Summary
The Cul4-Cdt2 (CRL4Cdt2) E3 ubiquitin ligase is a master regulator of cell cycle progression and genome stability. Despite its central role in the degradation of many cell-cycle regulators, e.g. Cdt1, p21 and Pr-Set7/Set8, little is known about the regulation of its activity. We report that Cdt2 is autoubiquitylated by the CRL4A E3 ubiquitin ligase. Cdt2 is additionally polyubiquitylated and degraded by Cul1-FBXO11 (CRL1FBXO11). CRL1FBXO11-mediated degradation of Cdt2 stabilizes p21 and Set8, and this is important during the response to TGF-beta, with the Set8 induction being important for turning off the activation of Smad2. The migration of epithelial cells is also stimulated by CRL1FBXO11-mediated downregulation of Cdt2 and the consequent stabilization of Set8. This is a novel example of cross-regulation between specific cullin 4 and cullin 1 E3 ubiquitin ligases and highlights the role of ubiquitylation in regulating cellular responses to TGF-beta and the migration of epithelial cells.
Keywords: Cdt2, Set8, CRL4, FBXO11, Cullin, Ubiquitylation
Introduction
Cullin-RING ligases (CRLs) constitute the largest subclass of RING domain containing E3 ubiquitin ligases in mammals, controlling the steady state levels of many protein substrates involved in various biological processes and diseases including cancer (Lipkowitz and Weissman, 2011; Petroski and Deshaies, 2005). The SKP1-CUL1-F-box protein (CRL1 or SCF) ligases work via at least 69 unique substrate receptors (F-box proteins) to regulate cell cycle progression, DNA replication, gene transcription and apoptosis.
The CRL4 ubiquitin ligases are also important for many physiological processes, particularly those related to chromatin regulation and genomic stability (Jackson and Xiong, 2009). CRL4 ligases resemble CRL1 ligases: the scaffold protein Cul4 (Cul4A or Cul4B) interacts at its C-terminal end with a small RING-finger protein (Rbx1/2) to recruit an E2 ubiquitin-conjugating enzyme. The N-terminus of CUL4A/B interacts with the adaptor protein damaged DNA binding protein 1 (DDB1), which bridges to a substrate receptor, DDB1 and CUL4 associated factor (DCAF). >90 DCAFs recruit various substrates for ubiquitylation by the CRL4 ligases (Angers et al., 2006; He et al., 2006; Higa et al., 2006b; Jin et al., 2006).
Cdt2 (Cdc10-dependent transcript 2; also known as DTL, DCAF2, L2DTL and RAMP) is a DCAF for CRL4 to form CRL4Cdt2, a critical regulator of cell cycle progression and genome stability (Abbas and Dutta, 2011). CRL4Cdt2 polyubiquitylates and targets for degradation the replication initiation factor Cdt1 (Cdc10-dependent transcript 1) (Higa et al., 2006a; Jin et al., 2006; Sansam et al., 2006; Senga et al., 2006). CRL4Cdt2 also targets for degradation the CDK inhibitor p21 and the histone H4 lysine 20 (H4K20) methyltransferase Pr-Set7/Set8. Degradation of these three substrates is important for cell cycle progression and PCNA-dependent DNA repair, chromatin condensation and gene expression, and prevention of aberrant DNA replication (Abbas and Dutta, 2011; Abbas et al., 2010; Abbas et al., 2008; Centore et al., 2010; Jorgensen et al., 2011; Kim et al., 2008; Nishitani et al., 2008; Oda et al., 2010; Tardat et al., 2010).
An unusual feature of the CRL4Cdt2 is that its activity requires the binding of many, but not all, of its substrates to chromatin-bound PCNA through a conserved and specialized PCNA-interacting peptide (PIP box), a condition only met during S-phase of the cell cycle and during the cellular response to DNA damage (Abbas and Dutta, 2011). Unlike many substrates of CRLs, no specific post-translational modifications, such as phosphorylation, is reported to be essential for ubiquitylation of CRL4Cdt2 substrates. The mere overexpression of Cdt2 in various human-derived cell lines is sufficient to decrease the stability of p21 (Abbas et al., 2008) and Set8 (this study). Cdt2 is elevated in liver, breast, gastric and colon cancers, and its elevation is associated with advanced cancer, metastasis and poor patient survival (Baraniskin et al., 2012; Li et al., 2009; Pan et al., 2006; Ueki et al., 2008). In addition, the Cdt2 gene is amplified in a subset of Ewing sarcomas (Mackintosh et al., 2012). Conversely, inhibition of CRL4Cdt2 is the major mechanism of action of a novel anti-cancer drug MLN4924 (Lin et al., 2010; Soucy et al., 2009).
Little is known about the regulation of CRL4Cdt2 activity or the factors involved in its assembly or disassembly. In this study, we investigated the role of ubiquitylation in regulating the steady state level of Cdt2 and found that, like many other cullin-scaffold substrate receptors, Cdt2 undergoes autoubiquitylation via the CRL4A ubiquitin ligase. Additionally, Cdt2 is ubiquitylated by the CRL1FBXO11 ubiquitin ligase. FBXO11 is an F-box protein substrate receptor for CRL1 that is a tumor suppressor with mutations in diffuse large B cell lymphomas (DLBCLs) (Duan et al., 2012). We found that FBXO11 downregulates the oncoprotein Cdt2 to restrain CRL4Cdt2 activity on its substrates p21 and Set8. The degradation of Cdt2, and the consequent stabilization of Set8 is important to curtail the phospho-Smad2 response to TGF-beta and to promote cell migration. The effects on cell migration may explain the developmental defects seen in mice with mutant FBXO11.
Results
Cul4A promotes the polyubiquitylation and degradation of Cdt2
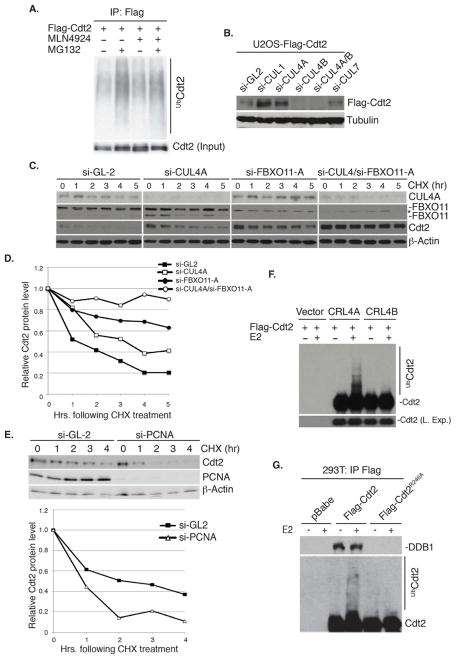
Incubation of the human osteosarcoma U2OS cells with the proteasome inhibitor MG132 resulted in the accumulation of polyubiquitylated Cdt2 (Figure 1A), suggesting that Cdt2 is degraded via the 26S proteasome. MLN4924, a potent inhibitor of the NEDD8-activating enzyme (NAE) that inhibits the cullins by preventing their neddylation (Pan et al., 2004; Podust et al., 2000; Read et al., 2000; Soucy et al., 2009), decreased the basal level of polyubiquitylated Cdt2 as well as the level of polyubiquitylated Cdt2 in cells treated with MG132 (Figure 1A). Therefore, Cdt2 may be polyubiquitylated through a cullin-dependent mechanism. Given that several substrate receptors of the cullin ubiquitin ligases undergo autoubiquitylation and degradation (Deshaies, 1999), we tested whether Cdt2 is similarly regulated by autoubiquitylation. U2OS cells stably expressing flag-tagged Cdt2 were used to eliminate secondary effects on Cdt2 protein due to transcriptional regulation of the Cdt2 promoter. Depletion of Cul4A by siRNA increased the flag-Cdt2 protein (Figure 1B). Interestingly, depletion of Cul4B alone or DDB1 decreased Cdt2 protein (Figure 1B and data not shown). Thus, Cul4B and DDB1 may both stabilize Cdt2 perhaps through interaction with Cdt2, while Cul4A may promote the degradation of Cdt2. Intriguingly, depletion of Cullin 1 (Cul1), but not cullin 3, 5 or cullin 7, also increased the Cdt2 protein (Figure 1B and data not shown).
Figure 1. CRL4A promotes the autoubiquitylation and degradation of Cdt2.
(A) Cdt2 is polyubiquitylated in vivo by cullin-dependent mechanism. Western blot of immunoprecipitated Cdt2 (IP: Flag). Both basal and MG132-stimulated polyubiquitylation of Cdt2 are repressed by MLN4924.
(B) Cul4A and Cul1 decrease the steady-state levels of Cdt2. Western blot of ectopic Cdt2 in U2OS depleted of the indicated cullins by si-RNA. Loading control: tubulin.
(C, D) Cul4A and FBXO11 destabilize Cdt2 protein (Also see Figure 2).
(C) Western blot of Cdt2 and FBXO11 in U2OS cells transfected with si-GL2 (luciferase gene, control) si-Cul4A or si-FBXO11-A as indicated. Cells harvested at indicated times following inhibition of new protein synthesis by cyclohexamide (CHX). Loading control: β-actin.
(D) Quantitation of Cdt2 from (C) normalized to the loading control and expressed relative to the 0 hr time point.
(E) PCNA is not required for the degradation of Cdt2. Top: Immunoblots of U2OS cells transfected with si-GL2 or si-PCNA and harvested at the indicated times following CHX. Bottom: Plot showing the quantitation of Cdt2 protein normalized to β-actin. Rest as in C and D.
(F, G) CRL4A, but not CRL4B promotes the autoubiquitylation of Cdt2 in vitro.
(F) Immunoblot of Cdt2 following incubation in vitro of immunopurified CRL4ACdt2 or CRL4BCdt2 from 293T cells with ATP and ubiquitin ± E2 ubiquitin-conjugating enzyme.
(G) Similar to (F) except Flag-Cdt2 or -Cdt2R246A was immunopurified with anti-Flag from 293T cells before incubation for in vitro ubiquitylation. Immunoblot of Cdt2 (bottom) and co-precipitated endogenous DDB1 (top).
To test whether Cul4A regulates the stability of endogenous Cdt2, we measured the half-life (t1/2) of Cdt2 following inhibition of new protein synthesis by cyclohexamide (CHX). Cdt2 has a t1/2 of 1.5–2 hr, while depletion of Cul4A increased its half-life to >3 hr (Figure 1C, D).
PCNA is critical for the activity of CRL4Cdt2 on several substrates (Abbas and Dutta, 2011). However, depletion of PCNA did not increase the level of Cdt2 and surprisingly, destabilized Cdt2 protein (Figure 1E). The decrease of Cdt2 is an indirect effect of PCNA depletion, because the cells stall in S/G2 phase of the cell cycle, in which phase the Cul1-dependent ubiquitin ligase is more active at degrading Cdt2 (data not shown). Therefore, the polyubiquitylation of Cdt2 by Cul4A does not require PCNA.
We next tested whether Cul4A polyubiquitylates Cdt2 in vitro. Transient overexpression of flag-Cdt2, myc-Cul4A, myc-DDB1 and flag-Rbx1 in 293T cells allowed us to immunoprecipitate the CRL4Cdt2 E3 ligase complex with anti-myc antibodies. The addition of E2, ubiquitin and ATP resulted in the polyubiquitylation of Cdt2 in the Cul4A and DDB1 immunocomplexes (Figure 1F). On the other hand, we did not detect significant Cdt2 polyubiquitylation when Cul4B was substituted for Cul4A (Figure 1F). To confirm that the polyubiquitylation was not due to the activity of other E3 ubiquitin ligases contaminating the Cul4A/DDB1 immunoprecipitates, we repeated the experiment with E3 ligase from anti-flag-Cdt2 immunoprecipitates. Wt-Cdt2 coimmunoprecipitated endogenous DDB1 and was significantly autoubiquitylated in vitro (Figure 1G). In contrast, Cdt2R246A, a mutant that does not bind to DDB1 and thus to Cul4 (Jin et al., 2006), was not polyubiquitylated (Figure 1G). Furthermore, si-RNA-mediated depletion of Cul4A reduced K-48 linked polyubiquitylation of Cdt2 in vivo (Figure 3E). Collectively, these results demonstrate that Cdt2 is autoubiquitylated and degraded via CRL4A ubiquitin ligase in a PCNA-independent reaction.
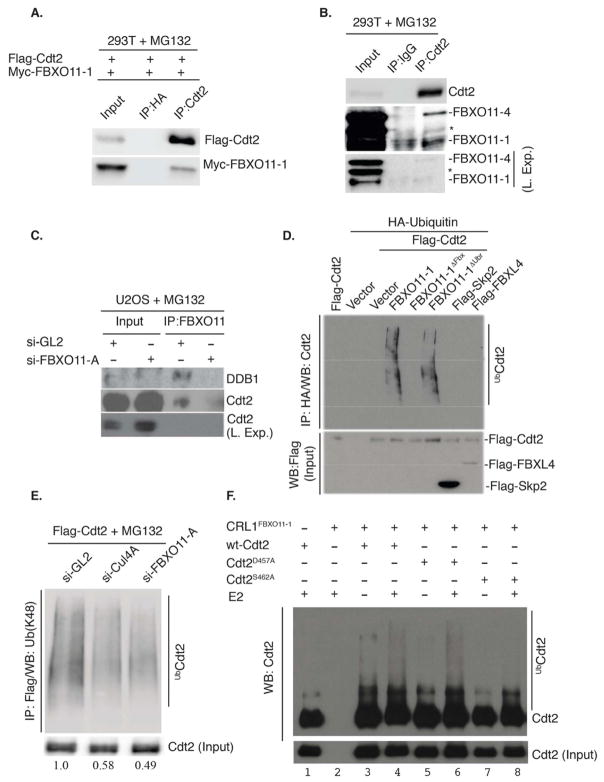
Figure 3. Cdt2 is a direct polyubiquitylation substrate of the CRL1FBXO11 E3 ligase.
(A) Immunoblot of Flag-FBXO11-1 co-immunoprecipitated with exogenous Cdt2 by anti-Cdt2 antibody from 293T cells transfected with plasmids expressing the indicated proteins. Cells were treated with MG132 for 3 hr prior to lysis.
(B) Similar to (A), except the interaction is observed with endogenous proteins. Both isoforms of FBXO11 (FBXO11-4 (upper) and FBXO11-1 (lower)) are detected in the anti-Cdt2 immunoprecipitates. Asterisk indicates a cross-reactive band in the anti-FBXO11 immunoblot. Two exposures are shown of the FBXO11 blot for clarity.
(C) Western blot showing that Cdt2 is detected in the anti-FBXO11 immunoprecipitates in control U2OS cells (si-GL2), but not in cells depleted of FBXO11 by si-FBXO11-A. DDB1, another component of the CRL4Cdt2 ubiquitin ligase, also co-immunoprecipitates with anti-FBXO11 in control cells, but not in cells depleted of FBXO11.
(D) FBXO11 promotes Cdt2 polyubiquitylation in vivo. Western blot analysis of polyubiquitylated Cdt2 (anti-Cdt2) in immunoprecipitated ubiquitylated proteins (anti-HA) from lysates of 293T cells transfected with the indicated F-box proteins, Flag-Cdt2 and HA-ubiquitin (see experimental procedure for details). Immunoblot of lysates with anti-Flag detects the expression of Flag-Cdt2, Flag-FBXL4 and Flag-Skp2 in the input.
(E) Depletion of 293T cells of FBXO11 or Cul4A decreases K-48-linked polyubiquitylation of Cdt2. 293T cells were transfected with flag-Cdt2 plasmid and subsequently transfected with the indicated si-RNA and treated with MG132 for 3 hr before lysis. Western blotting of anti-Flag-Cdt2 immunoprecipitates with anti-K48-linkage-specific-polyubiquitin antibody shows that both Cul4A and FBXO11 are required for maximal Cdt2 polyubiquitylation in vivo. Quantitation of polyubiquitylated Cdt2 relative to nonubiquitylated Cdt2 is shown below each lane.
(F) CRL1FBXO11 promotes the polyubiquitylation of Cdt2 in vitro. Immunoblot of Cdt2 immunoprecipitated from 293T cells transiently transfected with wt-Cdt2, Cdt2D457A or Cdt2S462A and incubated in an in vitro ubiquitylation reaction with separately immunopurified CRL1FBXO11-1 E3 ligase complex (prepared as described in Methods). Incubation was in the presence or absence of E2 ubiquitin-conjugating enzyme.
si-RNA screen identifies FBXO11 as a major regulator of Cdt2 stability
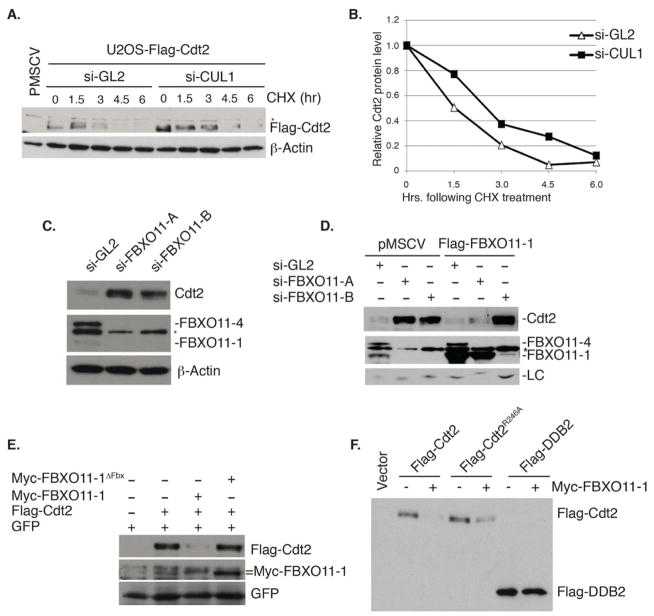
We next investigated how Cul1 regulates Cdt2 abundance (Figure 1B). Depletion of Cul1 increased the t1/2 of flag-Cdt2 (Figure 2A, B). The modest increase in the t1/2 may be explained if Cul1 associated with different F-box proteins with opposing effects on the stability of Cdt2. Therefore, we decided to identify the F-box protein that directly destabilizes Cdt2 by an siRNA screen of the various F-box proteins. Consistent with the notion that some CRL1 complexes may indirectly increase Cdt2, depletion of several F-box proteins, such as FBXW1, FBXO28 or FBXO32, decreased Cdt2 (Figure S1). On the other hand, depletion of Skp2 (FBXL1), FBXL19 or FBXO11 increased the level of Cdt2 (Figure S1). Further experiments in HeLa and U2OS cells however, demonstrated that only the depletion of FBXO11 by multiple siRNA oligonucleotides reproducibly increased endogenous Cdt2 (Figure 2C, and data not shown) or ectopic Cdt2 proteins (Figure 4D, lanes 1–4).
Figure 2. Cul1 and FBXO11 regulate the steady-state level of Cdt2.
(A) Immunoblot of ectopic Flag-Cdt2 from U2OS cells transfected with control si-GL2 or si-Cul1. Cells harvested at the indicated times following cycloheximide (CHX). Asterisk represents a cross-reactive band in the anti-Flag immunoblot. β-actin: loading control.
(B) Quantitation of the Cdt2 protein from (A) normalized to β-actin and expressed relative to the 0 hr time point.
(C) FBXO11 decreases the steady-state levels of Cdt2 (see also Figure 1C, D, and Figure S1). Immunoblot of Cdt2 and FBXO11 following transfection of two si-RNAs targeting distinct regions of the FBXO11 (si-FBXO11-A and si-FBXO11-B). Two isoforms of FBXO11 are detected (FBXO11-4 and FBXO11-1), with a cross-reactive band indicated by an asterisk. β-actin: loading control.
(D) Rescue of Cdt2 degradation by si-RNA resistant, ectopic FBXO11. Control U2OS (pMSCV) or U2OS stably expressing Flag-FBXO11-1 mutated at the target site of si-FBXO11-A, but not of si-FBXO11-B. Cells were transfected with indicated si-RNAs. Immunoblot of Flag-FBXO11-1 shows its sensitivity to si-FBXO11-B, but not si-FBXO11-A. The anti-Cdt2 blot shows restoration of Cdt2 degradation by Flag- FBXO11-1 in cells transfected with si-FBXO11-A but not si-FBXO11-B. Cross-reactive bands in the anti-Cdt2 blot (LC; loading control) and in the anti-FBXO11 blot (indicated with an asterisk) serve as loading controls.
(E)FBXO11 requires the F-box motif to destabilize Cdt2. Immunoblot of Flag-Cdt2 in 293T cells transiently transfected with the indicated plasmids. Cells were cotransfected with a plasmid expressing GFP to control for transfection efficiency.
(F) FBXO11 specifically destabilizes Cdt2 even when it is not associated with the rest of the CRL4 complex. Similar to (E): immunoblot of flag-wt-Cdt2, flag-Cdt2R246A and flag-DDB2, another CRL4 substrate receptor, in the absence or presence of co-transfected myc-FBXO11-1.
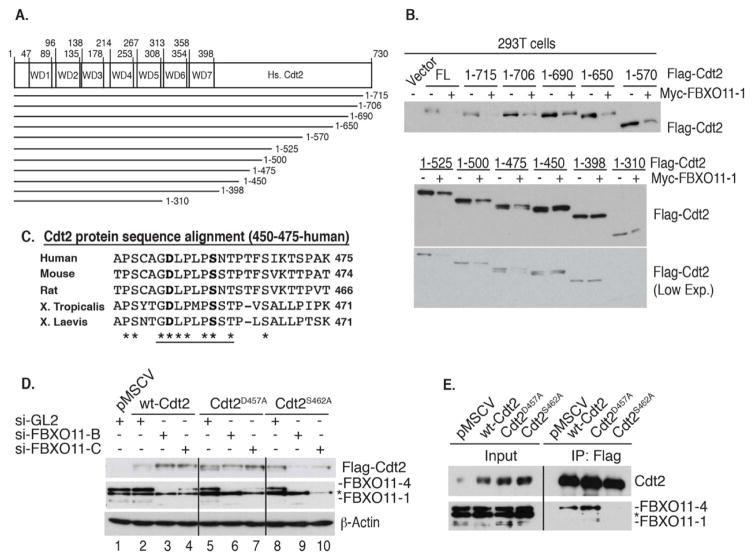
Figure 4. Identification of a 9 amino acid region within Cdt2 conferring sensitivity to FBXO11-mediated polyubiquitylation and degradation.
(A) A schematic of the human Cdt2 protein and various Cdt2 C-terminal truncations analyzed.
(B) A central fragment (amino acids 450–475) in human Cdt2 is required for FBXO11-mediated degradation. Ectopic expression of Cdt2 (wt and mutants) ± FBXO11-1 in 293T cells. Immunoblot of ectopic Cdt2. Low exposure (L. Exp.) of the lower immunoblot is also shown for clarity.
(C) Sequence alignment of the central fragment of human Cdt2 (450–475) with Cdt2 from the indicated species shows conservation of 9 amino acids (underlined). The two residues mutated in the subsequent studies are shown in bold.
(D) D457 and S462 of Cdt2 are both required for degradation by FBXO11. U2OS cells stably expressing wild type (wt) or mutated Cdt2 were transfected with indicated si-RNAs. si-FBXO11-B and si-FBXO11-C (targeting different regions of FBXO11) stabilize wt-Cdt2 compared to si-GL2. This stabilization is not seen for Cdt2D457A or Cdt2S462A. Asterisk represents a cross-reactive band in the anti-FBXO11 immunoblot. Loading control: β-Actin.
(E) Endogenous FBXO11 protein interacts with wt-Cdt2 or Cdt2D457A, but not with Cdt2S462A. U2OS cells stably expressing indicated ectopic Flag-Cdt2 proteins. Lysates (input) or anti-Flag immunoprecipitates of U2OS cells immunoblotted with anti-Cdt2 or anti-FBXO11. Asterisk represents a cross-reactive band in the anti-FBXO11 immunoblot.
Western blotting with anti-FBXO11 antibody detected two protein bands of FBXO11, −1 and −4, both of which are reduced by siRNA against FBXO11 (si-FBXO11-A; Figure 2C, D). The apparent molecular weights of these two bands correspond to two splice isoforms (ACCESSION #: NP_079409 and NP_001177203). Stable expression of the smaller isoform, FBXO11-1, resistant to si-FBXO11-A but not si-FBXO11-B, prevented the increase of Cdt2 in U2OS transfected with si-FBXO11-A, but not si-FBXO11-B (Figure 2D). Thus, the increase of Cdt2 by si-FBXO11 was not due to off-target effect of the si-RNA. The depletion of FBXO11 significantly increased the t1/2 of Cdt2 protein to >5 hr (Figure 1C, D). The depletion of FBXO11 stabilized Cdt2 much more than the depletion of Cul4A, and the co-knockdown of both proteins completely stabilized the Cdt2 protein (Figure 1C, D). Therefore, Cul4A and Cul1-FBXO11 independently destabilize Cdt2 with the FBXO11 pathway playing a more pronounced role.
We next tested whether FBXO11 was rate-limiting in cells for the degradation of Cdt2. Ectopic expression of wt-FBXO11 decreased the amount of co-expressed Cdt2 protein (Figure 2E). This was not seen with FBXO11without the F-box motif (FBXO11-1ΔFbx) demonstrating that FBXO11 has to interact with the SKP1-CRL1 ligase to destabilize Cdt2. FBXO11-1 also reduced the amount of Cdt2R246A (Figure 2F), showing that Cdt2 does not have to associate with the rest of the CRL4 ubiquitin ligase for it to be targeted by CRL1FBXO11, a difference from the targeting of Cdt2 by Cul4 (see Figure 1G). Finally, FBXO11 had no effect on DDB2 protein, another CRL4 substrate receptor, demonstrating its specificity in targeting Cdt2 (Figure 2F).
Cdt2 interacts with FBXO11 and is polyubiquitylated via CRL1FBXO11
To test whether Cdt2 is directly polyubiquitylated by CRL1FBXO11, we first tested whether Cdt2 interacts with FBXO11. Ectopic myc-FBXO11-1 coimmunoprecipitated with flag-Cdt2 in 293T cells treated with the proteasome inhibitor MG132 (Figure 3A). In addition, endogenous FBXO11-4 protein, and to a lesser extent the less detectable endogenous FBXO11-1 isoform, were detected in the immunoprecipitates of endogenous Cdt2 from MG132-treated 293T cells (Figure 3B). Reciprocally, endogenous Cdt2 and DDB1 are seen in the anti-FBXO11 immunoprecipitates in lysates from control (si-GL2 transfected), MG132-treated U2OS, but not after si-FBXO11 (Figure 3C).
We tested whether FBXO11 polyubiquitylates Cdt2 in vivo. Ectopic expression of wt-FBXO11-1 and flag-Cdt2 increased polyubiquitylated Cdt2 (Figure 3D). This was not seen with FBXO11-1ΔFbx, and thus is dependent on functional CRL1FBXO11. On the other hand, the C-terminal UBR domain of FBXO11, a domain essential for recognition of “N-degrons” (Tasaki et al., 2005), was dispensable for Cdt2 polyubiquitylation (Figure 3D). Furthermore, depletion of FBXO11, similar to the depletion of Cul4A, decreased K-48 linked polyubiquitylation of Cdt2, demonstrating that endogenous FBXO11 and Cul4A independently promote the polyubiquitylation of Cdt2 (Figure 3E). Finally, incubation of immunopurified Cdt2 with immunopurified CRL1FBXO11-1 complex from 293T cells promoted Cdt2 polyubiquitylation in vitro upon the addition of E2, ubiquitin, and ATP (Figure 3F, lane 4; also lane 6, which will be explained later). Therefore, CRL1FBXO11 interacts with and directly polyubiquitylates Cdt2.
FBXO11 interacts with Cdt2 through a conserved region located in a central fragment of Cdt2
Next, we mapped the region of Cdt2 required for its degradation via CRL1FBXO11. Ectopic expression of FBXO11-1 decreased full-length wt-Cdt2 (730 amino-acids; FL), as well as Cdt2 proteins with C-terminal deletions up to (Cdt2-1-475) (Figure 4A, B). On the other hand, Cdt2-1-450 or larger C-terminal deletion mutants were resistant to FBXO11-1-mediated destabilization (Figure 4B). Therefore, amino acids 450–475 of Cdt2 are required for FBXO11-mediated degradation.
Cdt2 is evolutionarily conserved in the N-terminally located WD40 repeats necessary for interaction with DDB1 (Figure 4A, and data not shown). Alignment of amino acids 450–475, however, demonstrates additional conservation of several amino acids in the 456–464 segment in various species (Figure 4C). Mutation of the conserved S462 to alanine revealed that S462 is important for FBXO11-mediated degradation. Unlike wt-Cdt2, Cdt2S462A was not increased by the depletion of cellular FBXO11 (Figure 4D, lanes 5–7). Additionally, unlike wt-Cdt2, Cdt2S462A fails to associate with endogenous FBXO11 (Figure 4E) and is not polyubiquitylated by CRL1FBXO11 in vitro (Figure 3F, lane 8). This proves that S462 is part of the degron on Cdt2 that is recognized by FBXO11.
The conserved D457 of Cdt2 is also important for the degradation of Cdt2 by FBXO11 in vivo, in that the level of Cdt2D457A was not increased when FBXO11 was knocked down (Figure 4D, lanes 8–10). Surprisingly however, Cdt2D457A associated with FBXO11 (Figure 4E) and was polyubiquitylated by CRL1FBXO11 in vitro (Figure 3F, lane 6). Thus, D457 of Cdt2, through a yet to be identified mechanism, is essential for FBXO11-mediated ubiquitylation and degradation in vivo without affecting interaction with CRL1FBXO11.
FBXO11 degrades Cdt2 to stabilize the CRL4Cdt2 substrates p21 and Set8
CRL4Cdt2 regulates the steady-state levels of p21, Set8 and Cdt1, both during the normal cell cycle and following DNA damage (Abbas and Dutta, 2011). We first determined that the depletion of mammalian cells of FBXO11 does not alter cell cycle distribution, nor affects their proliferation rate (Figure S2), suggesting that FBXO11 is dispensable for cell proliferation. Under rapid proliferative conditions in high serum (Figure 5A, C), the CRL4Cdt2 substrate p21 was destabilized upon FBXO11 depletion and attendant Cdt2 increase. In contrast, Cdt1 and Set8 were not destabilized by the depletion of FBXO11. Reducing cell proliferation by decreasing the serum concentration (Figure 5D) destabilized both p21 and Set8 upon FBXO11 depletion. The change in behavior of Set8 is because of the faster turnover of the protein in high serum concentration (see Discussion). The decrease in the t1/2 of p21 and Set8 under low serum concentration was also observed in two other cancer epithelial cell lines; A549 (Figure S3A) and HaCat cells (Figure S3B). The DNA replication factor, Cdt1 was unaffected by FBXO11 depletion (and Cdt2 increase) in undamaged cells, but this was expected because Cdt1 is degraded by CRL1Skp2 in the normal S phase (Nishitani et al., 2006; Senga et al., 2006) so that Cdt2 is not rate-limiting for Cdt1 turnover. In summary, FBXO11 is important for limiting the degradation of p21 and Set8, but not Cdt1, in undamaged cells.
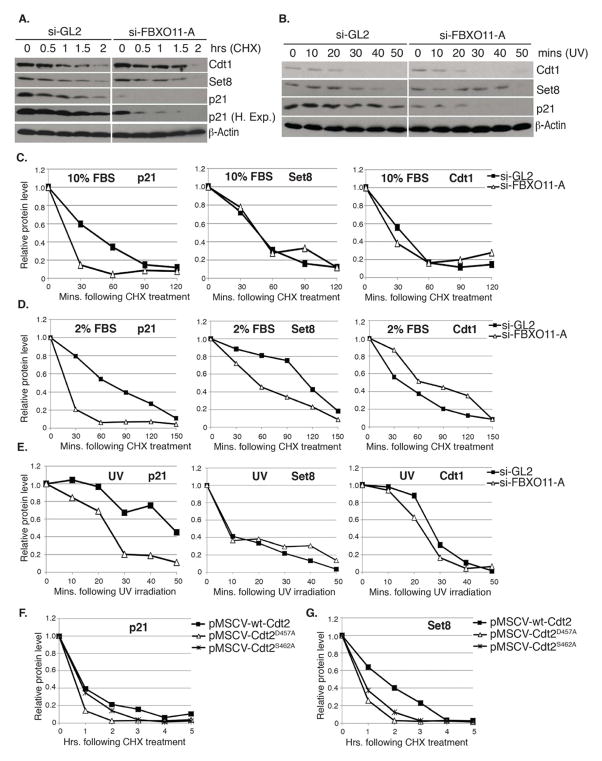
Figure 5. Inactivation of FBXO11-mediated degradation of Cdt2 increases the turnover of the Cdt2 substrates p21 and Set8.
(A–E) Measurement of the half-life (t1/2) of various CRL4Cdt2 substrate proteins following the depletion of FBXO11 from asynchronously proliferating U2OS cells cultured in 10% (A, C) or 2% serum (D) or in U2OS cells cultured in 10% serum and irradiated with 30J/m2 UV (B, E).
(A, B) Immunoblot of Cdt1, Set8 and p21 in extracts of U2OS cells cultured in 10% serum and transfected with indicated siRNAs following CHX addition for the indicated hours (A), or at the indicated time following UV irradiation (B). β-actin: loading control.
(C–E) t1/2 of p21, Set8 or Cdt1 in cells grown in 10% serum (C), 2% serum (D) or following UV irradiation (E). (C and E) Quantitation of indicated proteins shown in (A and B, respectively), normalized to β-actin and expressed relative to the 0 time point in each case. (D) Same as (C), except U2OS cells were cultured in 2% serum for 24 hr prior to the CHX addition. Western blots not shown. See also Figure S3.
(F, G) FBXO11-resistant Cdt2 proteins destabilize p21 and Set8. Quantitation of p21 (F) and Set8 (G) proteins in extracts of U2OS cells stably expressing wt-Cdt2, Cdt2D457A or Cdt2S462A from pMSCV-based retrovirus vectors. Values are normalized to those of β-actin and expressed relative to the 0 time point when CHX is added.
We next turned to the degradation of CRL4Cdt2 substrates following UV-induced DNA damage in U2OS cells growing in high serum (Figure 5B, E). Now Cdt1 is slightly destabilized and p21 significantly destabilized upon FBXO11 depletion. Again, Set8 has a rapid turnover upon UV irradiation in high serum and so Cdt2 is not rate-limiting for its half-life.
Because FBXO11 promotes the degradation of other proteins in addition to Cdt2, we analyzed whether these effects of FBXO11-depletion resulted from the specific stabilization of Cdt2. U2OS cells were made to stably express wt-Cdt2 or Cdt2 mutant proteins resistant to FBXO11 (Cdt2D457A and Cdt2S462A). After a few passages in culture, these cells proliferated with similar kinetics as parental U2OS cells or cells expressing wt-Cdt2 (data not shown). Cdt2D457A or Cdt2S462A decreased the t1/2 of p21 and Set8 proteins compared to wt-Cdt2 (Figure 5F, G). These findings demonstrate that the destabilization of p21 and Set8 in cells depleted of FBXO11 is due to increase of Cdt2 and not any other target of FBXO11.
FBXO11 degrades Cdt2 to upregulate Set8 and to limit the phospho-Smad2 in response to TGF-beta
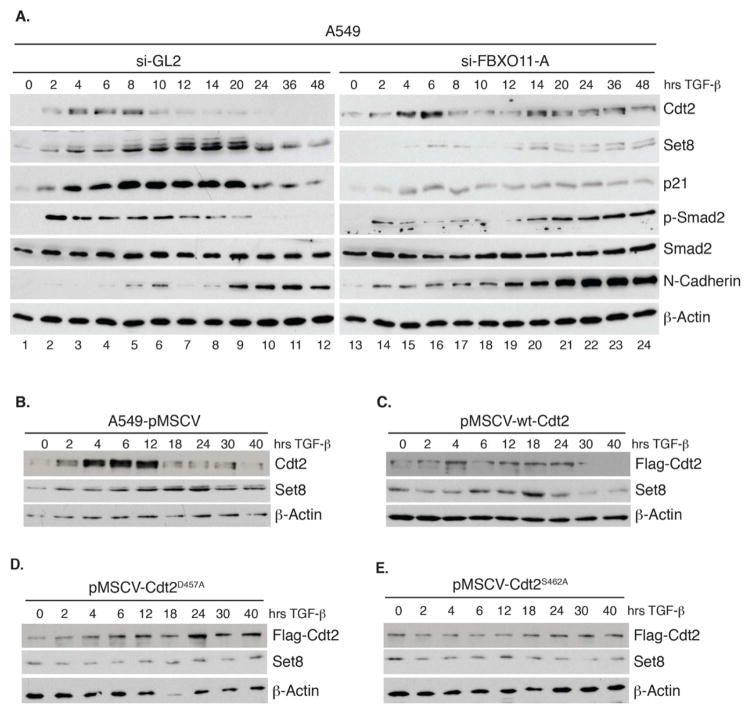
FBXO11 has been suggested to function as a regulator of the cell’s response to TGF-beta and mice with inactivated FBXO11 exhibited significant phospho-Smad2 (P-Smad2) signal and P-Smad2 nuclear staining in the epithelial cells of palatal shelves, eyelids and airways of the lung, potentially leading to the developmental defects of these tissues (Tateossian et al., 2009). Thus, we hypothesized that the degradation of Cdt2 by FBXO11 is important for the normal response of a cell to TGF-beta. For these experiments we used A549 lung carcinoma cells known to respond well to TGF-beta. In control (si-GL2) A549 cells (Figure 6A, left panel), the TGF-beta-induced P-Smad2 (Ser465/467) was seen for 2–20 hrs but subsequently declined to basal levels. Cdt2 protein was transiently induced at 4–8 hrs, but declined at 10 hr, and this decline was accompanied by elevation of CRL4Cdt2 substrates Set8 and p21 at 12–20 hr (Figure 6A, left panel). Although Cdt2 mRNA was reduced at these later time points (Figure S4A), depletion of FBXO11 resulted in the persistence of Cdt2 protein up to 48 hr post TGF-beta treatment (Figure 6A, right). This persistence of Cdt2 in FBXO11-depleted A549 cells was accompanied by low levels of Set8 and p21 at all times, but particularly in the 12–20 hr window (Figure 6A).
Figure 6. FBXO11 promotes the degradation of Cdt2 and the accumulation of p21 and Set8 and downregulates P-Smad2 in TGF-beta treated epithelial cells.
(A) Immunoblots of A549 lung epithelial cancer cells growing in 2% FBS and transfected with si-GL2 or si-FBXO11-A. Cells were harvested at indicated times following TGF-beta addition at 5 ng/ml. Lysates were immunoblotted with the indicated antibodies. A lighter exposure of the P-Smad2 is shown for the FBXO11-depleted cells to demonstrate further induction of the signal late in the time course.
(B–E) Similar to (A) except control A549 (pMSCV) or A549 cells stably expressing wt-Cdt2 or Cdt2 proteins resistant to FBXO11 (Cdt2D457A and Cdt2S462A) were used. Quantitation of the Set8 protein expression for this experiment is shown in Figure S4E. See also Figure S4.
As an aside, the induction of endogenous Cdt2 at the early times after TGF-beta induction (Figure 6A, left) is a post-transcriptional effect of TGF-beta on the 3′UTR of Cdt2 and is unrelated to FBXO11. This conclusion is based on the observations that: a) the initial induction of endogenous Cdt2 was still observed in cells depleted of FBXO11 (Figure 6A, right), b) exogenous Cdt2 protein, transcribed from a heterologous promoter and without the natural 3′UTR, is not induced by TGF-beta (Figure 6D–G), and c) there was no induction of endogenous Cdt2 mRNA by TGF-beta (Figure S4A).
To test whether FBXO11-mediated degradation of Cdt2 alone was sufficient for the induction of Set8 seen at 12–20 hr after TGF-beta, we repeated the TGF-beta treatment in control A549 (pMSCV) or in cells expressing wt-Cdt2 or FBXO11-resistant mutants of Cdt2 (Figure 6B–E). The expression of Cdt2 or mutant Cdt2 protein did not impact cell proliferation (Figure S4D). Consistent with the previous results, endogenous Cdt2 in control A549 cells (pMSCV) declined between 12 and 40 hr following TGF-beta treatment (Figure 6B). Ectopically expressed wt-Cdt2 persisted a little longer than endogenous Cdt2 protein but nonetheless declined over 24–40hr (Figure 6C). In contrast, both the FBXO11-resistant forms of Cdt2 remained constant through the entire 40 hrs after TGF-beta treatment, confirming that FBXO11 is responsible for the decline of Cdt2 in the late stages after TGF-beta treatment (Figure 6D, E). Consistent with the hypothesis that Cdt2 decline is important for elevating Set8 in the 12–20 hr window, the FBXO11-resistant forms of Cdt2 prevented the induction of Set8 during that window (Figure 6D, E, S4E). Thus, the decrease of endogenous Cdt2 at 12–20 hr is critical for the elevation of Set8 and this is mediated by CRL1FBXO11.
Signal transduction downstream from the TGF-beta receptor is mediated by phosphorylation of Smad2. Normally, the phosphorylation of Smad2 declines at 14–20 hr after TGF-beta treatment and does not reappear. Upon FBXO11 depletion, P-Smad2 is induced at the same early time point (2 hrs) following TGF-beta, but remains high at 14 – 48 hr post TGF-beta treatment (Figure 6A, right; lighter exposure shown to demonstrate the additional increase at 14–48 hr). This was accompanied by earlier and higher induction of N-cadherin, (Figure 6A, right). We hypothesized that in the absence of FBXO11-mediated destabilization of Cdt2, Set8 is not induced during the 12–20 hr window and this is responsible for the increase of P-Smad2 and N-cadherin. To test this, we depleted Set8 by siRNA and monitored the level of P-Smad2 early (2 hrs) and late (24 and 48 hrs) after TGF-beta treatment. Set8 depletion alone significantly increased the P-Smad2 levels even at 24–48 hr post-TGF-beta treatment (Figure S4B). Set8 depletion also induced N-cadherin (Figure S4C), suggesting that Set8 represses N-cadherin. These results indicate that FBXO11 degrades Cdt2 to induce Set8 at 12–20 hr post TGF-beta treatment, and the induction of Set8 during this critical period decreases both P-Smad2 and N-cadherin at 24–48 hr.
FBXO11 promotes cellular migration by downregulating Cdt2 and thus upregulating Set8
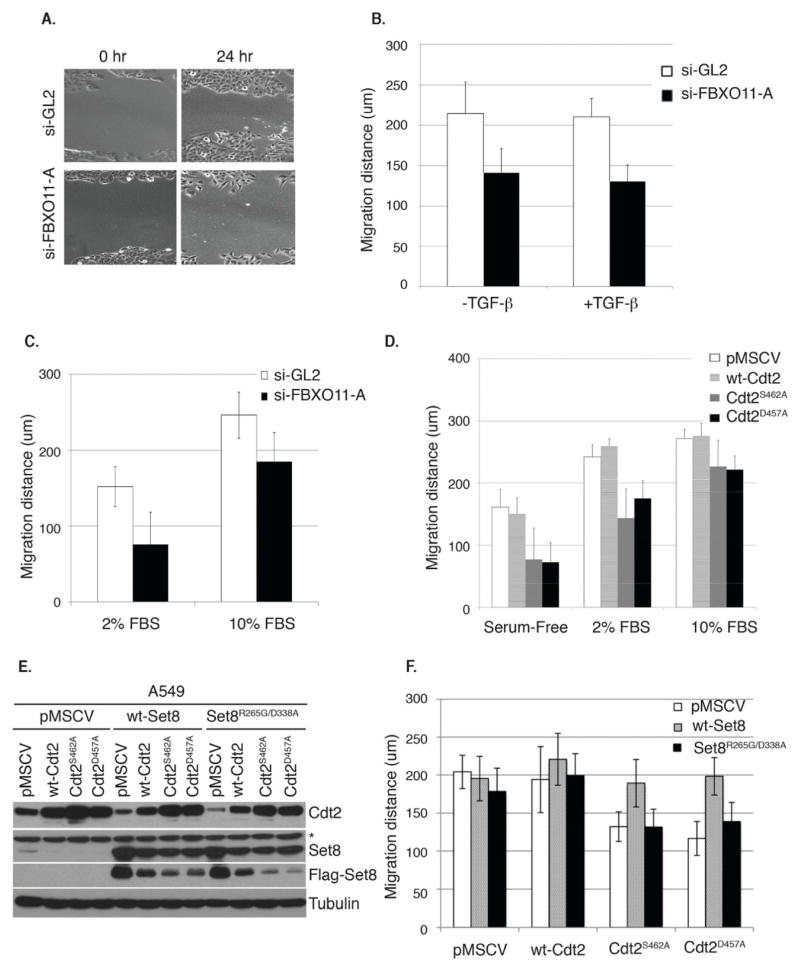
The histone monomethyl transferase Set8 has been implicated in regulating cell migration (Moustakas and Heldin, 2012; Yang et al., 2012). We thus investigated whether the migration of A549 cells, in standard wound-healing assays, is dependent on FBXO11-mediated decrease of Cdt2 and consequent increase of Set8. Depletion of FBXO11 reduced the migration of A549 cells (Figure 7A–C). FBXO11 was required for migration, both in TGF-beta-treated and untreated cells (Figure 7B). The migration defect of the FBXO11-depleted cells was more marked when serum was depleted (Figure 7C), a condition in which the lack of FBXO11 accelerated the turnover of Set8 (Figure 5D and Figure S3). Similarly, the stable expression of Cdt2D457A or Cdt2S462A, unlike wt-Cdt2, retarded the migration of A549 cells and this was similarly, more pronounced upon serum-depletion (Figure 7D). The FBXO11-resistant Cdt2 proteins also inhibited the migration of the human keratinocytes HaCat cells (Figure S5).
Figure 7. FBXO11 promotes Set8-dependent migration of epithelial cancer cells through the targeted degradation of Cdt2.
(A) Representative images of scratch or wound healing assay showing the migration of A549 cells transfected with si-GL2 or si-FBXO11. Cells migrate more efficiently in the presence of FBXO11 as demonstrated 24 hrs following the scratch.
(B) Quantitation of the migration distance traveled (in micrometers) in si-GL2 or si-FBXO11-trasnfected cells without or with TGF-beta for 48 hrs. Error bars represent the average +/− SD from 12 independent scratches (three fields per scratch).
(C) Similar to (B) except that the analysis was performed on A549 cells growing in 2% or 10% serum concentrations. Error bars represent the average +/− SD from 12 independent scratches (three fields per scratch).
(D) Similar to (B, C), but in control A549 cells (pMSCV) or in cells stably expressing wt-Cdt2 or Cdt2 proteins resistant to degradation by FBXO11 (Cdt2D457A and Cdt2S462A). See also Figure S5. Error bars represent the average +/− SD from 12 independent scratches (three fields per scratch).
(E, F) Set8 is required for the efficient migration of epithelial cells. (E) Immunoblot of lysates from A549 cell lines from (D) infected with a second retrovirus expressing either wt-Set8 or catalytically inactive Set8 (Set8R265G/D338A) to show the expression of the indicated Set8 proteins. Anti-flag immunoblot shows the expression of ectopic Set8 proteins. Asterisk represents a cross-reactive band in the anti-Set8 immunoblot. Loading control: tubulin. (F) Similar to (D) except the analysis was performed in the indicated A549 cell lines as described and shown in (E). Migration distance (in micrometers) was determined 48 hrs following the initial scratches. Error bars represent the average +/− SD from 12 independent scratches (three fields per scratch).
Finally, we tested whether the migration defect in cells with inactivated FBXO11-Cdt2 degradation pathway was due to excessive degradation of Set8. Over-expression of wt-Set8, but not catalytically inactive mutant Set8 (Set8R265G/D338A), reversed the migration inhibition caused by FBXO11-resistant Cdt2 (Figure 7E, F). Thus, FBXO11-mediated degradation of Cdt2 is critical for providing sufficient levels of catalytically active Set8 to promote cellular migration.
Discussion
Two ubiquitin ligases control the steady state levels of Cdt2
We identified two distinct pathways regulating the steady-state level of the CRL4 substrate receptor Cdt2 via ubiquitin-dependent degradation. Cdt2 undergoes autoubiquitylation via its association with Cul4A. Autoubiquitylation of substrate adaptors is seen with another substrate receptor of CRL4, DDB2 (Chen et al., 2001; Nag et al., 2001), and with substrate receptors of other cullins (Deshaies, 1999). As with DDB2, the autoubiquitylation of Cdt2 is mediated by Cul4A, but not Cul4B. In contrast, Cul4B and DDB1 are required for the stability of Cdt2, although it remains unclear how they stabilize Cdt2. Although further investigation is necessary to establish the basis of the functional difference between the two Cul4s, significant phenotypic differences have been described in mice null for Cul4A or Cul4B (Kopanja et al., 2011; Liu et al., 2012), consistent with the two proteins having different substrate specificities. We speculate that the ability of Cul4A to autoubiquitylate Cdt2 is critical for limiting the activity of CRL4Cdt2 following the degradation of its substrates.
The second pathway is the ubiquitin-dependent degradation of Cdt2 via CRL1FBXO11, providing an interesting example of cross regulation between the Cul4 and Cul1 ubiquitin ligases. To our knowledge, this represents the first report of a Cul4 substrate receptor regulation, and hence Cul4-specific ubiquitin ligase activity, by Cul1 ubiquitin ligases. This additional pathway for regulating the level of Cdt2 is independent of Cul4A and appears to be the dominant pathway for mediating the turnover of Cdt2.
FBXO11 interacts with Cdt2 via a conserved 9 amino-acid peptide located in a central fragment of Cdt2 (amino acids 456–464) and promotes the polyubiquitylation and degradation of Cdt2 in proliferating cells. We do not know whether this peptide can act as an independent “degron” when attached to heterologous proteins, but FBXO11 requires Ser462 in this peptide for interaction with Cdt2. Asp457 of Cdt2 on the other hand, is not required for FBXO11-Cdt2 interaction in vivo or for Cdt2 polyubiquitylation by CRL1FBXO11 in vitro. Therefore, the D457A mutation of Cdt2 affects degradation by CRL1FBXO11 by an unknown mechanism. We do not think that the failure to degrade Cdt2 D457A can be explained by a failure of the mutant to bind to chromatin because Cdt2D457A continues to degrade its own chromatin-associated substrates in vivo (p21, Set8). Most F-box-containing substrate receptors of CRL1 directly recognize and bind to one or more phosphorylations on the target substrate (Lipkowitz and Weissman, 2011). Mass-spectrometry analysis of wt-Cdt2 immunoprecipitated from asynchronously growing 293T cells did not detect phosphorylation on Ser462, although multiple serine and threonine residues located in the C-terminal half of the protein were phosphorylated (data not shown). However, pharmacological inhibitors against the kinases predicted to phosphorylate the mapped sites did not impact Cdt2 degradation by FBXO11 (data not shown). Thus, we suggest that Cdt2 does not require phosphorylation at the site recognized by the FBXO11. We cannot, of course, rule out a role of phosphorylation in the degradation without mutating all phosphorylation sites of Cdt2.
Importance of FBXO11-mediated restraint of CRL4Cdt2 in proliferating cells
In normal growth medium, the depletion of FBXO11 from asynchronously growing cells results in the accumulation of Cdt2 and reduction of CRL4Cdt2 substrate p21, but not of Set8 or Cdt1. The turnover of Set8 is significantly increased in cells depleted of FBXO11, but only if the culture medium contains low serum concentration. We propose that high serum activates other E3 ligases for degrading Set8, so that Cdt2 is not rate-limiting for the t1/2 of Set8. Indeed the t1/2 of Set8 in high serum (40 min) is less than in low serum (110 min) (Figure 5). The APCCdh1 and SCFSkp2 E3 ubiquitin ligases polyubiquitylate and promote the degradation of Set8 (Oda et al., 2010; Wu et al., 2010), and since both are cell cycle regulated, we suspect that they are more active in high serum concentration in the growth medium.
FBXO11 is necessary for the downregulation of Cdt2 to decrease the response to TGF-beta and for Set8-dependent cellular migration
FBXO11 regulates of the cell’s response to TGF-beta, but the mechanism by which it does so remained to be identified (Tateossian et al., 2009). In this study, we show that FBXO11 is essential to degrade the Cdt2 that is initially induced when TGF-beta is added, and thus stabilizes CRL4Cdt2 substrates p21 and Set8 during a critical window of 12–20 hr after TGF beta addition. FBXO11 is required for limiting Smad2 phosphorylation and N-cadherin induction at 24–48 hr after TGF-beta addition. The increased P-Smad2 signaling and N-cadherin in FBXO11-depleted cells is a consequence of failure to induce Set8 during the 12–20 hr window because siRNA-mediated depletion of Set8 had similar effects on phosphorylation of Smad2 and induction of N-cadherin. Our results are consistent with the reported enhanced P-Smad2 signaling in mice with inactivated FBXO11 (Tateossian et al., 2009). Our results also demonstrate a critical role for the FBXO11-mediated degradation of Cdt2 in cellular migration through the stabilization of catalytically active Set8 because overexpression of Set8 rescued the migration deficits produced by constitutively stable Cdt2. Thus, our studies establish a critical role for Set8 in cellular migration and demonstrate that FBXO11 ensures the existence of sufficient pools of Set8 to promote migration.
How Set8 decreases P-Smad2 or N-cadherin and promotes cellular migration remains to be investigated. We speculate that changes in gene expression from altered methylation of histone H4, or altered methylation of as yet unknown substrates of Set8, are involved. By identifying Cdt2 and Set8 as missing links in the chain from FBXO11 to both TGF-beta response and cell migration, our finding provides a mechanistic understanding of the factors underlying the developmental defects observed in mice with homozygous mutations of the FBXO11 gene. These mice have cleft-palate and eyes open at birth, developmental defects that often result from defects in cell migration (Hardisty-Hughes et al., 2006).
Finally, FBXO11 has recently been found to be a haplo-insufficient tumor-suppressor gene mutated or deleted in a subset of human diffuse large B-cell lymphomas (DLBCL) through its ability to promote the degradation of the tumor oncoprotein BCL6 (Duan et al., 2012). On the other hand, Cdt2 is overexpressed in human breast, liver and gastric cancers and is amplified in Ewing sarcomas, suggesting that Cdt2 may exhibit oncogenic activity (Abbas and Dutta, 2011). Thus, it will be interesting to see whether stabilization of the Cdt2 oncoprotein and decrease of Cdt2 substrates like p21 and Set8 contribute to the development of DLBCLs upon FBXO11 inactivation.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Reagents
U2OS, HeLa, 293T and A549 cells were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and antibiotics. HaCat cells (from Michele Pagano, NYU) were propagated similarly. Antibodies were purchased: anti-p21 (C-19), -PCNA (PC10), -MCM2, -N-cadherin, -Hemaglutinin (HA), -actin and -tubulin (Santa Cruz Biotechnology); anti-FBXO11 (Bethyl Laboratories); anti-Set8 and -phospho-Smad2 (Ser465/467) (Cell Signaling); anti-DDB1 (Invitrogen; Carlsbad, CA). The anti-Cdt2 antibody was described before (Abbas et al., 2008). Cells were lysed in Triton X-100 lysis buffer (50 mM Tris at pH 7.5, 250 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 50 mM NaF and protease inhibitor cocktail (Sigma)). Additional experimental procedures are available in the supplemental materials and methods.
In vivo and in vitro polyubiquitylation of Cdt2
For the in vivo ubiquitin labeling of Cdt2 (Figure 3D), 293T cells were transiently transfected with HA-ubiquitin and Flag-Cdt2 ± myc-tagged FBXO11 proteins or flag-tagged F-box proteins as indicated. Cells were treated with MG132 (20 μg/mL) for two hr prior to harvesting under denaturing conditions as described previously (Abbas et al., 2008). Immunoprecipitated ubiquitylated proteins (anti-HA) were fractionated on SDS-PAGE, transferred, and immunoblotted with anti-Cdt2 antibody. A similar approach was used in Figure 3E to detect ubiquitylated endogenous Cdt2 (without overexpression of ubiquitin) except cells were transfected with si-GL2, si-FBXO11 or si-Cul4A and the membranes blotted with anti-ubiquitin lys 48-specific antibody (Cell Signaling). The in vitro ubiquitin labeling of Cdt2 was done as described (Abbas et al., 2008), except overexpressed and immunoprecipitated Cdt2 protein was used as substrate and overexpressed and immunoprecipitated FBXO11 (along with Cul1, Skp1 and Rbx1) as enzyme in the reaction (Figure 3F). For in vitro autoubiquitylation of Cdt2, overexpressed and immunoprecipitated (anti-myc) CRL4Cdt2 (Myc-Cul4A (or Cul4B), Myc-DDB1, Flag-Cdt2 and Flag-Rbx1) was used without addition of substrate (Figure 1F). We also used overexpressed and immunoprecipitated (anti-flag) Flag-Cdt2 or Flag-Cdt2R246A without overexpression of any other component of the CRL4Cdt2 complex (Figure 1G).
Supplementary Material
Highlights.
Cullin 4A promotes the autoubiquitylation of Cdt2, a substrate adaptor of CRL4Cdt2.
CRL1FBXO11 also ubiquitylates and promotes the degradation of Cdt2.
FBXO11 degrades Cdt2 to stabilize Set8 and regulate the response to TGF-beta.
FBXO11 is similarly required to stabilize Set8 and promote cell migration.
Acknowledgments
We would like to thank Marty Mayo (University of Virginia) and Michele Pagano (New York University) for providing valuable reagents, and members of the Dutta lab for valuable discussions. This work was supported by NIH grants, R00CA140774 to T. A. and R01CA60499 to A. D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle. 2011;10:241–249. doi: 10.4161/cc.10.2.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, Zollner H, Munding J, Klein-Scory S, Reinacher-Schick A, Schwarte-Waldhoff I, Schmiegel W, Hahn SA. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting DTL. Carcinogenesis. 2012;33:732–739. doi: 10.1093/carcin/bgs020. [DOI] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem. 2001;276:48175–48182. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisty-Hughes RE, Tateossian H, Morse SA, Romero MR, Middleton A, Tymowska-Lalanne Z, Hunter AJ, Cheeseman M, Brown SD. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet. 2006;15:3273–3279. doi: 10.1093/hmg/ddl403. [DOI] [PubMed] [Google Scholar]
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Banks D, Wu M, Kobayashi R, Sun H, Zhang H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle. 2006a;5:1675–1680. doi: 10.4161/cc.5.15.3149. [DOI] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006b;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Eskildsen M, Fugger K, Hansen L, Larsen MS, Kousholt AN, Syljuasen RG, Trelle MB, Jensen ON, Helin K, Sorensen CS. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol. 2011;192:43–54. doi: 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22:2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopanja D, Roy N, Stoyanova T, Hess RA, Bagchi S, Raychaudhuri P. Cul4A is essential for spermatogenesis and male fertility. Dev Biol. 2011;352:278–287. doi: 10.1016/j.ydbio.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ng EK, Ng YP, Wong CY, Yu J, Jin H, Cheng VY, Go MY, Cheung PK, Ebert MP, et al. Identification of retinoic acid-regulated nuclear matrix-associated protein as a novel regulator of gastric cancer. Br J Cancer. 2009;101:691–698. doi: 10.1038/sj.bjc.6605202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yin Y, Li Y, Prevedel L, Lacy EH, Ma L, Zhou P. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 2012 doi: 10.1038/cr.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh C, Ordonez JL, Garcia-Dominguez DJ, Sevillano V, Llombart-Bosch A, Szuhai K, Scotlandi K, Alberghini M, Sciot R, Sinnaeve F, et al. 1q gain and CDT2 overexpression underlie an aggressive and highly proliferative form of Ewing sarcoma. Oncogene. 2012;31:1287–1298. doi: 10.1038/onc.2011.317. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Induction of epithelial-mesenchymal transition by transforming growth factor beta. Semin Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Nag A, Bondar T, Shiv S, Raychaudhuri P. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol Cell Biol. 2001;21:6738–6747. doi: 10.1128/MCB.21.20.6738-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan HW, Chou HY, Liu SH, Peng SY, Liu CL, Hsu HC. Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle. 2006;5:2676–2687. doi: 10.4161/cc.5.22.3500. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, Palombella VJ. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, Hopkins N, Lees JA. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 2006;20:3117–3129. doi: 10.1101/gad.1482106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem. 2006;281:6246–6252. doi: 10.1074/jbc.M512705200. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateossian H, Hardisty-Hughes RE, Morse S, Romero MR, Hilton H, Dean C, Brown SD. Regulation of TGF-beta signalling by Fbxo11, the gene mutated in the Jeff otitis media mouse mutant. Pathogenetics. 2009;2:5. doi: 10.1186/1755-8417-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Nishidate T, Park JH, Lin ML, Shimo A, Hirata K, Nakamura Y, Katagiri T. Involvement of elevated expression of multiple cell-cycle regulator, DTL/RAMP (denticleless/RA-regulated nuclear matrix associated protein), in the growth of breast cancer cells. Oncogene. 2008;27:5672–5683. doi: 10.1038/onc.2008.186. [DOI] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.