Abstract
There are two homologous membrane-bound enzymes in Escherichia coli that catalyze reversible conversion between succinate/fumarate and quinone/quinol. Succinate;ubiquinone reductase (SQR) is a component of aerobic respiratory chains, whereas quinol:fumarate reductase (QFR) utilizes menaquinol to reduce fumarate in a final step of anaerobic respiration. Although, both protein complexes are capable of supporting bacterial growth on either minimal succinate or fumarate media, the enzymes are more proficient in their physiological directions. Here we evaluate factors that may underlie this catalytic bias.
Keywords: Complex II, succinate:quinone oxidoreductase, quinol:fumarate reductase, protein electron transport, iron sulfur clusters, quinone binding sites
1. Introduction
Proteins comprising the complex II family are found in the inner mitochondrial membrane in eukaryotes and the plasma membrane of many aerobic or facultative bacteria [1, 2]. They catalyze reversible 2e−/2H+ transfer between the water soluble dicarboxylic acids succinate/fumarate and lipophilic quinone/quinol. Based on their role in cellular energetics, complex II enzymes are commonly classified as succinate dehydrogenases (Succinate:Quinone Reductases, SQRs) and fumarate reductases (Quinol:Fumarate Reductases, QFRs). SQRs, as a component of the tricarboxylic acid cycle, are predominately members of the aerobic respiratory chain where they are responsible for oxidation of succinate to fumarate (Em =+30 mV) and reducing ubiquinone to quinol (UQ, Em= +100mV). QFRs are found in anaerobic and facultative organisms such as bacteria, parasitic helminthes, and lower marine organisms which utilize low potential quinols (menaquinol (MQH2, Em=−74mV) or rhodoquinol (RQH2, Em =−63 mV)) to reduce fumarate in a final step of anaerobic respiratory chains [3]. In vivo electron flow between succinate/fumarate and quinone/quinol usually proceeds in a thermodynamically preferable direction which may be set by the availability and reduction state of the electron donor/acceptor. Organisms that are exposed to changes in environmental oxygen concentrations during their life cycle often regulate the properties of their respiratory chains by modulating expression of high and low potential quinones and adjusting the composition of the respiratory protein complexes [4].
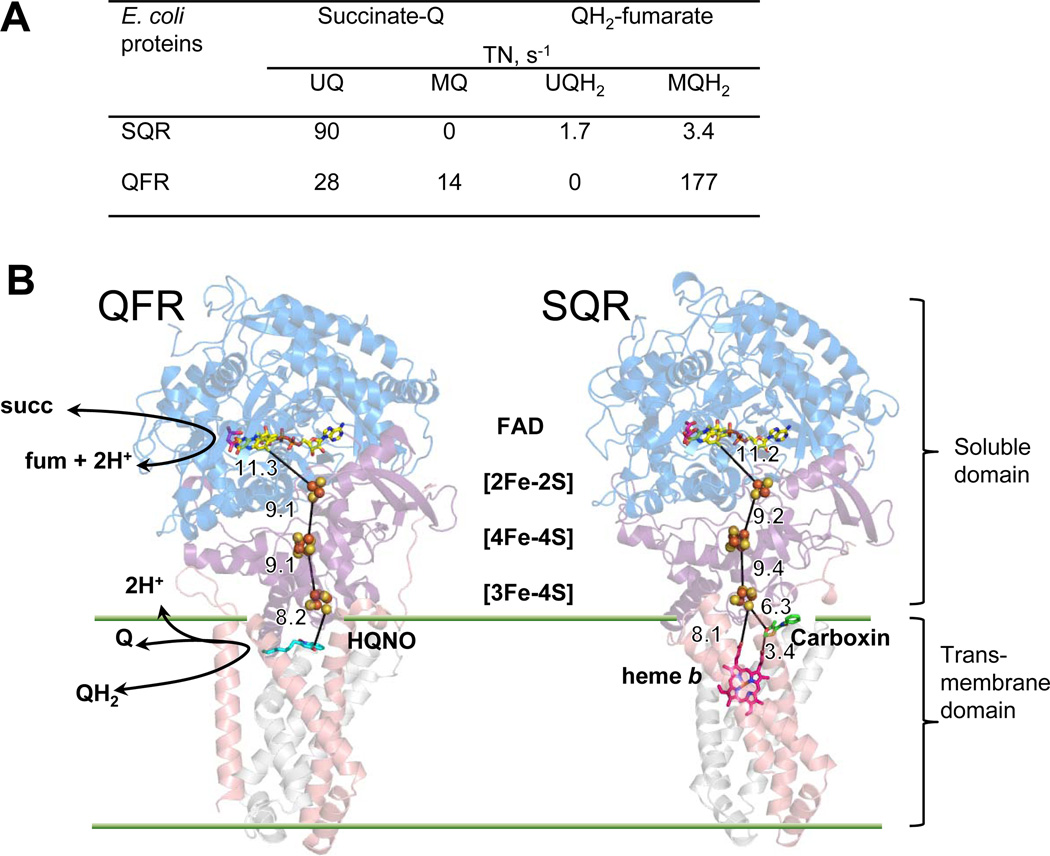
The genomes of many Gram-negative bacteria such as Escherichia coli contain two operons for complex II enzymes, one for SQR where expression is promoted by high environmental oxygen and the second for QFR which is predominantly expressed under microaerophilic and anaerobic conditions. Both enzymes share the ability to interact with UQ and MQH2 and can even replace each other in vivo if they can be expressed [5, 6]. Comparison of the rates of succinate-oxidation and fumarate-reduction reflects the physiological role SQR and QFR portray in cells. For E. coli QFR, the greater driving force for the MQH2 –fumarate reductase direction (ΔE=100 mV) correlates with a higher rate in this reaction compared to succinate-UQ reductase activity (ΔE=70 mV). However, the UQ-reductase activity of SQR is 20-fold higher than the MQH2-fumarate reductase activity (Fig. 1A). The accumulated body of data from early biophysical studies to recent progress in structural and mutagenic analysis of E. coli SQR and QFR provide insight into how these two enzymes are poised to be catalytically most efficient in their preferred physiological activity.
Figure 1. E. coli SQR and QFR.
(A) Catalytic activity (TN, turnover number) of SQR and QFR with UQ and MQ [43]. (B) The overall structures and spatial arrangement redox centers in E. coli QFR (pdb: 1KF6 [7], left) and SQR (pdb:2WDQ [39], right). The flavoprotein subunits (SdhA/FrdA) are shown in blue; the iron protein subunits (SdhB/FrdB) are in purple. The transmembrane subunits are in pink (SdhC/FrdC) and gray (SdhD/FrdD). The specific quinone site inhibitor 2-n-heptyl-4-hydroxyquinoline-N-oxide (HQNO) for QFR is shown in cyan and the specific quinone site inhibitor carboxin for SQR is shown in green. Oxaloacetate bound near the isoalloxazine ring of FAD is shown in magenta. The edge-to-edge distances (Å) of redox active centers are indicated. The partial reactions at the catalytic sites are diagrammed in the QFR structure.
2. Comparison of E. coli SQR and QFR
Since 1999 when the first x-ray structure of E. coli QFR was published [7], structures for five more complex II enzymes have become available; including QFR from Wolinella succinogenes [8] and Ascaris suum [9], as well as, SQRs from E. coli [10], pig [11], and chicken [12]. The overall structure of complex II enzymes is very similar (Fig. 1B) and is comprised of two domains which are independently stable. A soluble dehydrogenase fragment catalyzes reactions of succinate oxidation and fumarate reduction with appropriate artificial electron donors/acceptors. This domain consists of two subunits; a larger flavoprotein (Fp, SdhA or FrdA, ~65 kDa) containing a covalently bound FAD cofactor and an active site that binds the substrate for the reaction succinate or fumarate. A smaller iron-sulfur protein subunit (Ip, SdhB, FrdB, ~27 kDa), contains three distinct iron-sulfur clusters, [2Fe-2S]2+,1+; [4Fe-4S]2+,1+; and [3Fe-4S]1+,0, linearly arranged for electron transfer between the flavin and quinone catalytic sites. The Fp and Ip subunits share a high degree of sequence and structural homology within the complex II family [13]. In addition, Fp proteins are also structural homologues of single subunit soluble bacterial enzymes, a flavocytochrome fumarate reductase [14] and L-aspartate oxidase [15], which are able to reduce fumarate and to oxidize a dicarboxylate substrate. This is consistent with the structural similarity underlying the reversible mechanism for succinate/fumarate conversion [16] and also suggests that members of complex II family have evolved from a common evolutionary ancestor [1]. The soluble domain is attached to the membrane through a hydrophobic membrane anchor. The composition of the membrane-bound domain is the main basis for further subdivision of complex II enzymes into different types according to number of subunits (one or two), type of quinone preferentially utilized (ubiquinone, menaquinone, rhodoquinone) or number of heme b groups (none, one, or two) [17, 18]. In the enzymes that lack heme b (E. coli QFR), or have only a single heme, such as E. coli, avian, and mammalian SQR, the hydrophobic domain is comprised of two transmembrane subunits (SdhCD or FrdCD, ~12–15 kDa each). A single catalytically active quinone binding site is located at the cytoplasmic side of the membrane and proximal to the soluble domain. Therefore, the protons needed for the reactions at the dicarboxylate and quinone binding site are from the cytoplasm and neither succinate-quinone nor quinol-fumarate reductase reactions are coupled with a transmembrane proton gradient (Fig. 1B).
The simplified reversible catalytic turnover of the succinate-quinone reductase reaction may be described in several steps. First, flavin reduction occurs via the hydride transfer reaction from succinate to the N5 position on the isoalloxazine ring of the flavin [16]. FADH2 is recycled to FAD in a process that involves sequential transfer of single electrons to the adjacent [2Fe-2S] cluster through formation of the intermediate flavin radical. Each electron is further transferred to the bound quinone molecule via the array of the three iron-sulfur centers. Finally, two electron reduction of a quinone molecule proceeds via a stabilized semiquinone radical. It is often suggested that with such multi-centered redox proteins the formation or breaking of covalent hydrogen-bonds associated with the succinate/fumarate and quinone/quinol couples should be rate limiting. Therefore, the oxidoreductase reactions occurring at the dicarboxylate and quinone binding sites, as well as electron transfer via Fe-S clusters appear to be important in the control of the overall rate of catalysis of complex II enzymes.
3. Electron transfer pathways
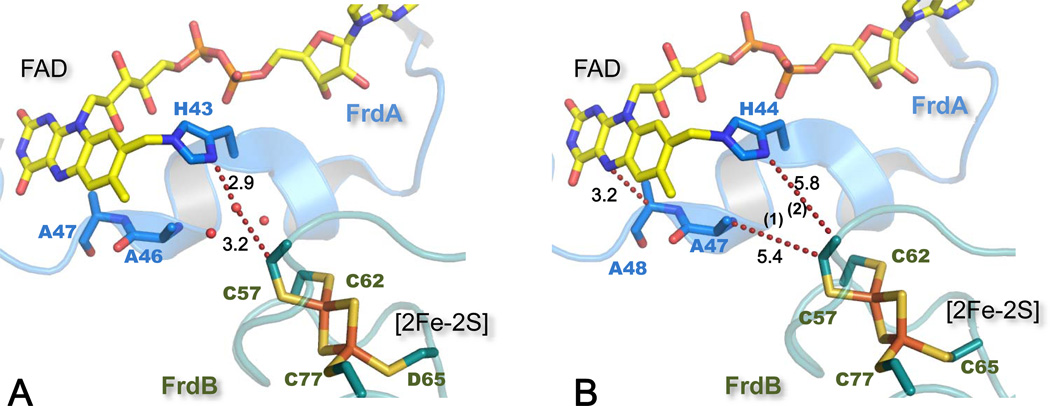
Within the soluble domain the longest edge-to-edge distance of ~11 Å is for intermolecular electron transfer (ET) between the FAD of the flavoprotein subunit and the [2Fe-2S] cluster of the iron-sulfur subunit (Fig. 1B). The 1.78 Å resolution structure of W. succinogenes QFR (Fig. 2A) showed the presence of several water molecules at the interface between the FrdA and FrdB subunits with one water hydrogen atom bonded to the Nδ of the FrdA His43 residue that covalently links the FAD moiety [8] [19]. This water was suggested to provide a bridge for through-space jumps between Nδ of His43 and a Cys residue, one of the ligands for the [2Fe-2S] cluster [8]. Most complex II structures show the presence of these conserved water molecules at the thin aqueous interface between the subunits, the lone exceptions being the relatively low resolution E. coli QFR structures. For the water depleted E. coli QFR structure a different ET pathway was calculated using GREENPATH software analysis [20]. In this case a through-space jump from the ligating Cys 57 to FrdA Ala47 is calculated followed by travel along the backbone to Ala 48 and then a jump to the flavin N5 atom (Fig. 2B). Residues Ala47 and Ala48 in FrdA subunit are not conserved among complex II enzymes and when calculations were done for an FrdA A47G mutation a path similar to that for W. succinogenes QFR was suggested, i.e., a jump from Cys57 to FrdA His44 Nδ. These calculations indicate that water can influence ET reactions rates by mediating the ET coupling pathway between FAD and the [2Fe-2S] cluster. Recent experimental observations and theoretical analyses confirm that a small number of structured waters between donor and acceptor cofactors may be essential for efficient electron tunneling [21]. Therefore, one may predict that improved resolution of E. coli QFR structure should reveal a similar interdomain aqueous layer.
Figure 2. Electron transfer pathways between FAD and [2Fe-2S] cluster.
(A) The structure of W. succinogenes QFR (pdb: 2BS2 [19]) demonstrates a water molecule hydrogen-bonded to His A45 and within van der Waals distance to Cys B55 [8]. This water mediates ET coupling pathway between FAD and the [2Fe-2S] cluster. (B) GREENPATH calculations outline the ET path in E. coli QFR (pdb:1KF6 [7]) according to [20]. Path 1 is calculated for the wild type structure and path 2 when Ala A47 was in silico mutated to Gly. SdhA/FrdA subunits and their residues are in blue and SdhB/FrdB in teal; the distances between the atoms (Å) are indicated next to the dashed lines.
The iron-sulfur subunit folds into two well defined domains. The [2Fe-2S] cluster is coordinated by the N-terminal domain and the [4Fe-4S] and [3Fe-4S] centers are harbored within the C-terminal domain (Fig. 3). The x-ray structures confirmed earlier work in which a systematic substitution of Cys to Ser residues in E. coli FrdB revealed that the [2Fe-2S] cluster is part of a more stable and separate protein fold [22–24]. Although all-cysteinyl coordination of the [2Fe-2S] cluster is most common for complex II enzymes, in E. coli SQR one of the Cys ligands is replaced by Asp A63 [10]. The influence of an Asp ligand on complex II [2Fe-2S] centers was examined using E. coli QFR in which the equivalent Frd B Cys 65 residue was substituted by Asp. It had no effect on the midpoint potential of the cluster and has little influence on the catalysis [25]. Interestingly, sequence analysis confirms that an Asp in this position is reasonably well conserved in bacterial complex II enzymes, e.g.,Micrococcus luteus SQR.
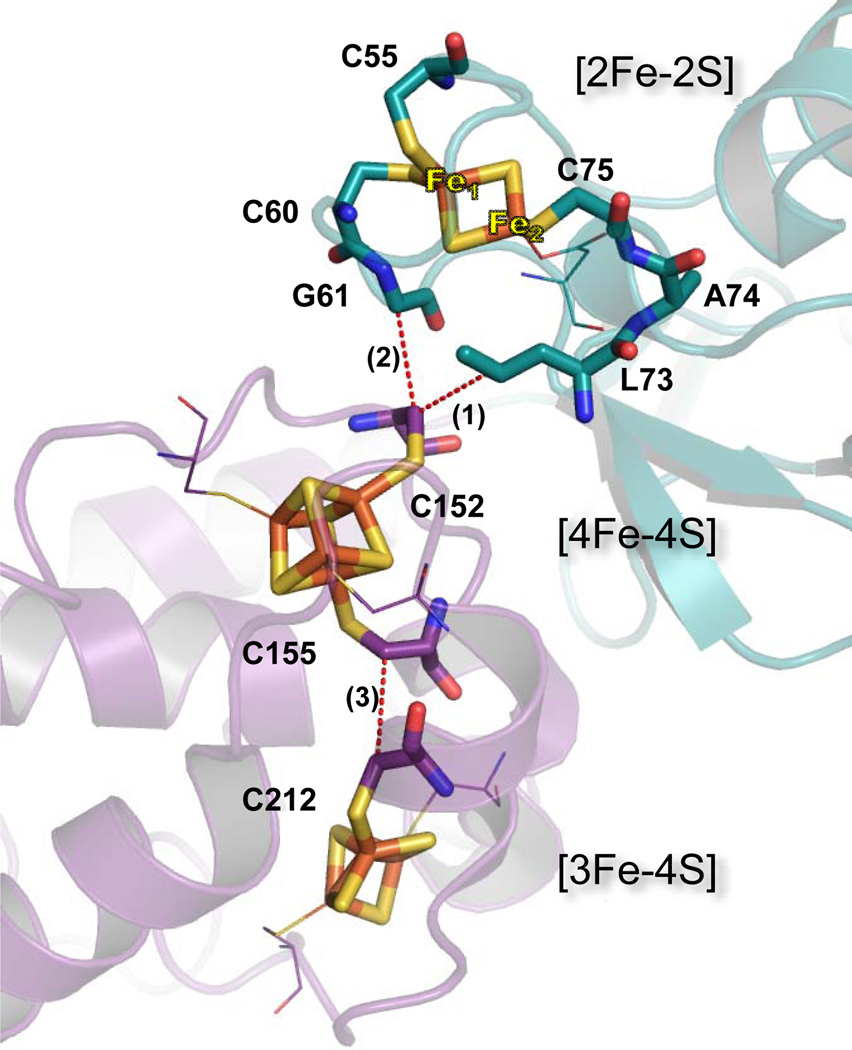
Figure 3.
Electron transfer pathways between iron-sulfur clusters in SdhB of E. coli SQR. The N-terminal [2Fe-2S] domain is shown in teal and C-terminal in purple. Dashed lines show through-space jumps. Path 1 (C75, A74, L73, and 3.8 Å jump to C152) and Path 2 (C60, G61, and 3.8 Å jump to C152) are for [2Fe-2S] and [4Fe-4S] centers. Path 3 (C155 and 3.8 Å jump to C152) is for [4Fe-4S] and [3Fe-4S]. Residues involved in ET are shown as sticks, while other ligating residues are shown as thin lines.
The interdomain ET between the [2Fe-2S] and [4Fe-4S] centers comes through surface contacts where water is not present in all high resolution complex II structures and the residues separating these iron-sulfur centers are highly conserved throughout the complex II family (Fig. 3). A possible ET pathway in W. succinogenes QFR was suggested to involve a mediating Leu B75 residue [8]. In E. coli SQR SdhB, Leu73 is in van der Waals contact with Cys B152, a ligand of the [4Fe-4S] cluster, and is connected by the polypeptide backbone to Cys 77, a ligand to the [2Fe-2S] center (Fig. 3, path 1). An alternative ET path is predicted between the [2Fe-2S] and [4Fe-4S] clusters by utilizing HARLEM (www.harlem.chem.cmu.edu) calculations to the E. coli SQR structure. This path includes Cys-B60, the neighboring Gly-B61 and a jump to Cys-B152 (Fig. 3, path 2). Werth et al., replaced individual Cys residues ligating the [2Fe-2S] cluster in E. coli QFR with Ser [24] and showed that FrdB Cys55 and FrdB Cys60 (in E. coli SQR numbering on Fig.3) affected the EPR characteristics of [2Fe-2S] cluster and drastically decreased its midpoint potential by more than −100 mV. This observation lead to the suggestion that these two Cys residues are the ligands to the valence-localized Fe(II) of the [2Fe-2S] cluster [20]. Also, the ET pathway analysis is consistent with the redox role of the Fe1 atom of the [2Fe-2S] cluster in the SQR structure by providing electron tunneling via Cys57 to FAD (Fig. 2) and Cys60 to the [4Fe-4S] cluster (Fig. 3). The arrangement of the [4Fe-4S] and [3Fe-4S] clusters shows orientation of ligating Cys B155 and Cys-B212 residues towards each other, a common structural arrangement for efficient ET found in respiratory enzymes (Fig. 3, path 3).
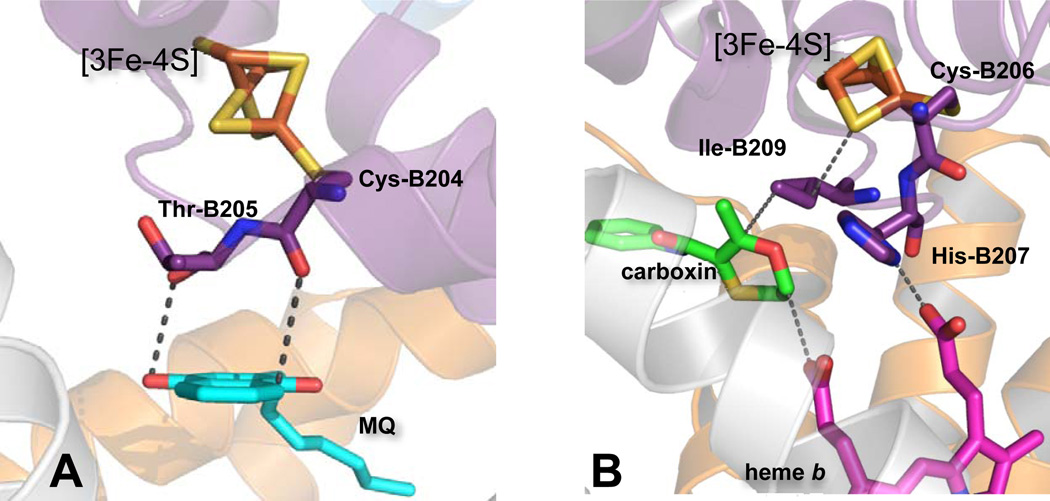
The [3Fe-4S] cluster is covered by a short amino acid loop, which is reminiscent of a classic ferredoxin fold, and points towards the central core of the hydrophobic domain; a four-helix bundle oriented perpendicular to the plane of the membrane. In E. coli QFR, a complex II (Fig.1B) not containing heme, the central position of the cytosolic side of the hydrophobic domain is occupied by the quinone-binding site formed by residues from the FrdB, C and D subunits (Fig. 4A). This catalytically active Q-binding site accommodates both benzo- and naphthoquinones which undergo two electron oxidation/reduction via single electron exchanges with the [3Fe-4S] cluster. The aromatic rings of these quinones and their analogues lie within the same plane and 3 to 4 Å from Thr-B205, a neighboring residue of the ligating Cys-204 of the [3Fe-4S] cluster. The peptide backbone of these residues comprises the main ET pathway. Several x-ray structures of the E. coli QFR protein show the presence of a second bound quinone molecule at the periplasmic side of the membrane, termed the distal Q-binding site. However, because of the large distance (~27 Å) between the proximal and distal quinone-binding sites and the absence of a redox candidate to mediate electron transfer between them, it would appear unlikely that the distal (Qd)-site is catalytically active [26].
Figure 4. Spatial arrangements of redox groups in the hydrophobic domain and [3Fe-4S] cluster in E. coli complex II enzymes.
(A) In QFR Cys B204 and Thr B205 mediate electron transfer between MQ and [3Fe-4S]. (B) His B207 in SQR provides ET coupling between [3Fe-4S] and heme b. Ile B209 is a residue mediating ET between UQ and [3Fe-4S]. Carboxin occupies a position deep in the UQ binding site and within van der Waals distance to the heme propionate. This would be similar to the position of UQ during catalysis and within effective ET distance to heme b.
Mammalian and E. coli SQR enzymes harbor a single low-spin heme b centrally located near the cytoplasmic side of the transmembrane domain, similar to the position of the functional Q-binding site in E. coli QFR (Fig. 1B). As a result, the quinone binding site in SQR is shifted toward the outer rim of the hydrophobic domain but remains within close proximity to the [3Fe-4S] cluster and heme b (Fig. 1B). The distances between all three redox centers are suitable for efficient electron tunneling [27]. The presence of the heme b and its catalytic role in complex II has been considered enigmatic mostly because of the well-studied bovine complex II with a low potential heme (Em = −185 mV) that is not reducible by succinate [28]. In contrast, the heme b of E. coli SQR is some 200 mV more positive (Em = +36mV) than its bovine counterpart and is reducible by succinate [29]. The presence of a quinone at the functioning quinone binding site is essential for fast heme b reduction with succinate [30], thus, indicating that heme b may not be an obligatory redox center to mediate ET between the [3Fe-4S] cluster and ubiquinone. Indeed, mutagenesis studies have shown that when either of the His residues coordinating the heme b is mutated to Tyr, E. coli SQR still assembles and retains both quinone-reductase and quinol-oxidase activity despite the total absence of the heme [31]. Although not essential for catalysis, the heme is important for stabilization of the transmembrane domain and the four-subunit complex [31]. SdhB His207, a residue homologous to the FrdB Thr205 position, is predicted by the HARLEM program to be involved in the ET relay from the [3Fe-4S] center to the heme, while the side chain of conserved Ile-B209 would mediate electron tunneling between the [3Fe-4S] and UQ (Fig. 4B).
The possible existence of a second quinone binding site in mammalian and similar bacterial SQRs requires a more detailed discussion. The EPR detectable ubisemiquinone radical in complex II enzymes was first observed in membrane-bound bovine protein [32] and its presence was later confirmed in the isolated enzyme [33]. The characteristics of the signal indicated that it might originate from the interaction of ubisemiquinone with a second paramagnetic species. At that time the spatial organization of complex II was not established. Among possible candidates for the partner interacting with UQ were a FAD radical, another UQ radical, and a Fe-S cluster which we now know would have been the [3Fe-4S] center. Although, the consistently observed association of the signal with the [3Fe-4S] cluster suggested its involvement in the interaction, computer simulation was in favor of UQ•− : semiquinone interaction over UQ•− : [3Fe-4S] interaction with a distance of ≤7.7 Å between the partners [32, 34, 35]. The hypotheses that a thermodynamically stable ubiquinone pair may function as the two-electron gating system between iron sulfur clusters of the soluble SdhAB and quinone pool, similar to the QA and QB pair of the bacterial photosynthetic center, became a popular mechanism for complex II function [36]. The remarkable structural similarities and conservation of the critical residues around the quinone binding site in SQRs enzymes suggests a similar molecular mechanism of catalysis. In E. coli and pig/avian structures the distance between the [3Fe-4S] center and ligands bound at the Q- binding site is less than 8 Å [11, 37]. The precise position of the UQ molecule at the Q-binding site varies in different structures. This could reflect the movement of the UQ molecule from the primary binding position at the opening of the cavity into a deeper position which is the actual catalytic site. Importantly, the Q-binding site is not large enough to accommodate a second ubiquinone molecule [10, 26, 38, 39]. Similarly to E. coli QFR, the SQR structures present a distal cavity at the periplasmic side of the hydrophobic domain and ~25 Å away from the Q-binding site. This cavity is fairly hydrophobic and several lipophilic molecules were found in it; cardiolipin in the E. coli enzyme and phosphatidyl-ethanolamine in avian and porcine SQRs. Also, the first structure of porcine SQR with theonyltrifluoroacetone (TTFA), a Q-site inhibitor, showed the presence of a second inhibitor molecule near the distal cavity, thus termed the distal Q-site. However, 20 different structures of Q-site inhibitors within the porcine structure are now available [40, 41] and all inhibitors are present only at the catalytic Q-site proximal to the [3Fe-4S]. Thus, existence of the second TTFA molecule in the initial structure most likely is the result of high concentrations of the inhibitor used during the crystallization. If electron transfer between the two quinone-binding sites occurs then the heme b should be an essential component for mediating the electron transfer pathway as it is for di-heme QFR from W. succinogenes [42] and SQR from Bacillus subtilis [18]. It is worth repeating that in E. coli SQR assembled without the heme catalysis in both directions was not significantly affected and, in addition, the heme-less enzymes retained the EPR detectable UQ•− radical with the same properties as in the wild type enzyme [31]. Therefore, in light of the recent structural and mutagenesis studies the paramagnetic partner that interacts with UQ•− radical observed in the bovine enzyme is likely to be the [3Fe-4S] center.
4. Kinetic properties of complex II enzymes
Kinetic analysis of complex II enzymes is usually limited to conventional steady-state assays using visible spectrophotometry. The physiologically relevant reactions are probed with different quinone analogs while partial reactions of succinate/fumarate oxidoreduction are performed with a number of artificial electron donors/acceptors [43]. The rates of succinate oxidation by SQR and QFR are similar when measured with UQ or an artificial electron acceptor. Measured rates are about 100 s−1 and 30 s−1 for E .coli SQR and QFR, respectively (Fig. 1A). The rates of the fumarate reductase reaction of E. coli QFR, as well as W. succinogenes QFR and B. subtilis SQR (enzymes whose natural electron donor is MQH2) are dictated by the reduction potential of the electron donor. Maximal rates for fumarate reduction are measured using low potential donors such as methyl viologen (Em =−395 mV) and benzyl viologen (Em =−359 mV). Using these donors E. coli QFR shows rates of fumarate reduction of ~400 to 800 s−1 [44]. This fast rate of fumarate reduction, however, is not found with enzymes classified as SQRs which utilize UQ as the natural electron acceptor. In SQRs there is a specific inhibitory effect of the increased thermodynamic driving force (i.e., low potential driving force greater than ~−70 mV) on the fumarate-reductase activity [45]. This will be discussed further in section 8 of this review.
In addition to the spectrophotometric methods discussed above, protein film voltammetry (PFV) has been used to measure ET rates within the soluble two subunit (Fp-Ip) dehydrogenase domain of complex II [44, 46, 47]. During PFV experiments the soluble enzyme is adsorbed on a pyrolytic graphite ‘edge’ electrode which replaces the quinone as the electron donor/acceptor. The assumption is that the electrons enter the enzyme via the relatively exposed [3Fe-4S] center. This method allows for the determination of the reduction potential of the redox centers (FAD and Fe-S clusters) including reactions with apparent catalytic ET rates up to 1500 s−1 for FAD reduction in the presence of fumarate [47]. Unden et al. [48] used succinate/fumarate and low potential naphthoquinol to determine rate constants of reduction/oxidation of the W. succinogenes QFR components using stopped-flow spectrophotometry for solution kinetics for heme(s) b and freeze-quench EPR for the Fe-S centers. The rate constants for the individual redox centers were found to be very close to that for the corresponding catalytic turnover (13 s−1 for succinate reduction and 133 s−1 for reduction with 2,3-dimethyl-1,4-naphthoquinol). Thus, indicating that the rate limiting steps are at the level of electron entry into the system, in other words, at the active sites.
One experimental obstacle in kinetic studies is the presence of inhibitory tightly bound oxaloacetate at the active site of many well studied complex II enzymes. Oxaloacetate dissociates very slowly from the complex with oxidized SQR/QFR enzymes and prevents interactions with succinate causing the major complication for kinetic experiments [49]. In order to remove oxaloacetate, complex II preparations have to be incubated with substrates or their analogs that compete for the enzyme’s substrate binding site. However, not all of the substrate analogs can be used: malate, for instance, can be desaturated into oxaloacetate at the active site of complex II enzymes [50]. Prolonged incubation with fumarate also results in apparent conversion of fumarate to malate-like intermediates, tautomeric forms of oxaloacetate involving the C2 and C3 atoms of four-carbon substrates [12, 14]. Malonate, a three-carbon dicarboxylic acid, was shown to resist such modifications and is found to be the best ligand for activation of complex II enzymes [12].
Recently, pulse radiolysis was used to introduce a single electron from a strongly reducing N-methylnicotinamide radical which reacts with proteins with second-order rate constants in excess of 108 M−1s−1, making it possible to follow characterize subsequent slower intermolecular ET [27]. It was suggested that [3Fe-4S] cluster or bound quinone were initially reduced followed by equilibrium with heme b, where reduction with rate constant of 7.2 × 10−4 s−1 (ubisemiquinone to the heme) and 1.2 × 10−4 s−1 ([3Fe-4S] to the heme) was monitored spectrophometrically These data showed that at least for E. coli SQR the high potential heme b is in electronic equilibrium with the quinone of complex II. Unfortunately, because of the limitations of the optical detection method, pulse radiolysis cannot be applied for determination of the rates between the Fe-S clusters. Nevertheless, the conventional steady-state kinetic methods are able to assess how changes in thermodynamic properties of individual Fe-S clusters affect the physiological reactions in complex II enzymes.
5. EPR application for complex II studies
Continuous wave EPR has been the basic technique used in the studies of the iron-sulfur clusters and radical species in complex II enzymes for detection, quantization, characterization of the redox properties, and interspin interactions [2, 51–53]. It still remains the only method that allows for the detection of the FAD and Q semiquinone species and is the most reliable approach for the determination of the reduction potential of the Fe-S clusters, especially in the membrane bound complex II enzymes. An important requirement for the analysis and assignment of specific experimental observations in biological samples is the ability to study the contributions individually arising from particular species. In proteins with multiple redox centers, such as complex II, the EPR spectral lines are often overlapped and broadened, thus, becoming more difficult to analyze. In many cases separation of the individual spectra from overlapping resonance lines may be achieved by biochemical methods and pulsed EPR methods exploring the temperature dependence of relaxation times.
Pulsed EPR techniques (such as Electron Spin Echo Envelope Modulation (ESEEM) and Electron-Nuclear Double Resonance (ENDOR)) that are able to probe interactions between the electron spin of a paramagnetic species and magnetic nuclei (such as 1H and 14N) of the protein can provide information about the local spatial and electronic structure relevant to its function, independent of a complete crystallographic structure [54–58]. The isotropic and anisotropic hyperfine, and nuclear quadrupole interactions with magnetic nuclei depend on the structure local to the paramagnetic center (iron-sulfur clusters, radical species), and its electronic state. ESEEM spectroscopy was used to study the reduced [2Fe-2S] and oxidized [3Fe-4S] clusters [53, 59–64]. The range of 14N nuclear frequencies observed in the three-pulse ESEEM spectra was similar between [2Fe-2S] and [3Fe-4S] giving the same quadrupole coupling constant e2qQ/h≈3.2–3.4 MHz indicative of peptide 14N(s). Hyperfine coupling with these nitrogens are ~ 1.0–1.2 MHz for [2Fe-2S] and two times smaller, 0.5–0.9 MHz for the [3Fe-4S] cluster [63]. The experimentally determined hyperfine and quadrupolar coupling constants are consistent with a peptide 14N hydrogen bonded via a Cys ligand or bridging sulfur atom rather than directly coordinated to the iron. Particularly, this data definitively excludes an imidazole ring as a ligand of the iron as in the case of Rieske proteins [53].
Thus, ESEEM experiments identified a presence of at least one hydrogen bond with peptide nitrogen around the reduced [2Fe-2S] and oxidized [3Fe-4S] cluster, but without any confirmed assignment to a particular residue in the protein structure. This has also precluded the detailed analysis of the path involved in transfer of the spin density creating the observed coupling. Even an attempt to identify H-bond donors around the [2Fe-2S] cluster using comprehensive 2D ESEEM spectroscopy was not able to unambiguously assign hydrogen bonded nitrogens because of the complex character of the orientation-selected spectra and dependence of the simulated spectra from a large number of parameters [65]. A more promising approach applied recently to iron-sulfur clusters [66, 67] and semiquinones [68, 69] is based on selective 15N isotope labeling of residues of interest using a set of auxotrophs in a commonly used E. coli expression strain C43(DE3), a derivative of E. coli BL21(DE3) [66, 67]. The nitrogen H-bond donors carry unpaired spin density producing an isotropic hyperfine coupling and were identified unambiguously using 15N selective labeling approach in conjunction with 2D ESEEM. This methodology could be a valuable tool for studying the immediate protein environment, especially the hydrogen bonding interaction with altered protein environments around Fe-S centers and quinone binding sites in complex II proteins.
6. Effect of the redox properties of the Fe-S clusters on the catalytic bias activity
It is generally recognized that potentials of the iron sulfur clusters in complex II enzymes corresponds to the physiological direction of the reaction and the partnering quinone. Table 1 combines thermodynamic data available for different complex II enzymes. The Em of flavin reversibly interacting with succinate/fumarate is similar for SQRs and QFRs. The midpoint potential of soluble FAD (Em =−219 mV) is significantly increased to about −80 mV in complex II enzymes as a result of a covalent attachment of FAD to SdhA His45/FrdA His44 (E. coli enzymes) via an 8α-[N(3) histidyl] linkage [70]. This allows FAD reduction with succinate and reoxidation by fumarate. The [2Fe-2S] center, the immediate electron partner to FAD, is more positive in all functional SQRs (regardless of their quinone partners) than in QFRs. While the [4Fe-4S] cluster was recognized as a part of SQR, it was not initially considered to be involved in the electron transfer because of its very low redox potential. Nevertheless, its location midway in the Fe-S electron transfer relay found in the complex II structures implies a direct participation in ET. Also, it is now recognized that electron tunneling will be efficiently accomplished through a low potential center in the preferred direction [71]. Interestingly, the Em values of the [3Fe-4S] center in complex II enzymes are commensurate with the preferable quinone substrate. The cluster exhibits higher potentials in enzymes whose natural quinone partner is UQ rather than MQ (Table 1). The correlation of generally higher Em potentials for SQRs than for QFRs has often been interpreted as an evolutionary adaptation that underlines the kinetic advantages for succinate:ubiquinone reductase over menaquinol:fumarate reductase direction. Yet the relationship between electron transfer and the reduction potentials of individual Fe-S clusters have been rarely established in biological systems. E. coli SQR and QFR, because of their sequence and structural homology and ease in genetic manipulation have become useful models for testing how redox properties of individual Fe-S centers influence the overall catalytic turnover in complex II enzymes.
Table 1.
Reduction potentials of the redox centers in complex II enzymes.
| Species | FAD | [2Fe-2S] | [4Fe-4S] | [3Fe-4S] | b heme | Reference |
|---|---|---|---|---|---|---|
| Em (mV) | ||||||
| SQR-UQ | ||||||
| E. coli | −80 | −15 to +10 | −213 to −175 | +55 to +70 | +15 to+30 | [72, 80, 88] |
| Bos taurus | −79 | 0 | −260 | +60 | −185 | [93–95] |
| Rhodobacter sphaeroides | +50 | −250 | +80 | [96] | ||
| SQR-MQ | ||||||
| Micrococcus luteus | +70 | −295 | +10 | [97] | ||
| Bacillus subtilis | +80 | −240 | −25 | bh +65 bl −95 |
[98, 99] | |
| QFR | ||||||
| E. coli | −50 to−90 | −79 | −300 to−333 | −70 | none | [23, 44, 88] |
| Wolinella succinogenes | −59 | −250 | −24 | bh +20 bl −200 |
[48] | |
Amino acid substitutions in E. coli QFR have been made both to verify ligands of the cluster [24, 25], including those involved in the [3Fe-4S] to [4Fe-4S] conversion [22], and to change residues surrounding cluster with the intent to affect its redox properties. Known thermodynamic properties of the redox cofactors in complex II enzymes are given in Table 1. As noted, the Em value of the [4Fe-4S] cluster in E. coli QFR is about 145 mV lower than in E. coli SQR (Table 1). To better understand the role this low potential cluster plays in catalytic activity different residues have been targeted for mutagenesis in both QFR [44] and SQR [72] but with mixed results. For example in the FrdB subunit Leu153 and Tyr155 were substituted with Cys and Ser residues respectively, that occupy homologous positions in SQR [44]. The structure of E. coli SQR shows that SdhB C154 and SdhB S156 each provide a H-bond to different ligating cysteinyls. Presence of hydrogen bonds from side chain residues to the bridging sulfur atoms of Fe-S cluster or coordinated cysteine residues stabilizes the reduced form and, thus increases the midpoint potential of a cluster. Studies on the Rieske [2Fe-2S] cluster, a redox center of bc1 complex, have shown that changes in the observed redox potentials of the cluster are a direct result of individual H-bonds where each cause about a 45 to 140 mV positive shift in the Em of the cluster [73–77]. A positive shift of +54 mV in a single mutant FrdB L153C correlates well with the Rieske protein data. However, an FrdB Y155S variant produced a negative shift of similar amplitude (−46mV). Neither of these mutant enzymes was crystallized so the structural impact of the mutations may only be projected; from small, which likely put the side chain of L153C in appropriate position for donation of a hydrogen bond to the cluster, to more significant alterations in case of Y155S. The role of [4Fe-4S] cluster as a factor governing the efficacy of long-range electron transfer was examined in protein film voltammetry (PFV) using the soluble FrdAB catalytic fragment of the mutant enzymes [44]. No significant differences for the rates of catalysis were observed upon 100 mV change in reduction potential of the [4Fe-4S] cluster, supporting the suggestion that as long as distances between the centers and driving force remains the same, the rates of ET should not be affected [78]. One interesting observation related to [4Fe-4S] properties was noticed in that study. Since PFV could extend the range of potentials beyond those of natural substrates, applied potentials that maintain the [4Fe-4S] cluster in the reduced state boost catalysis by up to 50%. This was explained as a possible thermodynamic advantage to boost FAD reduction due to increased electron supply at both the [2Fe-2S] and [4Fe-4S] cluster [44].
In the SQR study, non-polar residues SdhB Ile150 and Leu220 were chosen for mutagenesis based on their relative proximity to the [4Fe-4S] cluster [72]. The L220S substitution caused a pronounced decrease in enzymatic activities in both directions and a shift by −70 mV of the Em of the [3Fe-4S] center without affecting the reduction potential of the [4Fe-4S]. The Ile150 residue was substituted by Asp and His residues. Introduction of a positive charge from the His side chain next to Fe-S clusters had also been a successful approach to raise the potential of clusters by 100–200 mV in bacterial ferredoxin [79]. Surprisingly, both mutations of SdhB Ile150 decreased the Em values of the [4Fe-4S] cluster by as much as −120 mV with similarly reduced activity (by more than 50%) for both succinate oxidation and fumarate reduction. It was concluded that the Em values of the [Fe-S] clusters in SQR indicate a preference for the forward reaction of succinate oxidation over the fumarate reduction reaction. However, since structural changes caused by a protein alteration were not monitored it appears that this may be a critical factor in interpretation of the consequences of mutations.
In E. coli SQR/QFR the [3Fe-4S] centers show ~140 mV difference in redox potentials. In SQR the Em= +70 mV is near the potential of its electron acceptor UQ, and in QFR (Em=−70 mV) it is almost isopotential with its donor MQH2 (Table 1). This suggested that tuning of the reduction potentials of the immediate electron donor/acceptor with quinone may have been evolutionary selected. Recently a study was undertaken to evaluate (a) the role of His or Thr residues occupying the equivalent position in E. coli SQR and QFR [80]. There is a short loop of five residues between two of the Cys ligands to the [3Fe-4S] cluster. This loop covers the cluster and provides a similar environment for H-bonding contact patterns from the backbone nitrogen to the sulfur atoms of the cluster for both enzymes. The side chain of a residue that is adjacent to the liganding Cys at the start of this loop is involved in interactions with quinones in both SQR and QFR (Fig. 4.). The substitution of SdhB His207 with Thr (the residue that occupies the same position in FrdB) caused a −70 mV negative shift of the reduction potential of the [3Fe-4S] cluster [80]. The converse substitution FrdB T205H raised the Em value of the cluster by +82 mV. In spite of the modest changes in Em values of the cluster, the catalytic rates remained basically unaffected. The x-ray structure of the SdhBH207T variant showed only minor changes caused by the substitution with no apparent effect on the redox properties of heme b and no effect of UQ-reductase and quinol oxidase activities. A more pronounced inhibition of catalysis in FrdB T205H is most likely associated with the introduction of the bulkier side chain into the Q-binding cavity (Fig. 4A) rather than with the change in the reduction potential of the [3Fe-4S] cluster caused by mutation.
It is striking that those two studies that report negative, approximately −70 mV, shifts in Em values of the [3Fe-4S] cluster in E. coli SQR conclude different functional outcomes. While SdhB H207T mutation causes 10% and 30% decrease the activity in succinate:UQ and MQH2:fumarate reductase reaction, respectively [80], in the SdhB L220S substitution the succinate oxidize activity is reduced by 80% and fumarate reductase by 90% [72]. Protein alteration may cause a variety of short and long distant perturbations in the protein scaffold and it is crucial to enhance (whenever possible) mutagenesis studies with structural evaluation. Work by Ruprecht et al. [80] represents the most comprehensive study thus far correlating changes in the reduction potential of a Fe-S cluster with catalysis in complex II enzymes.
It is also appears that conservative substitutions that retain a spatial occupation of the side chain cause minor changes in protein structure. Further, it has been long recognized that hydrogen bonding to both ligating and bridging sulfur atoms increases Em values of Fe-S clusters. It is well illustrated by the studies on Rieske proteins where observed decreases in midpoint potentials of the [2Fe-2S] clusters resulted from the mutation of Ser and Tyr residues providing hydrogen bonds to the sulfur atoms of the cluster [75]. The crystallographic studies confirmed that Ser to Ala and Tyr to Phe substitutions have no impact on the protein scaffold and changes in redox potential of the cluster were a direct consequence of the removal of individual hydrogen bonds. In the cytochrome bc1 complex the Reiske [2Fe-2S] cluster is thermodynamically poised to set the quinol oxidation rate. The Rieske high potential [2Fe-2S] cluster is an essential component of the respiratory bc1 and photosynthetic b6f complexes, in which it is the primary electron acceptor and drives the reaction by oxidizing ubiquinol and transferring one electron to cytochrome c1, while the second electron is used to reduce the low potential b heme, thus, making the reaction practically irreversible [73, 76]. Mutagenesis studies on several Rieske proteins are consistent and demonstrate that the decrease in the reduction potential of [2Fe-2S] cluster also reduces catalytic turnover [73–76]. On the average, a −60 mV shift in Em values of the [2Fe-2S] cluster caused about a 40% decrease in activity while in proteins with changes that exceeded −100 mV differences less than 10% of activity is retained.
In contrast to the Rieske protein, in complex II enzymes the [3Fe-4S] cluster is in electron equilibrium with quinone/quinol and redox reactions of quinones are readily reversible. There is no clear indication that changes in the Em values of individual Fe-S centers could differently affect succinate-UQ and MQH2 reductase activities. However, we should emphasize that the standard assays used for complex II activity are performed using conditions that provide a significant electron driving force for each direction due to the high reduction and oxidation states for the exogenously added electron donors and acceptors, respectively. These assays may mask the physiologically relevant change in activity when reduction states of the substrates reflect different metabolic conditions. Thus, modification of kinetic methods may be very informative for understanding the relationship between the reduction potentials of the Fe-S centers and catalysis.
7. Role of the quinone-binding site in reactions with UQ and MQ in E. coli SQR
Recent progress in understanding of the mechanism of redox reactions of E. coli complex II enzymes with ubi- and menaquinones at a single Q-binding site has been reviewed [26]. The structural and kinetic data all indicate that the methoxy substitutents of the benzoquinone ring play an important role in catalysis in general and could be a major factor that discriminates mechanisms of complex II reactions with UQ versus MQ. The methoxy groups not only have an impact on the redox potential of the quinone and on catalytic activity with proteins [81], but also appear to be important for movement into the catalytic position within the active site and also stabilization of the semiquinone intermediates during redox reactions [82] Opposite to most fumarate reductases, where rates of quinol-fumarate reductase reactions are set by the thermodynamic properties of the UQ and MQ derivatives [83, 84], E. coli SQR shows low rates of the MQH2-reductase reaction which is only twice higher than with UQH2 (Fig. 5C). Interestingly, the fumarate reductase reaction of E. coli SQR with plumbagin (PB, 5-hydroxy-2-methyl-1,4-naphthoquinone) was significantly higher (17 s−1). PBH2 (Em =−40mV) has been shown to be a convenient substrate for several bacterial anaerobic reductases [85].
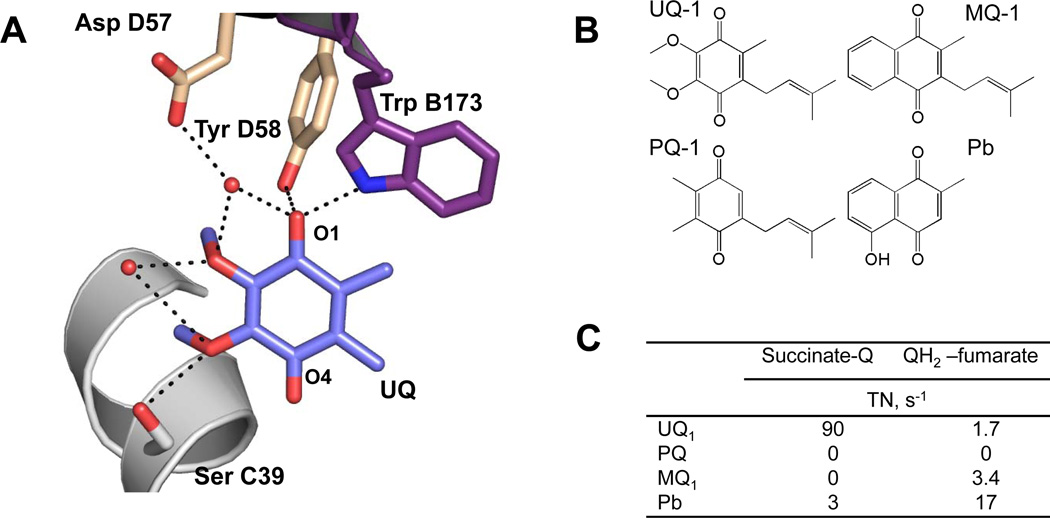
Figure 5. Role of the hydrophilic substitutions in quinone rings for SQR catalytic activity.
(A) Ubiquinone at the active site of avian SQR (pdb:1YQ3 [37]). (B) Chemical structures of quinones substrates for SQR. (C) Catalytic activity of E. coli SQR in succinate-oxidase and fumarate-reductase reactions with different quinones [30, 43].
The precise positions where ubiquinone intermediates are stabilized during the catalysis are not known, but a conserved Tyr residue from SdhD is found hydrogen bonded to the O1-carbonyl of UQ in most x-ray structures (Fig. 5A). A pendulum like movement of the quinone into the deeper part of the cavity to establish a H-bonding contact of O4 carbonyl with the side chain of Ser C39 has been postulated that facilitates proton exchange with ubiquinone and is critical for activity [26]. Earlier studies investigated the interaction of bovine SQR with UQ analogues have shown a complete loss of the activity with substrates in which 2- and 3- methoxy groups of the benzoquinone ring were substituted with a methyl group [86]. The structure of avian SQR with bound ubiquinone shows two water molecules hydrogen-bonded to the protein and methoxy groups of UQ molecule (Fig. 5A). That suggests that the key role of the methoxy groups for catalysis may be associated with their involvement in a proton shuttle for redox reactions with quinones. Figure 5C demonstrates that another natural quinone plastoquinone (PQ) where methoxy groups are substituted with methyl groups shows no activity with SQR. Of note, absence the 6-methyl group in PQ molecule does not allow significantly efficient quinone reduction in complex II [86]. It appears that one of the ways that makes SQR a predominant succinate-ubiquinone reductase is by restricting a productive interaction with menaquinol.
8. Inhibition of fumarate production in SQR at low reduction potentials
The dicarboxylate active site where succinate and fumarate interact with flavin also provides an opportunity to control electron flow in complex II enzymes. There is a unique phenomenon related to catalysis at the dicarboxylate binding site and is associated with functional SQR enzymes. It can be observed in conventional steady-state kinetic experiments with low potential electron donors as rates of fumarate reduction are accelerated with a decrease in reduction power of the electron donor [87]. The same effect was observed in PFV experiments of the soluble dehydrogenase fragment of E. coli SQR [45, 46]. These electrochemical properties of SQRs have been described as similar to that of a tunnel diode, which is a device displaying negative resistance in a certain range of electrochemical potentials. The succinate dehydrogenase enzyme is proficient in fumarate reduction above the potential of −70 mV with catalysis severely restricted at the potentials below this value. As a result, the driving force set for a productive fumarate reduction in QFR would impair the same reaction in SQR. The molecular mechanism for the tunnel-diode effect in SQR is yet to be understood. However, flavin has been identified as the most plausible candidate to exert this behavior [46]. There is a remarkable structural homology surrounding FAD and the dicarboxylate binding site with one notable exception. In the same position that puts a side chain within 5Å from the N5 position of the flavin there is a negative Glu45 in SdhA and neutral Gln44 in FrdA. Even though the reciprocal substitution of the residues did not induce diode-like behavior in QFR mutant, it significantly perturbed the overall reactions of both SQR and QFR enzymes [88]. The overall activity dropped for both enzymes. Nevertheless, QFR and SQR appeared to be more efficient succinate oxidases when Gln was in the target position and better fumarate reductases when Glu was present.
In addition, there is another essential property of complex II enzymes that relates to the flavin and distinguishes E. coli QFR from SQR, i.e., the ability to generate reactive oxygen species (ROS). Oxygen toxicity in cells are usually due to two major ROS products, hydrogen peroxide and superoxide and some evidence shows that mammalian complex II along with other respiratory chain proteins plays a role in cellular oxidative stress [89, 90]. It was shown that E. coli QFR produces a mixture of H2O2 and O2•− and the rate of generation and proportion of these species depends on the reduction levels of the flavin [91]. At low potentials, two electron autoxidation of FADH2 proceeds with significant rates of H2O2 production, while lower reduction levels promote monovalent oxidation and O2•− formation. In contrast to QFR, E. coli SQR does not generate H2O2 and superoxide is the predominant product. This reaction also occurs at much lower rates in comparison with E. coli QFR. The observed differences in ROS production by E. coli enzymes was linked to apparently higher flavin exposure to solvent in QFR that allows high rates of ROS production [91]. Still, there is no crystallographic support for this notion. The predominant production of superoxide by SQR may be attributed to enhanced stabilization of the flavin radical in SQR due to a neighboring high potential [2Fe-2S] center.
9. Conclusion
Here we focused on several factors that may govern the efficacy of E. coli SQR and QFR enzymes in succinate-UQ and MQH2-fumarate reductase reactions. Clearly, the catalysis at the active sites largely determines and controls the rate of enzymatic turnover. While QFR has no restrictions against interaction with succinate/fumarate and UQ/MQ, SQR has developed mechanisms that prevent proficient reaction in the MQH2-fumarate reductase direction and involve both catalytic sites. Yet, the role by which reduction potentials of the Fe-S clusters affects electron transfer rates through the enzymes remains uncertain. Based on mutagenesis data obtained for both enzymes it seems that catalytic activity is resistant to changes if a single cluster is involved. It is, however, possible that “it takes two to tango” and unidirectional simultaneous changes in redox properties of the [2Fe-2S] and [3Fe-4S] clusters may affect the physiological turnover. The flavin in E. coli QFR is a powerful ROS generator and evolutionary adaptation that raises the oxidizing potential of Fe-S centers may be achieved in aerobic SQRs in order to decrease the reduction state of FAD and, thus, oxidative stress. Further studies will be necessary to fully understand the physiological and evolutionary influence of the structural differences in complex II enzymes. Moreover, study of complex II has sparked renewed interest because of its role in cellular physiology and increased associations with diseases in humans. To date more than 400 unique DNA variants in complex II genes have been associated with tumorogenesis and neurodegeneration [92] which is an area of intense investigation. Understanding electron flow and catalysis may aid in providing insight into the role of complex II and mitochondrial function in disease.
Highlights.
An overview on the role of the redox properties of Fe-S clusters in the catalysis of complex II enzymes.
Described mechanisms that prevent SQR from efficient fumarate reduction.
The role of polar groups of the quinoid ring of ubiquinone in reactions with complex II.
Acknowledgments
The work from the authors laboratories was supported, in whole or in part by the Department of Veterans Affairs, Biomedical Laboratory Research and Development, and the National Institutes of Health Grant GM61606 (to G. Cecchini) and by the National Institutes of Health Grant GM62954 and Grant DE-FG02-08ER15960 from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Sciences, U.S. Department of Energy (to S.A. Dikanov). The authors gratefully acknowledge the critical reading of the manuscript and useful comments by Brian Ackrell and Alex Taguchi.
Abbreviations
- SQR
succinate:quinone oxidoreductase
- QFR
quinol:fumarate reductase
- ESEEM
Electron Spin Echo Envelope Modulation
- ENDOR
Electron-Nuclear Double Resonance
References
- 1.Cecchini G. Function and structure of complex II of the respiratory chain. Annu Rev Biochem. 2003;72:77–109. doi: 10.1146/annurev.biochem.72.121801.161700. [DOI] [PubMed] [Google Scholar]
- 2.Ackrell BA, Johnson MK, Gunsalus RP, Cecchini Cary. Chemistry and Biochemistry of Flavoenzymes. London: CRC Press; 1992. Structure and function of succinate dehydrogenase and fumarate reductase; pp. 229–297. [Google Scholar]
- 3.van Hellemond JJ, van der Klei A, van Weelden SW, Tielens AG. Biochemical and evolutionary aspects of anaerobically functioning mitochondria. Philos Trans R Soc Lond B Biol Sci. 2003;358:205–213. doi: 10.1098/rstb.2002.1182. discussion 213-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwata F, Shinjyo N, Amino H, Sakamoto K, Islam MK, Tsuji N, Kita K. Change of subunit composition of mitochondrial complex II (succinate-ubiquinone reductase/quinol-fumarate reductase) in Ascaris suum during the migration in the experimental host. Parasitol Int. 2008;57:54–61. doi: 10.1016/j.parint.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Guest JR. Partial replacement of succinate dehydrogenase function by phage- and plasmid-specified fumarate reductase in Escherichia coli. J Gen Microbiol. 1981;122:171–179. doi: 10.1099/00221287-122-2-171. [DOI] [PubMed] [Google Scholar]
- 6.Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol. 1998;180:5989–5996. doi: 10.1128/jb.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. Structure of the Escherichia coli fumarate reductase respiratory complex. Science. 1999;284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 8.Lancaster CR, Kroger A, Auer M, Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu H, Osanai A, Sakamoto K, Inaoka DK, Shiba T, Harada S, Kita K. Crystal structure of mitochondrial quinol-fumarate reductase from the parasitic nematode Ascaris suum. J Biochem. 2012;151:589–592. doi: 10.1093/jb/mvs051. [DOI] [PubMed] [Google Scholar]
- 10.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 11.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Huang LS, Shen JT, Wang AC, Berry EA. Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the "oxaloacetate-inhibited" state. Biochim Biophys Acta. 2006;1757:1073–1083. doi: 10.1016/j.bbabio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iverson TM, Maklashina E, Cecchini G. Structural Basis for Malfunction in Complex II. J Biol Chem. 2012 doi: 10.1074/jbc.R112.408419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor P, Pealing SL, Reid GA, Chapman SK, Walkinshaw MD. Structural and mechanistic mapping of a unique fumarate reductase. Nat Struct Biol. 1999;6:1108–1112. doi: 10.1038/70045. [DOI] [PubMed] [Google Scholar]
- 15.Mattevi A, Tedeschi G, Bacchella L, Coda A, Negri A, Ronchi S. Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure. 1999;7:745–756. doi: 10.1016/s0969-2126(99)80099-9. [DOI] [PubMed] [Google Scholar]
- 16.Doherty MK, Pealing SL, Miles CS, Moysey R, Taylor P, Walkinshaw MD, Reid GA, Chapman SK. Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and crystallographic study. Biochemistry. 2000;39:10695–10701. doi: 10.1021/bi000871l. [DOI] [PubMed] [Google Scholar]
- 17.Cecchini G, Maklashina E, Yankovskaya V, Iverson TM, Iwata S. Variation in proton donor/acceptor pathways in succinate:quinone oxidoreductases. FEBS Lett. 2003;545:31–38. doi: 10.1016/s0014-5793(03)00390-9. [DOI] [PubMed] [Google Scholar]
- 18.Hagerhall C. Succinate: quinone oxidoreductases. Variations on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 19.Madej MG, Nasiri HR, Hilgendorff NS, Schwalbe H, Lancaster CR. Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J. 2006;25:4963–4970. doi: 10.1038/sj.emboj.7601361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews FS, Cunane L, Durley RCE. Flavin Electron Transfer Proteins. In: Holzenburg A, Scrutton NS, editors. Enzyme-catalyzed electron and radical transfer. New York: Kluwer Academic/Plenum; 2000. [Google Scholar]
- 21.Lin J, Balabin IA, Beratan DN. The nature of aqueous tunneling pathways between electron-transfer proteins. Science. 2005;310:1311–1313. doi: 10.1126/science.1118316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manodori A, Cecchini G, Schroder I, Gunsalus RP, Werth MT, Johnson MK. [3Fe-4S] to [4Fe-4S] cluster conversion in Escherichia coli fumarate reductase by site-directed mutagenesis. Biochemistry. 1992;31:2703–2712. doi: 10.1021/bi00125a010. [DOI] [PubMed] [Google Scholar]
- 23.Kowal AT, Werth MT, Manodori A, Cecchini G, Schroder I, Gunsalus RP, Johnson MK. Effect of cysteine to serine mutations on the properties of the [4Fe-4S] center in Escherichia coli fumarate reductase. Biochemistry. 1995;34:12284–12293. doi: 10.1021/bi00038a024. [DOI] [PubMed] [Google Scholar]
- 24.Werth MT, Cecchini G, Manodori A, Ackrell BA, Schroder I, Gunsalus RP, Johnson MK. Site-directed mutagenesis of conserved cysteine residues in Escherichia coli fumarate reductase: modification of the spectroscopic and electrochemical properties of the [2Fe-2S] cluster. Proc Natl Acad Sci U S A. 1990;87:8965–8969. doi: 10.1073/pnas.87.22.8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werth MT, Sices H, Cecchini G, Schroder I, Lasage S, Gunsalus RP, Johnson MK. Evidence for non-cysteinyl coordination of the [2Fe-2S] cluster in Escherichia coli succinate dehydrogenase. FEBS Lett. 1992;299:1–4. doi: 10.1016/0014-5793(92)80086-v. [DOI] [PubMed] [Google Scholar]
- 26.Maklashina E, Cecchini G. The quinone-binding and catalytic site of complex II. Biochim Biophys Acta. 2010;1797:1877–1882. doi: 10.1016/j.bbabio.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson RF, Hille R, Shinde SS, Cecchini G. Electron transfer within complex II. Succinate:ubiquinone oxidoreductase of Escherichia coli. J Biol Chem. 2005;280:33331–33337. doi: 10.1074/jbc.M506002200. [DOI] [PubMed] [Google Scholar]
- 28.Hatefi Y, Galante YM. Isolation of cytochrome b560 from complex II (succinateubiquinone oxidoreductase) and its reconstitution with succinate dehydrogenase. J Biol Chem. 1980;255:5530–5537. [PubMed] [Google Scholar]
- 29.Kita K, Vibat CR, Meinhardt S, Guest JR, Gennis RB. One-step purification from Escherichia coli of complex II (succinate: ubiquinone oxidoreductase) associated with succinate-reducible cytochrome b556. J Biol Chem. 1989;264:2672–2677. [PubMed] [Google Scholar]
- 30.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006;281:32310–32317. doi: 10.1074/jbc.M607476200. [DOI] [PubMed] [Google Scholar]
- 31.Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. Escherichia coli succinate dehydrogenase variant lacking the heme b. Proc Natl Acad Sci U S A. 2007;104:18007–18012. doi: 10.1073/pnas.0707732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruzicka FJ, Beinert H, Schepler KL, Dunham WR, Sands RH. Interaction of ubisemiquinone with a paramagnetic component in heart tissue. Proc Natl Acad Sci U S A. 1975;72:2886–2890. doi: 10.1073/pnas.72.8.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miki T, Yu L, Yu CA. Characterization of ubisemiquinone radicals in succinate-ubiquinone reductase. Arch Biochem Biophys. 1992;293:61–66. doi: 10.1016/0003-9861(92)90365-4. [DOI] [PubMed] [Google Scholar]
- 34.Salerno JC, Blum H, Ohnishi T. The orientation of iron-sulfur clusters and a spin-coupled ubiquinone pair in the mitochondrial membrane. Biochim Biophys Acta. 1979;547:270–281. doi: 10.1016/0005-2728(79)90010-0. [DOI] [PubMed] [Google Scholar]
- 35.Salerno JC, Harmon HJ, Blum H, Leigh JS, Ohnishi T. A transmembrane quinone pair in the succinate dehydrogenase--cytochrome b region. FEBS Lett. 1977;82:179–182. doi: 10.1016/0014-5793(77)80579-6. [DOI] [PubMed] [Google Scholar]
- 36.Salerno JC, Ohnishi T. Studies on the stabilized ubisemiquinone species in the succinate-cytochrome c reductase segment of the intact mitochondrial membrane system. Biochem J. 1980;192:769–781. doi: 10.1042/bj1920769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang LS, Sun G, Cobessi D, Wang AC, Shen JT, Tung EY, Anderson VE, Berry EA. 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J Biol Chem. 2006;281:5965–5972. doi: 10.1074/jbc.M511270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006;281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- 39.Ruprecht J, Yankovskaya V, Maklashina E, Iwata S, Cecchini G. Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site. J Biol Chem. 2009;284:29836–29846. doi: 10.1074/jbc.M109.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Zhai Y, Lou J, Liu M, Pang X, Sun F. Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature. Protein Cell. 2011;2:531–542. doi: 10.1007/s13238-011-1079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harada S, Sasaki T, Shindo M, Kido Y, Inaoka DK, Omori J, Osanai A, Sakamoto K, Mao J, Matsuoka S, Inoue M, Honma T, Tanaka A, Kita K. 18 Crystal structures of porcine heart mitochondrial complex II with different Q-site inhibitors. 2011 in, www.pdb.org. [Google Scholar]
- 42.Lancaster CR, Herzog E, Juhnke HD, Madej MG, Muller FG, Paul R, Schleidt PG. Electroneutral and electrogenic catalysis by dihaem-containing succinate:quinone oxidoreductases. Biochem Soc Trans. 2008;36:996–1000. doi: 10.1042/BST0360996. [DOI] [PubMed] [Google Scholar]
- 43.Maklashina E, Cecchini G. Comparison of catalytic activity and inhibitors of quinone reactions of succinate dehydrogenase (Succinate-ubiquinone oxidoreductase) and fumarate reductase (Menaquinol-fumarate oxidoreductase) from Escherichia coli. Arch Biochem Biophys. 1999;369:223–232. doi: 10.1006/abbi.1999.1359. [DOI] [PubMed] [Google Scholar]
- 44.Hudson JM, Heffron K, Kotlyar V, Sher Y, Maklashina E, Cecchini G, Armstrong FA. Electron transfer and catalytic control by the iron-sulfur clusters in a respiratory enzyme, E. coli fumarate reductase. J Am Chem Soc. 2005;127:6977–6989. doi: 10.1021/ja043404q. [DOI] [PubMed] [Google Scholar]
- 45.Sucheta A, Ackrell BA, Cochran B, Armstrong FA. Diode-like behaviour of a mitochondrial electron-transport enzyme. Nature. 1992;356:361–362. doi: 10.1038/356361a0. [DOI] [PubMed] [Google Scholar]
- 46.Pershad HR, Hirst J, Cochran B, Ackrell BA, Armstrong FA. Voltammetric studies of bidirectional catalytic electron transport in Escherichia coli succinate dehydrogenase: comparison with the enzyme from beef heart mitochondria. Biochim Biophys Acta. 1999;1412:262–272. doi: 10.1016/s0005-2728(99)00066-3. [DOI] [PubMed] [Google Scholar]
- 47.Leger C, Heffron K, Pershad HR, Maklashina E, Luna-Chavez C, Cecchini G, Ackrell BA, Armstrong FA. Enzyme electrokinetics: energetics of succinate oxidation by fumarate reductase and succinate dehydrogenase. Biochemistry. 2001;40:11234–11245. doi: 10.1021/bi010889b. [DOI] [PubMed] [Google Scholar]
- 48.Unden G, Albracht SPJ, Kroger A. Redox potentials and kinetic properties of fumarate reductase complex from Vibrio Succinogenes. Biochim Biophys Acta. 1984;767:460–469. [Google Scholar]
- 49.Ackrell BA, Kearney EB, Singer TP. Mammalian succinate dehydrogenase. Methods Enzymol. 1978;53:466–483. doi: 10.1016/s0076-6879(78)53050-4. [DOI] [PubMed] [Google Scholar]
- 50.Panchenko MV, Vinogradov AD. Direct demonstration of enol-oxaloacetate as an immediate product of malate oxidation by the mammalian succinate dehydrogenase. FEBS Lett. 1991;286:76–78. doi: 10.1016/0014-5793(91)80944-x. [DOI] [PubMed] [Google Scholar]
- 51.Hederstedt L, Ohnishi T. Progress in succinate:quinone oxidoreductase research. In: Ernster L, editor. Moleclular Mechanisms in bioenergetics. Elsevier; 1992. pp. 163–198. [Google Scholar]
- 52.Cammack R, Gay E, Shergill JK. Studies of hyperfine interactions in [2Fe–2S] proteins by EPR and double resonance spectroscopy. Coord. Chem. Rev. 1999;192:1003–1022. [Google Scholar]
- 53.Cammack R, MacMillan F. Electron magnetic resonance of iron-sulfur proteins in electron-transfer chains: resolving complexity. Biol. Magn. Reson. 2010;29:11–44. [Google Scholar]
- 54.Schweiger A, Jeschke G. Principles of pulsed electron paramagnetic resonance. Oxford: University press; 2001. [Google Scholar]
- 55.Van Doorslaer S. Continuous wave and pulsed EPR analyses of metalloproteins. Electron Paramagn. Reson. 2008;21:162–183. [Google Scholar]
- 56.Van Doorslaer S, Vinck E. The strength of EPR and ENDOR techniques in revealing structure-function relationships in metalloproteins. Phys Chem Chem Phys. 2007;9:4620–4638. doi: 10.1039/b701568b. [DOI] [PubMed] [Google Scholar]
- 57.Dikanov SA, Crofts AR. Electron Paramagnetic Resonance Spectroscopy. In: Vij DR, editor. Handbook of Applied Solid State Spectroscopy. Berlin: Springer; 2006. pp. 97–149. [Google Scholar]
- 58.Dikanov SA, Tsvetkov YD. Electron Spin Echo Envelope Modulation (ESEEM) Spectroscopy. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- 59.Lobrutto R, Haley PE, Yu CA, Ohnishi T, Leigh JS. Electron-paramagnetic resonance and electron-spin echo studies of iron–sulfur clusters-S-1 and clusters-S-2 in beef heart succinate dehydrogenase. Biophys. J. 1986;49:A327–A327. [Google Scholar]
- 60.Cammack R, Chapman A, McCracken J, Cornelius JB, Peisach J, Weiner JH. Electron spin-echo spectroscopic studies of Escherichia coli fumarate reductase. Biochim Biophys Acta. 1988;956:307–312. doi: 10.1016/0167-4838(88)90148-3. [DOI] [PubMed] [Google Scholar]
- 61.Shergill JK, Cammack R, Weiner JH. Electron spin-echo envelope modulation and spin–spin interaction studies of the iron–sulphur clusters in fumarate reductase of Escherichia coli. J. Chem. Soc., Faraday Trans. 1991;87:3199–3202. [Google Scholar]
- 62.Shergill JK, Cammack R. ESEEM studies of the iron-sulphur clusters of succinate dehydrogenase in Arum maculatum spadix mitochondrial membranes. Biochim Biophys Acta. 1994;1185:43–49. doi: 10.1016/0005-2728(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 63.Hung SC, Grant CV, Peloquin JM, Waldeck AR, Brit RD, Chan SI. ESEEM studies of succinate:ubiquinone reductase from Paracoccus denitrificans. J Biol Inorg Chem. 2000;5:593–602. doi: 10.1007/s007750000142. [DOI] [PubMed] [Google Scholar]
- 64.Ackrell BA, Kearney EB, Mims WB, Peisach J, Beinert H. Iron-sulfur cluster 3 of beef heart succinate-ubiquinone oxidoreductase is a 3-iron cluster. J Biol Chem. 1984;259:4015–4018. [PubMed] [Google Scholar]
- 65.Dikanov SA, Samoilova RI, Kappl R, Crofts AR, Huttermann J. The reduced [2Fe-2S] clusters in adrenodoxin and Arthrospira platensis ferredoxin share spin density with protein nitrogens, probed using 2D ESEEM. Phys Chem Chem Phys. 2009;11:6807–6819. doi: 10.1039/b904597j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwasaki T, Fukazawa R, Miyajima-Nakano Y, Baldansuren A, Matsushita S, Lin MT, Gennis RB, Hasegawa K, Kumasaka T, Dikanov SA. Dissection of Hydrogen Bond Interaction Network around an Iron-Sulfur Cluster by Site-Specific Isotope Labeling of Hyperthermophilic Archaeal Rieske-Type Ferredoxin. J Am Chem Soc. 2012;134:19731–19738. doi: 10.1021/ja308049u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin MT, Sperling LJ, Frericks Schmidt HL, Tang M, Samoilova RI, Kumasaka T, Iwasaki T, Dikanov SA, Rienstra CM, Gennis RB. A rapid and robust method for selective isotope labeling of proteins. Methods. 2011;55:370–378. doi: 10.1016/j.ymeth.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin MT, Samoilova RI, Gennis RB, Dikanov SA. Identification of the nitrogen donor hydrogen bonded with the semiquinone at the Q(H) site of the cytochrome bo3 from Escherichia coli. J Am Chem Soc. 2008;130:15768–15769. doi: 10.1021/ja805906a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin E, Baldansuren A, Lin TJ, Samoilova RI, Wraight CA, Dikanov SA, O'Malley PJ. Hydrogen Bonding between the Q(B) Site Ubisemiquinone and Ser-L223 in the Bacterial Reaction Center: A Combined Spectroscopic and Computational Perspective. Biochemistry. 2012 doi: 10.1021/bi300834w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker WH, Singer TP, Ghisla S, Hemmerich P. Studies on succinate dehydrogenase. 8 -Histidyl-FAD as the active center of succinate dehydrogenase. Eur J Biochem. 1972;26:279–289. doi: 10.1111/j.1432-1033.1972.tb01766.x. [DOI] [PubMed] [Google Scholar]
- 71.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 72.Cheng VW, Ma E, Zhao Z, Rothery RA, Weiner JH. The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J Biol Chem. 2006;281:27662–27668. doi: 10.1074/jbc.M604900200. [DOI] [PubMed] [Google Scholar]
- 73.Denke E, Merbitz-Zahradnik T, Hatzfeld OM, Snyder CH, Link TA, Trumpower BL. Alteration of the midpoint potential and catalytic activity of the rieske iron-sulfur protein by changes of amino acids forming hydrogen bonds to the iron-sulfur cluster. J Biol Chem. 1998;273:9085–9093. doi: 10.1074/jbc.273.15.9085. [DOI] [PubMed] [Google Scholar]
- 74.Schroter T, Hatzfeld OM, Gemeinhardt S, Korn M, Friedrich T, Ludwig B, Link TA. Mutational analysis of residues forming hydrogen bonds in the Rieske [2Fe-2S] cluster of the cytochrome bc1 complex in Paracoccus denitrificans. Eur J Biochem. 1998;255:100–106. doi: 10.1046/j.1432-1327.1998.2550100.x. [DOI] [PubMed] [Google Scholar]
- 75.Kolling DJ, Brunzelle JS, Lhee S, Crofts AR, Nair SK. Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters. Structure. 2007;15:29–38. doi: 10.1016/j.str.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guergova-Kuras M, Kuras R, Ugulava N, Hadad I, Crofts AR. Specific mutagenesis of the rieske iron-sulfur protein in Rhodobacter sphaeroides shows that both the thermodynamic gradient and the pK of the oxidized form determine the rate of quinol oxidation by the bc(1) complex. Biochemistry. 2000;39:7436–7444. doi: 10.1021/bi992491+. [DOI] [PubMed] [Google Scholar]
- 77.Leggate EJ, Hirst J. Roles of the disulfide bond and adjacent residues in determining the reduction potentials and stabilities of respiratory-type Rieske clusters. Biochemistry. 2005;44:7048–7058. doi: 10.1021/bi050189x. [DOI] [PubMed] [Google Scholar]
- 78.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim Biophys Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 79.Chen K, Bonagura CA, Tilley GJ, McEvoy JP, Jung YS, Armstrong FA, Stout CD, Burgess BK. Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential. Nat Struct Biol. 2002;9:188–192. doi: 10.1038/nsb751. [DOI] [PubMed] [Google Scholar]
- 80.Ruprecht J, Iwata S, Rothery RA, Weiner JH, Maklashina E, Cecchini G. Perturbation of the quinone-binding site of complex II alters the electronic properties of the proximal [3Fe-4S] iron-sulfur cluster. J Biol Chem. 2011;286:12756–12765. doi: 10.1074/jbc.M110.209874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prince RC, Dutton PL, Bruce JM. Electrochemistry of ubiquinones, menaquinones and plastoquinones in aprotic solvents. FEBS Lett. 1983;160:273–276. [Google Scholar]
- 82.Wraight CA, Vakkasoglu AS, Poluektov Y, Mattis AJ, Nihan D, Lipshutz BH. The 2-methoxy group of ubiquinone is essential for function of the acceptor quinones in reaction centers from Rba. sphaeroides. Biochim Biophys Acta. 2008;1777:631–636. doi: 10.1016/j.bbabio.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 83.Kroger A, Biel S, Simon J, Gross R, Unden G, Lancaster CR. Fumarate respiration of Wolinella succinogenes: enzymology, energetics and coupling mechanism. Biochim Biophys Acta. 2002;1553:23–38. doi: 10.1016/s0005-2728(01)00234-1. [DOI] [PubMed] [Google Scholar]
- 84.Lemma E, Hagerhall C, Geisler V, Brandt U, von Jagow G, Kroger A. Reactivity of the Bacillus subtilis succinate dehydrogenase complex with quinones. Biochim Biophys Acta. 1991;1059:281–285. doi: 10.1016/s0005-2728(05)80213-0. [DOI] [PubMed] [Google Scholar]
- 85.Rothery RA, Chatterjee I, Kiema G, McDermott MT, Weiner JH. Hydroxylated naphthoquinones as substrates for Escherichia coli anaerobic reductases. Biochem J. 1998;332(Pt 1):35–41. doi: 10.1042/bj3320035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gu LQ, Yu L, Yu CA. Effect of substituents of the benzoquinone ring on electron-transfer activities of ubiquinone derivatives. Biochim Biophys Acta. 1990;1015:482–492. doi: 10.1016/0005-2728(90)90082-f. [DOI] [PubMed] [Google Scholar]
- 87.Ackrell BA, Armstrong FA, Cochran B, Sucheta A, Yu T. Classification of fumarate reductases and succinate dehydrogenases based upon their contrasting behaviour in the reduced benzylviologen/fumarate assay. FEBS Lett. 1993;326:92–94. doi: 10.1016/0014-5793(93)81768-u. [DOI] [PubMed] [Google Scholar]
- 88.Maklashina E, Iverson TM, Sher Y, Kotlyar V, Andrell J, Mirza O, Hudson JM, Armstrong FA, Rothery RA, Weiner JH, Cecchini G. Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain. J Biol Chem. 2006;281:11357–11365. doi: 10.1074/jbc.M512544200. [DOI] [PubMed] [Google Scholar]
- 89.Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moreno-Sanchez R, Hernandez-Esquivel L, Rivero-Segura NA, Marin-Hernandez A, Neuzil J, Ralph SJ, Rodriguez-Enriquez S. Reactive Oxygen Species Are Generated by the Respiratory Complex II: Evidence for Lack of Contribution of the Reverse Electron Flow in Complex I. FEBS J. 2012 doi: 10.1111/febs.12086. [DOI] [PubMed] [Google Scholar]
- 91.Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- 92.Iverson TM, Maklashina E, Cecchini G. Structural basis for malfunction in complex II. J Biol Chem. 2012;287:35430–35438. doi: 10.1074/jbc.R112.408419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohnishi T, Lim J, Winter DB, King TE. Thermodynamic and EPR characteristics of a HiPIP-type iron-sulfur center in the succinate dehydrogenase of the respiratory chain. J Biol Chem. 1976;251:2105–2109. [PubMed] [Google Scholar]
- 94.Ohnishi T, Salerno JC. Thermodynamic and EPR characteristics of two ferredoxin-type iron-sulfur centers in the succinate-ubiquinone reductase segment of the respiratory chain. J Biol Chem. 1976;251:2094–2104. [PubMed] [Google Scholar]
- 95.Maguire JJ, Johnson MK, Morningstar JE, Ackrell BA, Kearney EB. Electron paramagnetic resonance studies of mammalian succinate dehydrogenase. Detection of the tetranuclear cluster S2. J Biol Chem. 1985;260:10909–10912. [PubMed] [Google Scholar]
- 96.Ingledew WJ, Prince RC. Thermodynamic resolution of the iron-sulfur centers of the succinic dehydrogenase of Rhodopseudomonas sphaeroides. Arch Biochem Biophys. 1977;178:303–307. doi: 10.1016/0003-9861(77)90195-3. [DOI] [PubMed] [Google Scholar]
- 97.Crowe BA, Owen P, Cammack R. Study of the respiratory chain in Micrococcus luteus (lysodeikticus) by electron-spin-resonance spectroscopy. Eur J Biochem. 1983;137:185–190. doi: 10.1111/j.1432-1033.1983.tb07813.x. [DOI] [PubMed] [Google Scholar]
- 98.Maguire JJ, Magnusson K, Hederstedt L. Bacillus subtilis mutant succinate dehydrogenase lacking covalently bound flavin: identification of the primary defect and studies on the iron-sulfur clusters in mutated and wild-type enzyme. Biochemistry. 1986;25:5202–5208. doi: 10.1021/bi00366a033. [DOI] [PubMed] [Google Scholar]
- 99.Hagerhall C, Aasa R, von Wachenfeldt C, Hederstedt L. Two hemes in Bacillus subtilis succinate:menaquinone oxidoreductase (complex II) Biochemistry. 1992;31:7411–7421. doi: 10.1021/bi00147a028. [DOI] [PubMed] [Google Scholar]