Vogelstein B, Miyoshi Y, Miki Y, Ando H, Horii A. Mutations of the APC (adenomatous polyposis coli) gene in FAP (familial polyposis coli) patients and in sporadic colorectal tumors. Tohoku J Exp Med. 1992;168:141–147. doi: 10.1620/tjem.168.141. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura Y, Nishisho I, Kinzler KW, Vogelstein B, Miyoshi Y, Miki Y, Ando H, Horii A, Nagase H. Mutations of the adenomatous polyposis coli gene in familial polyposis coli patients and sporadic colorectal tumors. Princess Takamatsu Symp. 1991;22:285–292. [PubMed] [Google Scholar]
- 17.Bodmer WF, Bailey CJ, Bodmer J, Bussey HJ, Ellis A, Gorman P, Lucibello FC, Murday VA, Rider SH, Scambler P, et al. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature. 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 18.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 19.Joslyn G, Carlson M, Thliveris A, Albertsen H, Gelbert L, Samowitz W, Groden J, Stevens J, Spirio L, Robertson M, et al. Identification of deletion mutations and three new genes at the familial polyposis locus. Cell. 1991;66:601–613. doi: 10.1016/0092-8674(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 20.Levy DB, Smith KJ, Beazer-Barclay Y, Hamilton SR, Vogelstein B, Kinzler KW. Inactivation of both APC alleles in human and mouse tumors. Cancer Res. 1994;54:5953–5958. [PubMed] [Google Scholar]
- 21.Jen J, Powell SM, Papadopoulos N, Smith KJ, Hamilton SR, Vogelstein B, Kinzler KW. Molecular determinants of dysplasia in colorectal lesions. Cancer Res. 1994;54:5523–5526. [PubMed] [Google Scholar]
- 22.Smith AJ, Stern HS, Penner M, Hay K, Mitri A, Bapat BV, Gallinger S. Somatic APC and K-ras codon 12 mutations in aberrant crypt foci from human colons. Cancer Res. 1994;54:5527–5530. [PubMed] [Google Scholar]
- 23**.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. This review centers on the question of how the deregulation of β-catenin leads to malignant development in the mammalian gut epithelium upon loss of APC.
- 24.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 25.Luongo C, Moser AR, Gledhill S, Dove WF. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994;54:5947–5952. [PubMed] [Google Scholar]
- 26.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 27.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 28.Garber JE, Goldstein AM, Kantor AF, Dreyfus MG, Fraumeni JF, Jr., Li FP. Follow-up study of twenty-four families with Li-Fraumeni syndrome. Cancer Res. 1991;51:6094–6097. [PubMed] [Google Scholar]
- 29.Pretlow TP, Brasitus TA, Fulton NC, Cheyer C, Kaplan EL. K-ras mutations in putative preneoplastic lesions in human colon. J Natl Cancer Inst. 1993;85:2004–2007. doi: 10.1093/jnci/85.24.2004. [DOI] [PubMed] [Google Scholar]
- 30*.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. The first report demonstrating that the loss of APC in differentiated cells is not sufficient to promote tumorigenesis, rather loss of APC in stem cells or transient amplifying cells promotes tumorigenesis.
- 31*.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. The cell of origin in colon adenomas is a stem cell.
- 32.Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 34.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 35.Yeh A, Wei M, Golub SB, Yamashiro DJ, Murty VV, Tycko B. Chromosome arm 16q in Wilms tumors: unbalanced chromosomal translocations, loss of heterozygosity, and assessment of the CTCF gene. Genes Chromosomes Cancer. 2002;35:156–163. doi: 10.1002/gcc.10110. [DOI] [PubMed] [Google Scholar]
- 36.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 37.Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19:95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- 38*.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. This study shows that genes abberrantly methylated in cancer are also methylated in normal aged colonic tissue.
- 39.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 40.Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, Van Tornout JM, Jones PA. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 42.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, Knuchel R, Klopocki E, Sauter G, Simon R, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26:5680–5691. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- 44.Steegenga WT, de Wit NJ, Boekschoten MV, Ijssennagger N, Lute C, Keshtkar S, Grootte Bromhaar MM, Kampman E, de Groot LC, Muller M. Structural, functional and molecular analysis of the effects of aging in the small intestine and colon of C57BL/6 J mice. BMC Med Genomics. 2012;5:38. doi: 10.1186/1755-8794-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Decottignies A, d’Adda di Fagagna F. Epigenetic alterations associated with cellular senescence: a barrier against tumorigenesis or a red carpet for cancer? Semin Cancer Biol. 2011;21:360–366. doi: 10.1016/j.semcancer.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 47.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 48*.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. The use of 5-aza-2′-deoxycytidine or the selective depletion of DNMT1 reactivates silenced tumor-suppressor genes. Suggesting that DNMT1 is necessary and sufficient to maintain global methylation and aberrant CpG island methylation in human cancer cells.
- 49*.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. The first report showing that changes in methylation in cancer are not restricted to gene promoters but are also observed at CpG island shores. Furthermore, these methylation changes were highly variable between tumor subtypes.
- 50*.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. This study shows that stochastic methylation variation at previously defined cancer DMRs distinguishes adenomas from normal tissue. Furthermore, this paper is the first to show that large blocks of the genome either loose or acquire methylation suggesting a loss of DNA methylation boundary function.
- 51.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 55.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 56.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 58.Boumber YA, Kondo Y, Chen X, Shen L, Guo Y, Tellez C, Estecio MR, Ahmed S, Issa JP. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 2008;4:e1000162. doi: 10.1371/journal.pgen.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 61.Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A. 1996;93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, Hwang KS, Kim HY, Lee YS, Kang GH, Jeoung DI. Promoter hypomethylation of a novel cancer/testis antigen gene CAGE is correlated with its aberrant expression and is seen in premalignant stage of gastric carcinoma. Biochem Biophys Res Commun. 2003;307:52–63. doi: 10.1016/s0006-291x(03)01121-5. [DOI] [PubMed] [Google Scholar]
- 64.Nakamura N, Takenaga K. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin Exp Metastasis. 1998;16:471–479. doi: 10.1023/a:1006589626307. [DOI] [PubMed] [Google Scholar]
- 65.Jette C, Peterson PW, Sandoval IT, Manos EJ, Hadley E, Ireland CM, Jones DA. The tumor suppressor adenomatous polyposis coli and caudal related homeodomain protein regulate expression of retinol dehydrogenase L. J Biol Chem. 2004;279:34397–34405. doi: 10.1074/jbc.M314021200. [DOI] [PubMed] [Google Scholar]
- 66.Nadauld LD, Phelps R, Moore BC, Eisinger A, Sandoval IT, Chidester S, Peterson PW, Manos EJ, Sklow B, Burt RW, et al. Adenomatous polyposis coli control of C-terminal binding protein-1 stability regulates expression of intestinal retinol dehydrogenases. J Biol Chem. 2006;281:37828–37835. doi: 10.1074/jbc.M602119200. [DOI] [PubMed] [Google Scholar]
- 67.Nadauld LD, Sandoval IT, Chidester S, Yost HJ, Jones DA. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J Biol Chem. 2004;279:51581–51589. doi: 10.1074/jbc.M408830200. [DOI] [PubMed] [Google Scholar]
- 68.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rai K, Sarkar S, Broadbent TJ, Voas M, Grossmann KF, Nadauld LD, Dehghanizadeh S, Hagos FT, Li Y, Toth RK, et al. DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell. 2010;142:930–942. doi: 10.1016/j.cell.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanc V, Henderson JO, Newberry RD, Xie Y, Cho SJ, Newberry EP, Kennedy S, Rubin DC, Wang HL, Luo J, et al. Deletion of the AU-rich RNA binding protein Apobec-1 reduces intestinal tumor burden in Apc(min) mice. Cancer Res. 2007;67:8565–8573. doi: 10.1158/0008-5472.CAN-07-1593. [DOI] [PubMed] [Google Scholar]
- 71.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 72.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 73.Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, Ljungman M, Merajver SD, Kleer CG. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu J, Cao Q, Mehra R, Laxman B, Tomlins SA, Creighton CJ, Dhanasekaran SM, Shen R, Chen G, Morris DS, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 75.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 76.Behrens J. The role of the Wnt signalling pathway in colorectal tumorigenesis. Biochem Soc Trans. 2005;33:672–675. doi: 10.1042/BST0330672. [DOI] [PubMed] [Google Scholar]
- 77.de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 78.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 79.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jass JR. Colorectal cancer: a multipathway disease. Crit Rev Oncog. 2006;12:273–287. doi: 10.1615/critrevoncog.v12.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 81.Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, et al. Wnt Signaling through Inhibition of beta-Catenin Degradation in an Intact Axin1 Complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 82.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 83.Anderson CB, Neufeld KL, White RL. Subcellular distribution of Wnt pathway proteins in normal and neoplastic colon. Proc Natl Acad Sci U S A. 2002;99:8683–8688. doi: 10.1073/pnas.122235399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blaker H, Scholten M, Sutter C, Otto HF, Penzel R. Somatic mutations in familial adenomatous polyps. Nuclear translocation of beta-catenin requires more than biallelic APC inactivation. Am J Clin Pathol. 2003;120:418–423. doi: 10.1309/4E4W-G3AY-GJNC-D11P. [DOI] [PubMed] [Google Scholar]
- 85.Phelps RA, Broadbent TJ, Stafforini DM, Jones DA. New perspectives on APC control of cell fate and proliferation in colorectal cancer. Cell Cycle. 2009;8:2549–2556. doi: 10.4161/cc.8.16.9278. [DOI] [PubMed] [Google Scholar]
- 86.Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, Broadbent T, Sarkar S, Burt RW, Jones DA. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, Weichert J, Haag JD, Chen KS, Waller JL, Gould MN, et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc Natl Acad Sci U S A. 2007;104:4036–4041. doi: 10.1073/pnas.0611690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Janssen KP, Alberici P, Fsihi H, Gaspar C, Breukel C, Franken P, Rosty C, Abal M, El Marjou F, Smits R, et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology. 2006;131:1096–1109. doi: 10.1053/j.gastro.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 89.Sansom OJ, Meniel V, Wilkins JA, Cole AM, Oien KA, Marsh V, Jamieson TJ, Guerra C, Ashton GH, Barbacid M, et al. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci U S A. 2006;103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]