Abstract
Normal cellular division requires that the genome be faithfully replicated to ensure that unaltered genomic information is passed from one generation to the next. DNA replication initiates from thousands of origins scattered throughout the genome every cell cycle; however, not all origins initiate replication at the same time. A vast amount of work over the years indicates that different origins along each eukaryotic chromosome are activated in early, middle or late S phase. This temporal control of DNA replication is referred to as the replication-timing program. The replication-timing program represents a very stable epigenetic feature of chromosomes. Recent evidence has indicated that the replication-timing program can influence the spatial distribution of mutagenic events such that certain regions of the genome experience increased spontaneous mutagenesis compared to surrounding regions. This influence has helped shape the genomes of humans and other multicellular organisms and can affect the distribution of mutations in somatic cells. It is also becoming clear that the replication-timing program is deregulated in many disease states, including cancer. Aberrant DNA replication timing is associated with changes in gene expression, changes in epigenetic modifications and an increased frequency of structural rearrangements. Furthermore, certain replication timing changes can directly lead to overt genomic instability and may explain unique mutational signatures that are present in cells that have undergone the recently described processes of “chromothripsis” and “kataegis”. In this review, we will discuss how the normal replication timing program, as well as how alterations to this program, can contribute to the evolution of the genomic landscape in normal and cancerous cells.
1. Introduction
In order to divide a eukaryotic cell must undergo precise DNA replication to ensure that an exact copy of its genetic content is passed on to its daughter cells. This process occurs during S phase and proceeds via the coordinated initiation of DNA replication at hundreds of replication origins scattered throughout the length of each chromosome [1]. Interestingly, the cell begins preparation for DNA synthesis in telophase of the prior cell cycle [2]. This is when the pre-replicative complex (pre-RC) begins to form on each potential origin of replication. However, not all pre-RCs will go on to become active replication origins. In mid-G1, at the origin decision point (ODP), some pre-RCs are chosen to become initiators of DNA replication while others remain inactive throughout S-phase [3,4]. The addition of other replication factors to a subset of the pre-RCs transforms them into pre-initiation complexes (pre-ICs) [5]. Shortly after the pre-IC is formed, DNA polymerase and primase are recruited to each origin and DNA synthesis begins in a bidirectional manner. DNA replication proceeds from each origin until the replication forks from two neighboring origins meet and the nascent DNA strands are ligated [6].
While DNA replication can initiate from any active origin within a given S-phase, the timing at which initiation takes place can vary widely between origins. Adjacent origins tend to initiate DNA replication at the same time resulting in large, synchronously replicating chromosomal domains called “replicon clusters” [7,8]. Some replicon clusters begin replication at the onset of S-phase while others begin later during the middle or near the end of S-phase. This coordination of the temporal control of DNA replication is referred to as the replication-timing program. The replication-timing program is established shortly after mitosis at a point in the G1 phase, preceding the ODP, called the timing decision point (TDP) [9,10]. The TDP is established coincidently with a global reorganization of chromatin into specified regions within the nucleus [10].
The replication-timing program is mitotically stable, heritable and subject to differential regulation during differentiation and development, making it a robust epigenetic feature of all eukaryotic chromosomes [11]. The biological significance of this replication-timing program is currently unknown; however, the existence of aberrant replication timing in many different genetic diseases suggests that it is a vital cellular process [12,13,14,15,16]. Not surprisingly, the 3-dimensional chromosome architecture in the nucleus is highly coordinated with DNA replication timing. In most, if not all, eukaryotic organisms, early-replicating DNA resides in the interior of the nucleus while the later-replicating regions remain at the nuclear periphery or near the nucleolus [10,17,18]. Molecular analysis has also revealed that late-replicating regions tend to cluster with other late-replicating regions in the nucleus and vice-versa [19]. Additional complex associations have been observed with genome sequence, structure and replication timing. For example, early-replicating regions tend to positively correlate with gene expression, G+C rich sequences, light-staining Giemsa bands, and active chromatin marks, while late-replicating regions tend to be gene-poor, A+T rich, and have repressive chromatin marks [17,20,21]. It should be pointed out that while these correlations are significant they are not absolute, as some expressed genes with transcriptional active chromatin marks reside in late-replicating regions [8].
DNA synthesis occurs in replication factories within the 3-dimensional space of the nucleus. In these factories, regions of similar replication timing cluster together in the nucleus, with early-replicating regions residing in the nuclear interior and late-replicating regions remaining at the nuclear periphery or near the nucleolus [9,17,19,22]. Additionally, replication-timing changes that occur during development are accompanied by changes in nuclear architecture, indicating that these two features are very closely linked [23]. Therefore, regions that replicate at comparable times in S phase tend to have a closer spatial association than regions that replicate at different times. This association has been highlighted using the HiC method, which probes the three-dimensional architecture of whole genomes by coupling proximity-based ligation with massively parallel sequencing [19,24].
One prominent disease that is characterized by replication-timing aberrations is cancer. Cancer develops when normal cells acquire genetic and epigenetic alterations that lead to uncontrolled growth and the ability to evade cell death. These genetic and epigenetic alterations are generally thought to drive carcinogenesis by deregulating key pathways that control cell growth and proliferation [25]. Genetic alterations can arise during cancer progression through normal cellular processes, induced or spontaneous mutagenesis, or as a result of genomic instability. Mutagenesis refers to the process by which genetic changes occur, either spontaneously or as a consequence of exposure to mutagens, resulting in a change in the DNA sequence. Genomic instability, on the other hand, refers to an increase in the rate of mutagenesis per unit time. While normal cells have a very low intrinsic mutation rate, any mechanism that increases the mutation rate can be said to cause genomic instability. Current models suggest that an underlying genomic instability is responsible for the rapid accumulation of the genetic and epigenetic changes that affect gene function in cancer [25]. Therefore, it is very difficult to understand cancer development without understanding the mechanisms that cause genomic instability.
In this review, we highlight research suggesting that the normal DNA replication-timing program has a profound impact on the distribution of mutations that arise during the evolution of species as well as during the evolution of cancer. Aberrant DNA replication timing is associated with altered gene expression, mutagenesis and genomic instability. Furthermore, we propose that certain DNA replication-timing aberrations can explain the newly described processes of “chromothripsis” and “kataegis”, which have been found to generate unique genomic signatures in the genomes of some tumor cells [26,27].
2. DNA Replication Timing and the Evolution of the Genomic Landscape
The conventional view of evolution assumes that DNA mutations occur randomly throughout the genome and the eventual presence or absence of those DNA changes in the population is determined through the process of natural selection. While natural selection remains the most potent force shaping the evolution of the genomic landscape, the notion that DNA mutations occur randomly in the genome has become outdated. We now know that mutation rate varies widely throughout the genome and is influenced by many local genetic and epigenetic features such as recombination rate, CpG content, transcriptional status, repetitive-sequence content and chromatin conformation [28,29,30]. Although it was observed more than 20 years ago [31], a wealth of recent experimental data has confirmed that DNA replication timing is also a potent force that influences mutation rates.
An elegant series of experiments in yeast established that late-replicating regions of the genome have higher rates of spontaneous mutagenesis than early-replicating regions. By inserting an exogenous sequence into different regions of a chromosome and calculating the rate of mutations occurring in that sequence, Lang et al. demonstrated that there was a strong positive correlation between the time of replication and the rate of mutation [32]. Furthermore, by deleting an early-replicating origin, and consequently delaying replication, near one of these exogenous sequences they observed a slight increase in the rate of mutagenic events. This indicates that delaying the initiation of DNA replication at a particular sequence is sufficient to increase its mutation rate [32].
Other experiments have demonstrated that endogenous loci from many different organisms show a similar correlation of mutagenesis and replication timing. Regions of single-nucleotide diversity in mice and humans are enriched in late-replicating regions [14,33,34,35]. When comparing the human genome to multiple non-human primate genomes it was also observed that areas of single-nucleotide divergence between species disproportionately lie in late-replicating regions [33,34]. A parallel correlation between divergence and late replication was also seen when comparing the genomes of mice and rats [34]. Similarly, regions that have a high density of duplications tend to be late replicating in flies [36]. And duplication hotspots that are shared between different species of flies also reside preferentially in regions of late replication [37]. Genomic domains prone to duplication events are also hotspots for neutral point mutations [38,39], further supporting the idea that mutational events occur in spatial proximity to one another (i.e. in late-replicating genomic regions). Interestingly, a correlation between early replication timing and deletion variation in flies was also observed, and it will be interesting to determine if this holds true for other eukaryotes as well [36]. These data indicate that timing of DNA replication may be a driving force in copy number and single-nucleotide polymorphism (SNP) diversity that is observed within a species and between species. There have been several suggestions for why replication timing and neutral mutation rate correlate so closely. Most involve differential repair mechanisms being used at different times during S phase such that error-prone DNA repair pathways are utilized more frequently during late S phase [32,34].
Late-replicating regions aren’t the only replication-associated sites of genomic change. There is also evidence that replication transition regions are also hotspots for spontaneous mutagenesis. Replication transition regions are areas that lie between early-replicating DNA and late-replicating DNA and, therefore, often replicate in mid to late S phase. Transition regions are void of origins and are passively replicated by a uni-directional fork that initiates at an adjacent early origin. This single fork replicates the entire transition region until it reaches the replication fork of an adjacent late origin [23]. One consequence of such a large replicon is an increase in the probability of replication fork stalling and DNA damage [23,40]. Indeed, a survey of SNPs on human chromosomes 11 and 21 indicated that there is a higher density of SNPs in replication transition regions than there are in early-replicating regions [14]. In addition, frequent gene amplifications on these same chromosomes also lie within these replication transition regions [41]. The frequent gene amplification that is seen in late-replicating and transition regions may have functional implications during evolution, as gene duplications are considered to be an important factor during speciation [42]. Additionally, syntenic breakpoints in the mouse and human genomes appear to occur predominantly in transition regions [41], indicating that these regions may be sources of breakage during the generation of new chromosomes during evolution. Of course more comprehensive studies will be needed to confirm these findings, but these observations nevertheless implicate DNA replication timing as a potent regulator of mutational dynamics.
Now that DNA replication timing is increasingly implicated in establishing a gradient of mutagenesis such that late replicating and transition regions have a higher rate of mutation than early replicating regions, it is important to understand why a replication timing program exists at all. It is well established that most regions of active gene transcription are early replicating, whereas silenced genes, intergenic regions and repetitive sequences are late replicating [43]. Most silenced genes (late replicating) tend to be tissue-specific and only become expressed (early replicating) in the tissue where they function [20,44]. Incidentally, many tissue-specific genes, e.g. receptors involved in sensing environmental changes and during the immune response, are much more divergent between species than genes involved in essential cell functions like metabolism and transcription [38,45]. It is tempting to speculate that replication timing might be a way to optimize the intrinsic mutation rate such that housekeeping genes incur fewer mutations while tissue-specific genes, that are generally under greater pressure to adapt, receive an increased mutational load. Interestingly, it was found that different classes of genes tend to reside in regions with differential substitution rates between mouse and human [38]. Genes with “receptor type” functions (cell adhesion, immune function, olfactory receptors) generally reside in regions of high mutation density while genes involved in RNA binding, kinase activity and metabolism reside in regions of low mutation density [38]. As we get deeper into the age of genomics it will be interesting to see what other trends emerge with respect to the evolution of the genomic landscape.
3. DNA Replication Timing and the Evolution of the Cancer Genome
The observations described above indicate that replication timing influences the mutation rate of different genomic regions in the germline, and over long periods of time differences in replication timing can contribute to the genetic variation within and between species. However, there is also increasing evidence that replication timing influences the mutation rate in somatic cells and may be a contributing factor to the distribution of genomic changes that arise during cancer development.
3.1. The Role of Normal DNA Replication Timing During Cancer Mutagenesis
An extensive sequence-based analysis of many different human tumors has revealed an increase in single-nucleotide variations (SNVs) and somatic copy number alterations (SCNAs) in late replicating regions of the genome [22,46]. Interestingly, in these tumor samples, genomic deletions are enriched in late replicating regions whereas amplifications are enriched in early replicating regions, which is opposite of what is seen in the germline [22]. Other groups have found that genomic rearrangements in cancer correlate differently with replication timing depending on the type of tumor studied. For example, rearrangement breakpoints in breast cancer and neuroblastoma tend to lie within early-replicating regions, whereas breakpoints in colorectal cancer and melanoma tend to reside in late replicating regions [47,48]. This variation highlights the epigenetic heterogeneity of different tumor types and may be due to differential selective constraints imposed during tumor evolution or differential deregulation of DNA repair pathways.
Similar to what is seen in the germline, transition regions seem to be hotspots of copy number alterations in human cancers [22,41]. As mentioned above, the complicated nature of DNA replication in these regions makes them especially susceptible to fork stalling and DNA damage [40]. Accordingly, many fusion genes and recurrent chromosome aberrations found in cancer lie within or near transition regions [14,49]. Not only are these regions prone to mutagenesis in cancer, but also harbor a higher proportion of genes with oncogenic and tumor-suppressing functions [14,22,41,49]. It has been proposed that the spatial proximity between regions of similar replication timing can influence translocation and rearrangement sites in the genome [22,50]. This is seen in many cancer cells where regions that cluster next to one another in the nucleus are more likely to undergo translocation events than more distant regions [22,51,52,53]. Not surprisingly, many recurrent and oncogenic translocations occur between regions of similar replication timing and nuclear proximity [51,54,55].
An additional unstable feature of most if not all mammalian chromosomes is the presence of chromosome fragile sites (CFSs) [56,57]. Common CFSs are discrete regions of chromosomes that are prone to breakage during times of replication stress [58]. CSFs have been found to lie at the interface of R and G chromosome bands [59,60], which is a hallmark of replication transition regions [40]. This indicates that transition regions and CFSs may represent the same genomic feature. Accordingly, CFSs are common points of chromosomal breakage in tumors and CFS instability is often seen in the early stages of carcinogenesis [61]. Since some cancer-related genes lie within fragile sites, CFS instability can directly deregulate some oncogene/tumor-supressor functions [61]. Thus, not only does DNA replication timing influence the rate of mutagenesis, it can also bias the location of rearrangement breakpoints.
The above data compare replication-timing profiles in normal cells to the acquisition of mutations in cancer cells, with the observation that at least some of the mutagenesis observed in cancer is collateral damage of having a normal replication-timing program. However, there is accumulating evidence that there are numerous alterations to the normal replication-timing program during carcinogenesis. In fact, there is abundant evidence indicating that changes in replication timing often accompany cancer development. While the extent to which these replication-timing changes influence the transformation process is still largely unknown, the presence of these changes in many different types of tumors indicates that altered replication timing may be an important component in tumor development.
3.2. Aberrant DNA Replication Timing in Cancer
DNA replication is a highly regulated process. For most of the genome, homologous loci replicate at the same time during S phase in a highly coordinated manner. Exceptions to this rule are represented by loci that display a mono-allelic gene expression pattern. Thus, mono-allelically expressed genes such as imprinted genes, allelically excluded genes and genes on female X chromosomes replicate asynchronously with one allele replicating before the other. This replication pattern is very stable in normal cells, and is independent of transcription [62].
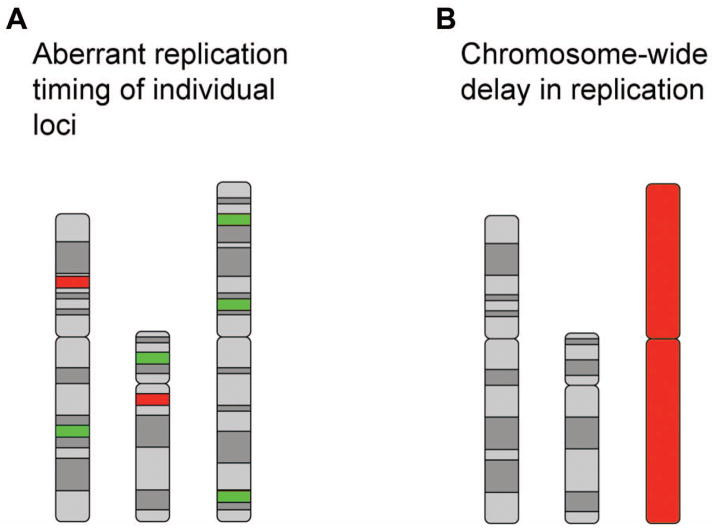
One well-documented change in cancer cells is the aberrant asynchronous replication of loci that normally replicate synchronously [63,64,65]. The early studies that looked at individuals with cancer assayed individual loci and, therefore, gave no indication of how widespread this aberrant asynchronous replication was throughout the genome. In contrast, a recent whole-genome replication timing study indicated that 9–18% of the genome undergoes a change in replication timing in leukemia cells compared to normal controls ([16]; see Fig. 1A). Changes in replication timing were detected on all chromosomes and were evenly distributed throughout the genome. Although there were slight differences between different types of leukemias, many of the changes in replication timing were common to all samples, suggesting that altered replication at specific locations is an early epigenetic event in cancer development [16]. In addition, many but not all of the replication-timing changes occurred near sites of genomic rearrangement. Indeed, a replication-timing change was found at a common site of translocation in leukemia cells [16]. However, all leukemia cells studied displayed this replication-timing change, but only a few displayed the actual translocation, indicating that replication-timing changes may predispose the cell to certain translocation events [16]. In addition, instead of genomic rearrangements correlating with small local changes in DNA replication timing, the changes were extensive and extended hundreds of kilobases beyond the site of rearrangement. Similar to the studies mentioned previously, the replication timing changes were generally from late-replicating regions replicating earlier, and fewer early-replicating regions replicating later [16]. This study was not calibrated to detect replication asynchrony, so it is unclear whether these loci that change replication timing replicate synchronously or asynchronously. However, it is likely that some of the site-specific replication asynchrony that was observed in the above reports is reflected in this genome-wide analysis. It will be interesting to determine if the replication-timing changes that occur genome-wide in different types of cancers are the result of a specific chromosomal feature (e.g. asynchronous replication) or if this deregulation represents a more heterogenous, nonspecific change.
Figure 1.
Acquired alterations in DNA replication timing in cancer cells. A) Examples of individual loci that display a shift in replication timing. Loci that shift to an earlier time of replication are indicated in green, and regions that shift to a later time of replication are indicated in red. Three different chromosomes are shown. B) An example of an individual chromosome with a chromosome-wide delay in replication (red). Two chromosomes with normal replication timing are shown in grey.
Surprisingly, the asynchronous replication pattern observed in cancer patients is not restricted to tumor tissue but also occurs in noncancerous cells as well [66,67,68,69,70]. This is best exemplified by the presence of aberrant asynchronous replication between alleles in the peripheral lymphocytes of individuals with solid tumors [66,67]. This replication asynchrony has been documented at multiple loci for many cancer-related genes and many other genomic locations indicating that this is not just the deregulation of a single locus or a single chromosome, but a widespread phenomenon [66,68,71]. Interestingly, this altered replication-timing pattern is present in pre-malignant cells, in individuals pre-disposed to cancer, and in individuals living in polluted areas with a high likelihood of getting cancer, suggesting that this may be an early event during carcinogenesis [63,65,67,72]. The replication asynchrony observed in cancerous tissue and normal cells in individuals with cancer is generally a result of the earlier replication of one of the alleles [67,70,72], however, in some cases the delayed replication of one allele has been detected [71]. This aberrant asynchronous replication is heritable (i.e. the earlier-replicating allele will be earlier replicating in all subsequent generations) but not dependent on parental origin and therefore resembling the process of human X chromosome inactivation and/or allelic exclusion [73]. Chemotherapy is not sufficient to correct the cancer-associated replication asynchrony detected in lymphocytes of cancer patients [70], but allogeneic stem cell transplantation, in vitro fusion to normal cells, or inhibiting DNA methylation by treating cells with 5-azacytidine can switch replication back to the normal state [69,73,74,75].
The replication-timing abnormalities mentioned above effect loci present on many different chromosomes and are detected sporadically throughout the genome. They generally result in the advancement of the replication-timing program such that one or both alleles will replicate earlier than normal. In contrast, a functionally distinct replication timing aberration has also been observed in tumor-derived cells. In 1967, Dr. Harald zur Hausen documented a delay in replication timing of individual chromosomes in cultured leukemia cells ([76]; see Fig. 1B). This chromosome-wide replication delay has since been observed in many different tumor-derived cell lines and primary tumor samples [77,78]. These studies indicated that some tumor cells contain individual chromosomes that are delayed in initiation and completion of DNA replication by 2–3 hours along their entire length [78,79]. This replication delay affects the entire chromosome but does not disrupt the replication timing of other chromosomes in the cell, and is therefore controlled by a cis acting mechanism. Whole chromosomes that are delayed in DNA replication timing also exhibit a delay in mitotic chromosome condensation and are associated with highly aneuploid karyotypes [78,80,81]. In addition, it has been demonstrated that this chromosome-wide replication delay increases the rate of secondary chromosomal rearrangement on the affected chromosome by 30–80 fold, indicating that this chromosome-wide replication delay causes genomic instability [81]. Unlike the genome-wide replication-timing changes mentioned previously, which tend to be a very stable feature of cancer cells during the progression of the disease, chromosome-wide replication delay is a more transient feature. The inherent instability of the delayed chromosomes makes them prone to extreme fragmentation occurring over a relatively few cell divisions, which eventually results in highly rearranged chromosomes that no longer display replication delay [78]. The transient existence of chromosome-wide replication delay in cancer cells makes it an underappreciated, yet potentially important force driving mutagenesis in cancer cells.
It is apparent that chromosome-wide replication delay can have a profound impact on the structural stability of individual chromosomes by increasing the rate of chromosome rearrangements [81]. In contrast, it is unclear whether the other sporadic genome-wide replication-timing changes that occur at specific loci are contributing to instability or are merely correlative. Due to the early onset of genome-wide replication-timing changes in cancer development (in some cases preceding malignancy) it is likely that this deregulation is directly or indirectly linked to transformation. As discussed below, replication-timing changes can be generated by different mechanisms and can lead to genetic and epigenetic changes within the cell. In the right context, any of these changes has the potential to influence cell growth and survival.
3.3. Causes and Consequences of Aberrant DNA Replication Timing
Depending on the context, some examples of aberrant DNA replication timing appear to have direct consequences on mutagenesis; however, in many cases the temporal order of events has not been established. Consequently, we are left with indirect conclusions to establish a timeline of events. The interconnectedness of many cellular properties like chromatin modifications, replication timing, transcription, and nuclear positioning complicates a cause and effect analysis, because the experimental manipulation of one will, in many cases, affect all three. This has led to the proposal that these properties are interdependent, such that a change in any one of them will have an effect on the others [17,82,83]. While keeping this in mind, we have highlighted some examples that indicate a direct relationship between specific cellular events and changes in replication timing.
A) Gene Expression Changes
DNA replication is a complicated cellular process involving the coordinated action of many different gene products. The deregulated expression of certain genes involved in DNA synthesis can cause defects in replication timing. For example, mutations in ORC proteins [84], cyclins [85], CDKs [86], nucleotide reductases [87], and other proteins involved in DNA structure checkpoints [86,88,89] have all been shown to cause abnormal DNA replication-timing patterns. Consistent with a relationship between chromatin modifications and DNA replication timing, the deregulation of many chromatin-modifying enzymes can impact the temporal replication of loci throughout the genome [90,91,92,93,94,95]. Disruption of these genes has a trans-acting effect, meaning it impacts the replication timing of distant loci on multiple chromosomes. Accordingly, deregulated HP1 gene expression was found to change the replication timing of 5–10% of genomic loci, suggesting a widespread effect [94]. In some cases, the replication timing changes that result from the deregulation of trans-acting factors resembles the genome-wide replication-timing changes seen in some cancers [16], which indicates that deregulation of replication components and chromatin modifiers may be one cause of abnormal replication timing in cancer cells.
One unique aspect of changes in replication timing is that the consequences on transcription are only observed in cis. For example, it was found that the advanced replication timing of loci on the inactive X chromosome was critical for their escape from gene inactivation [96]. Other studies have indicated that replication timing changes at specific loci can occur upstream of gene transcription changes [97,98]. However, it should be noted that a change in the replication timing of a particular gene does not always cause a change in transcription [71]. This has led to the idea that a change in replication timing is not sufficient to cause a change in transcription, and that other factors, such as the presence and activity of transcriptional activators, are required. Thus, replication timing may not affect transcription directly but, rather, affect transcriptional competence [11].
B) Epigenetic Changes
Although DNA replication timing is strongly associated with gene expression, the association is even stronger with some epigenetic modifications. Certain chromatin and DNA methylation states appear to have a substantial impact on replication timing. Tethering a histone acetylase to a late replicating origin is sufficient to convert it to early replicating, and the opposite is true when a histone deacetylase is brought to an early replicating origin [99]. Many different groups have observed that histone acetylation changes can precede changes in replication timing [100,101,102,103], which is consistent with histone deacetylation occurring at the G1/S transition prior to late-origin replication [104]. Changes in histone methylation have also been implicated in causing changes in DNA replication timing [91,95], bolstering the concept that chromatin accessibility and DNA replication timing go hand-in-hand. Similar to many histone modifications, changes in DNA methylation have also been found to precede changes in replication timing [92,105], and manipulation of DNA methylation can even reverse aberrant replication timing under certain conditions [69,73,74].
The studies mentioned above indicate that changes in histone modifications can switch the replication timing of an origin, but some chromatin modifiers can also inhibit the firing of an origin altogether [93]. Indeed, the addition or subtraction of origins can affect the replication timing of adjacent regions by decreasing or increasing the time it takes the replication fork to reach them [97,106,107,108]. Since most loci are replicated by clusters of origins, the addition or deletion of one origin generally won’t have much of an impact on replication timing. However, if a change in origin usage occurs in a region that is devoid of origins, like a transition region, then the impact can be large. This differential origin usage model has been used to describe why transition regions are so prone to drastic changes in replication timing [40]. In support of this model, the addition of an origin at the Igh locus, which lies in a transition region, was found to coincide with a shift from late to early replication [97].
In keeping with the interdependent theme of this section, changes in replication timing can also precede changes in epigenetic modifications. Studies in Dr. Howard Cedar’s lab found that plasmid DNA injected into a cell in early S-phase will be packaged into acetylated chromatin while DNA injected into cells in late S phase will be associate with hypoacetylated chromatin [109]. This was followed by the demonstration that a plasmid containing hypoacetylated histones injected into cells in early S phase will become remodeled with acetylated histones during replication and vice versa [110]. This suggests that the time of DNA replication within S phase can dictate the acetylation state of histones that are loaded onto DNA. This has led some to speculate that the differential association of various chromatin modifiers with DNA throughout S phase is responsible for the close association between DNA replication timing and chromatin modifications [83]. This notion is supported by the demonstration that some repressive histone modifiers and transcriptional repressors only localize to replication foci in mid-late S phase [111,112,113]. Therefore, a change in replication timing can change the chromatin landscape, and transcriptional competence, of a particular region by dictating which chromatin modifiers can associate during replication. DNA replication-timing changes have also been observed to occur before DNA methylation changes, indicating that the temporal order of replication can also affect the methylation status of DNA [114,115].
Although the cause and effect relationship between DNA replication timing and chromatin/DNA modifications has been studied extensively, there is still much more to learn. Furthermore, we still lack a good understanding of how DNA replication timing and 3-dimensional nuclear structure affect one another. Some studies have indicated that a change in nuclear position is not sufficient to change DNA replication timing [18,102,116], which would suggest that DNA replication timing determines nuclear position. However, other studies have suggested that the presence of replication foci in specific nuclear compartments dictates the temporal order of replication [117]. It is likely that both models are correct in certain contexts. As is the case with transcription and epigenetic modifications, DNA replication timing seems to be controlled by as many cellular events as it controls.
The effects of aberrant DNA replication timing on chromatin structure can extend beyond S phase as well. It has been observed that chromosome-wide delayed DNA replication can lead to abnormal mitotic chromosome condensation in early mitosis [78,84]. This delay in condensation coincides with a delay in the recruitment of Aurora B kinase resulting in a delay in the mitosis-specific phosphorylation of histone H3 [78,79]. Therefore, delayed replication can lead to chromosomes that are in an “interphase state” of condensation during mitosis [78].
C) Genetic Changes
Perhaps it is no surprise that genetic changes can be the cause of aberrant DNA replication timing, but the varied types of genetic damage and replication-timing changes discussed in this section indicate that this relationship is far from straightforward. Treating cells with various DNA damaging agents, such as ionizing radiation, hydrogen peroxide, and mitomycin C can lead to aberrant replication timing [72,118]. Furthermore, double-strand breaks caused by site-specific recombination, ionizing radiation and endonuclease digestion can induce a chromosome-wide delay in replication [80,81], indicating that DNA damage can cause different types of replication-timing defects.
Different types of DNA damaging agents can produce different types of mutations, and different types of mutations have been implicated in replication-timing changes. For example, nucleotide substitution at CTCF binding sites can deregulate allele-specific replication in imprinted regions [119]. In addition, telomere shortening can advance the replication timing of telomeric origins [120]. It has been known for some time that the juxtaposition of genetic regions to non-native loci can cause replication-timing changes [121,122,123]. One common type of genomic rearrangement in cancer is inter-chromosomal translocation, which brings together two regions from two different chromosomes. It has been observed that chromosomal translocations often accompany replication-timing changes [124,125,126]. Many of these replication-timing defects result from the newly acquired replication of homologous loci due to the juxtaposition of one locus to another [125,126]. In fact, most translocations that juxtapose regions of differential replication timing result in the earlier or later replication of at least one of the translocated alleles [16]. Because these abrupt replication-timing changes occur by juxtaposing an early-replicating region with a late-replicating region, it should be noted that a translocation involving regions of similar replication timing would not be expected to result in a replication-timing change [16]. These studies indicate that the majority of translocation events cause replication-timing changes that are relatively minor, and only affect local sequences or domains (Fig. 1A).
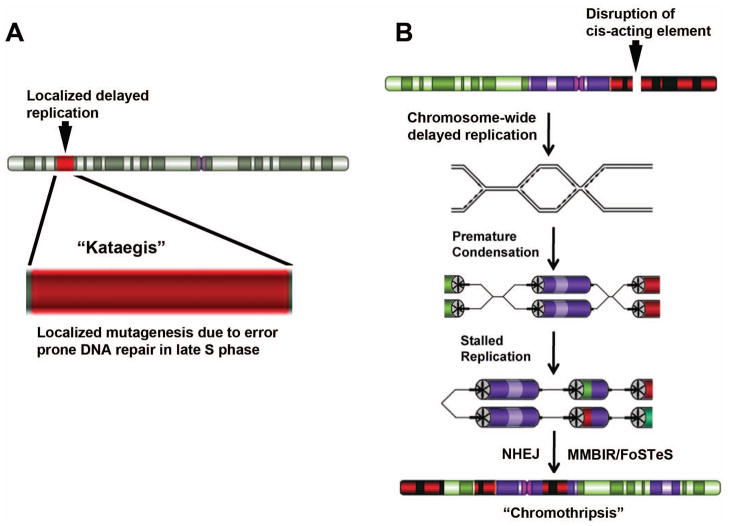
A recent phenomenon of localized hypermutation, termed “kataegis,” was observed in some cancer cells [26]. Kataegis is characterized by an increase in the frequency of SNVs in a particular region of the genome. Regions of kataegis differed between cancers, but usually colocalized with somatic rearrangements. While the mechanisms responsible for kataegis remain unknown, we propose that the localized replication-timing changes that occur near or prior to translocation breakpoints [16] could be responsible for the localized mutagenesis observed in the localized regions with kataegis (Fig. 2A).
Figure 2.
Models for localized genomic instability in cancer cells. A) Aberrant late replication model for kataegis. A localized region of a chromosome has acquired abnormally late replication (red) either as a result of chromosome rearrangement or as a result of a localized shift in the replication timing program [16]. Increased mutagenesis is induced in the late replicating region due to error prone repair mechanisms functioning during late replication. B) Aberrant late replication model for Chromothripsis. Disruption of discrete cis-acting loci result in a chromosome-wide delay in replication timing. Mitotic chromosome condensation initiates on the delayed chromosome prior to completion of DNA synthesis resulting in premature chromosome condensation, stalled replication forks, and rearrangement of the affected chromosomes via non-homologous end joining (NHEJ), microhomology mediated break induced replication (MMBIR) and fork stalling and template switching (FoSTeS) mechanisms. The resulting chromosome contains numerous structural alterations (translocations, deletions, inversions, and duplications).
In addition, genomic rearrangements can have larger effects than just the change in temporal replication of a specific locus or domain. Unlike juxtaposition-induced replication asynchrony, where the rearranged locus is the only site of aberrant replication timing, certain chromosomal rearrangements cause a chromosome-wide delay in replication timing of the entire chromosome [78,81,127,128,129]. This chromosome-wide effect is a result of the disruption of cis-acting elements that normally act to ensure the proper replication-timing program of individual chromosomes. A recent series of “chromosome-engineering” studies led to the identification of a discrete cis-acting locus that controls chromosome-wide replication timing and structural stability of human chromosome 6 [127]. Molecular characterization of this chromosome 6 locus identified a large intergenic non-coding RNA gene, which was named asynchronous replication and autosomal RNA on chromosome 6 (ASAR6). Cre/loxP-mediated disruption of the ASAR6 gene results in extremely late replication, an under-condensed appearance during mitosis, and structural instability of human chromosome 6 [81,127]. In a separate series of experiments, it was found that disruption of the large non-coding RNA gene Xist, results in extremely late replication, abnormal chromatin structure and instability of the X chromosome [128,129]. The Xist gene resides within the X inactivation center, and is known to participate in the silencing of genes during dosage compensation in female cells [130]. Interestingly, ASAR6 shares many characteristics with Xist, including random mono-allelic expression, asynchronous replication timing, and regulation of the expression of linked mono-allelic genes [127].
Furthermore, this chromosome-wide delayed replication timing phenotype has been detected on chromosome rearrangements involving many different human and mouse chromosomes [78,80,81,127]. Therefore, it seems likely that all mammalian chromosomes contain loci that function to regulate chromosome-wide replication timing, mitotic condensation and stability of individual chromosomes. Given the similarities in structure and function of the two loci characterized to date, Xist and ASAR6, it was proposed that all mammalian chromosomes contain functional chromosome “inactivation/stability centers” that act to maintain proper replication timing and structural stability of individual chromosomes [127]. Under this scenario every mammalian chromosome contains four cis-acting elements, origins of replication, centromeres, telomeres, and ‘inactivation/stability centers’, all functioning to ensure proper replication, segregation and stability of individual chromosomes [131].
Chromosome-wide delay in replication timing results in at least two distinct types of genomic instability. The first is chromosome instability (CIN), which is characterized by an increase in the rate at which cells gain or lose entire chromosomes [132]. Thus, cells with chromosome-wide delayed replication timing of individual chromosomes display frequent gains or losses of entire chromosomes resulting in dramatic aneuploidy affecting the entire karyotype [78,79]. In addition, cells containing chromosome-wide delayed replication contain abnormal mitotic spindles, abnormal centrosome number, and an increased frequency of endoreduplication [79]. It is unclear how chromosome-wide replication delay on one chromosome is causing these events, but these factors can certainly explain the CIN observed in cells with individual chromosomes with the delayed replication phenotype. The second type of instability observed in cells with chromosome-wide delayed replication is chromosome structure instability, which is characterized by an increase in the rate that new chromosomal rearrangements occur [81].
This structural instability is primarily observed on the affected chromosome, but other chromosomes can participate in inter-chromosomal translocations with the delayed chromosome, indicting that delayed replication on one chromosome can destabilize the structural integrity of all chromosomes within the cell [81]. This structural instability of individual chromosomes is reminiscent of the newly described phenomenon “chromothripsis”, which is present in some but not all cancers [26,27]. Chromothripsis appears to be a cataclysmic event in which one or a few chromosomes or chromosome arms are fragmented and then reassembled in a haphazard manner. The sequences at the junctions showed either a lack of homology or microhomology between the joined segments, suggesting that the ends were joined by non-homologous end joining (NHEJ) pathway. In addition, the complex chromosome rearrangements associated with genomic disorders in humans were recently found to resemble chromothripsis [135,136]. Sequencing the breakpoints at these complex rearrangements identified characteristic features, including small templated insertions of nearby sequences and microhomologies, suggestive of replicative processes. These observations led the Lupski group to propose the term “chromoanasynthesis” as an alternative descriptor to chromothripsis for the shattering and reassembly of chromosomes via replicative mechanisms [135]. The Lupski group proposed a microhomology mediated break induced replication (MMBIR) and a related fork stalling and template switching (FoSTeS) model for the origin of these complex rearrangements [137]. The distinction between MMBIR/FoSTeS and NHEJ is that the microhomology junctions in MMBIR/FoSTeS are followed by stretches of DNA sequence derived from elsewhere, usually nearby. The MMBIR/FoSTeS models involve stalled DNA replication forks that are resolved by replication restart using short stretches of homology [137]. Thus, the stalled replication forks of the MMBIR/FoSTeS pathways could potentially be caused by the premature condensation of partially replicated chromosomes as they enter mitosis. Thus, our model for the instability of individual chromosomes includes: 1) delayed replication timing of individual chromosomes caused by genetic disruption of an “inactivation/stability center”, 2) delayed recruitment of Aurora B resulting in delayed mitotic chromosome condensation, 3) delayed mitotic spindle attachment leading to chromosome missegregation and the formation of micronuclei, 4) checkpoint adaptation and the onset of mitotic chromosome condensation prior to the completion of DNA synthesis leading to stalled replication forks, and 5) multiple rearrangements generated at the stalled replication forks via NHEJ and/or MMBIR/FoSTeS type mechanisms (Figure 2B).
4. Concluding Remarks
Changes in the replication timing of individual loci throughout the genome in cancer cells can occur in two ways: either the advanced replication of individual loci or the delayed replication of individual loci. It currently appears that the advanced replication of individual loci is more common than delayed replication and it is unclear why this is. Furthermore, the change in replication timing of individual loci generally appears to result in aberrant asynchronous replication, but it is not known if this is always the case. Regardless, it is likely that the change in replication timing of individual loci sporadically throughout the genome can be caused by two different mechanisms. The first, which is highlighted by the Ryba et al. study, is genomic rearrangements resulting in a local change in replication timing [16]. In this scenario, regions of divergent replication timing are juxtaposed following a rearrangement and one of those regions changes its replication timing in response to its new environment. This can explain a switch in replication timing of an individual locus and aberrant asynchronous replication. However, this study also found replication-timing changes that did not coincide with rearrangement breakpoints [16]. Therefore, localized changes in replication timing may actually precede the rearrangement events at specific loci.
Delayed replication of individual chromosomes is a functionally distinct phenomenon from the aberrant asynchronous replication of individual loci. Although chromosome-wide replication delay does result in replication asynchrony, the asynchrony is at the chromosome level rather than at specific loci scattered throughout the genome. This replication delay affects the entire chromosome but the replication timing of all the other chromosomes within the cell remain normal. It is currently unknown whether chromosome-wide replication delay has any effect on gene expression. However, it is likely that gene expression is affected by delaying the replication timing of an entire chromosome. Thus, this process closely resembles X chromosome inactivation in female mammalian cells, where the inactive X chromosome undergoes gene inactivation and a chromosome-wide delay in replication timing while the active X chromosome remains earlier replicating [133].
In summary, DNA replication timing has helped shape the genomic landscape of many, if not all, eukaryotic organisms. By separating the genome into regions prone to hypomutability (early-replicating) and hypermutability (late-replicating and transition regions), the replication-timing program dictates that the mutation rate is higher in some regions than in others. The reason for this is currently unclear, however, it most likely results from the predominant use of different DNA repair pathways in early versus late S phase [32,34]. On one hand, it seems detrimental to have a high rate of mutagenesis anywhere in the genome, as most mutations either have no effect on fitness or are detrimental to the organism [134]. And if the cell uses error-free DNA repair pathways in early S phase, then why doesn’t it also use these same pathways in late S phase? A closer look reveals that it may be slightly beneficial to keep a higher mutation rate in late-replicating regions. Thus, most repetitive sequences tend to be late replicating and since many of these sequences are leftover viral or transposon integrations, this might be one way to ensure that non-native sequences mutate more frequently. This would result in a more rapid accumulation of mutations that inactivate viral or transposon gene products. Furthermore, tissue-specific genes are more likely to reside in late-replicating regions and since these genes are under greater pressure to adapt, a slightly higher mutation rate in those genes would provide the organism with a greater ability to respond to a changing environment.
Although there can be some benefits to having hypermutable regions of the genome in germ cells, it is hard to explain the presence of this phenomenon in somatic cells. Thus, the mutagenesis associated with the normal replication-timing program does appear to be contributing to the accumulation of mutations during cancer development. Although the normal replication-timing program may be responsible for an increased mutation rate in certain regions of the genome, the selective pressure to maintain early and late replicating regions ensures proper epigenetic regulation of gene expression and helps maintain genome stability.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 2.Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, Gilbert DM. Mammalian nuclei become licensed for DNA replication during late telophase. J Cell Sci. 2002;115:51–59. doi: 10.1242/jcs.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JR, Gilbert DM. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- 4.Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Semin Cell Dev Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Teer JK, Dutta A. Regulation of S phase. Results Probl Cell Differ. 2006;42:31–63. doi: 10.1007/b137221. [DOI] [PubMed] [Google Scholar]
- 6.Diffley JF, Labib K. The chromosome replication cycle. J Cell Sci. 2002;115:869–872. doi: 10.1242/jcs.115.5.869. [DOI] [PubMed] [Google Scholar]
- 7.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 8.Hiratani I, Takebayashi S, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect--part II. Curr Opin Genet Dev. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert DM. Cell fate transitions and the replication timing decision point. J Cell Biol. 2010;191:899–903. doi: 10.1083/jcb.201007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiratani I, Gilbert DM. Replication timing as an epigenetic mark. Epigenetics. 2009;4:93–97. doi: 10.4161/epi.4.2.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbosa AC, Otto PA, Vianna-Morgante AM. Replication timing of homologous alpha-satellite DNA in Roberts syndrome. Chromosome Res. 2000;8:645–650. doi: 10.1023/a:1009246327122. [DOI] [PubMed] [Google Scholar]
- 13.State MW, Greally JM, Cuker A, Bowers PN, Henegariu O, et al. Epigenetic abnormalities associated with a chromosome 18(q21-q22) inversion and a Gilles de la Tourette syndrome phenotype. Proc Natl Acad Sci U S A. 2003;100:4684–4689. doi: 10.1073/pnas.0730775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe Y, Fujiyama A, Ichiba Y, Hattori M, Yada T, et al. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: disease-related genes in timing-switch regions. Hum Mol Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- 15.Korenstein-Ilan A, Amiel A, Lalezari S, Lishner M, Avivi L. Allele-specific replication associated with aneuploidy in blood cells of patients with hematologic malignancies. Cancer Genet Cytogenet. 2002;139:97–103. doi: 10.1016/s0165-4608(02)00610-6. [DOI] [PubMed] [Google Scholar]
- 16.Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, et al. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res. 2012;22:1833–1844. doi: 10.1101/gr.138511.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, et al. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 18.Heun P, Laroche T, Raghuraman MK, Gasser SM. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goren A, Cedar H. Replicating by the clock. Nat Rev Mol Cell Biol. 2003;4:25–32. doi: 10.1038/nrm1008. [DOI] [PubMed] [Google Scholar]
- 21.Belyaev ND, Keohane AM, Turner BM. Histone H4 acetylation and replication timing in Chinese hamster chromosomes. Exp Cell Res. 1996;225:277–285. doi: 10.1006/excr.1996.0177. [DOI] [PubMed] [Google Scholar]
- 22.De S, Michor F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol. 2011;29:1103–1108. doi: 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 26.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prendergast JG, Campbell H, Gilbert N, Dunlop MG, Bickmore WA, et al. Chromatin structure and evolution in the human genome. BMC Evol Biol. 2007;7:72. doi: 10.1186/1471-2148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, et al. Why do human diversity levels vary at a megabase scale? Genome Res. 2005;15:1222–1231. doi: 10.1101/gr.3461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green P, Ewing B, Miller W, Thomas PJ, Green ED. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 31.Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- 32.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CL, Rappailles A, Duquenne L, Huvet M, Guilbaud G, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui P, Ding F, Lin Q, Zhang L, Li A, et al. Distinct contributions of replication and transcription to mutation rate variation of human genomes. Genomics Proteomics Bioinformatics. 2012;10:4–10. doi: 10.1016/S1672-0229(11)60028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardoso-Moreira MM, Long M. Mutational bias shaping fly copy number variation: implications for genome evolution. Trends Genet. 2010;26:243–247. doi: 10.1016/j.tig.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardoso-Moreira M, Emerson JJ, Clark AG, Long M. Drosophila duplication hotspots are associated with late-replicating regions of the genome. PLoS Genet. 2011;7:e1002340. doi: 10.1371/journal.pgen.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang JH, Li H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2004;2:E29. doi: 10.1371/journal.pbio.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S, Yu T, Chen Z, Yuan S, Chen S, et al. More single-nucleotide mutations surround small insertions than small deletions in primates. Hum Mutat. 2012;33:1099–1106. doi: 10.1002/humu.22085. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe Y, Maekawa M. Spatiotemporal regulation of DNA replication in the human genome and its association with genomic instability and disease. Curr Med Chem. 2010;17:222–233. doi: 10.2174/092986710790149756. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe Y, Ikemura T, Sugimura H. Amplicons on human chromosome 11q are located in the early/late-switch regions of replication timing. Genomics. 2004;84:796–805. doi: 10.1016/j.ygeno.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 43.Schwaiger M, Schubeler D. A question of timing: emerging links between transcription and replication. Curr Opin Genet Dev. 2006;16:177–183. doi: 10.1016/j.gde.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, et al. Global organization of replication time zones of the mouse genome. Genome Res. 2008;18:1562–1570. doi: 10.1101/gr.079566.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- 46.Woo YH, Li WH. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun. 2012;3:1004. doi: 10.1038/ncomms1982. [DOI] [PubMed] [Google Scholar]
- 47.Janoueix-Lerosey I, Hupe P, Maciorowski Z, La Rosa P, Schleiermacher G, et al. Preferential occurrence of chromosome breakpoints within early replicating regions in neuroblastoma. Cell Cycle. 2005;4:1842–1846. doi: 10.4161/cc.4.12.2257. [DOI] [PubMed] [Google Scholar]
- 48.Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2012 doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe Y, Abe T, Ikemura T, Maekawa M. Relationships between replication timing and GC content of cancer-related genes on human chromosomes 11q and 21q. Gene. 2009;433:26–31. doi: 10.1016/j.gene.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Soutoglou E, Misteli T. On the contribution of spatial genome organization to cancerous chromosome translocations. J Natl Cancer Inst Monogr. 2008:16–19. doi: 10.1093/jncimonographs/lgn017. [DOI] [PubMed] [Google Scholar]
- 51.Engreitz JM, Agarwala V, Mirny LA. Three-dimensional genome architecture influences partner selection for chromosomal translocations in human disease. PLoS One. 2012;7:e44196. doi: 10.1371/journal.pone.0044196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fudenberg G, Getz G, Meyerson M, Mirny LA. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lukasova E, Kozubek S, Kozubek M, Kjeronska J, Ryznar L, et al. Localisation and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukaemia. Hum Genet. 1997;100:525–535. doi: 10.1007/s004390050547. [DOI] [PubMed] [Google Scholar]
- 56.Hecht F, Sutherland GR. Detection of fragile sites on human chromosomes. Clin Genet. 1985;28:95–96. doi: 10.1111/j.1399-0004.1985.tb01227.x. [DOI] [PubMed] [Google Scholar]
- 57.Laird C, Jaffe E, Karpen G, Lamb M, Nelson R. Fragile sites in human chromosomes as regions of late-replicating DNA. Trends in Genetics. 1987;3:274–281. [Google Scholar]
- 58.Arlt MF, Casper AM, Glover TW. Common fragile sites. Cytogenet Genome Res. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- 59.Debatisse M, El Achkar E, Dutrillaux B. Common fragile sites nested at the interfaces of early and late-replicating chromosome bands: cis acting components of the G2/M checkpoint? Cell Cycle. 2006;5:578–581. doi: 10.4161/cc.5.6.2574. [DOI] [PubMed] [Google Scholar]
- 60.El Achkar E, Gerbault-Seureau M, Muleris M, Dutrillaux B, Debatisse M. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc Natl Acad Sci U S A. 2005;102:18069–18074. doi: 10.1073/pnas.0506497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ozeri-Galai E, Bester AC, Kerem B. The complex basis underlying common fragile site instability in cancer. Trends Genet. 2012;28:295–302. doi: 10.1016/j.tig.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Goldmit M, Bergman Y. Monoallelic gene expression: a repertoire of recurrent themes. Immunol Rev. 2004;200:197–214. doi: 10.1111/j.0105-2896.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- 63.Amiel A, Kolodizner T, Fishman A, Gaber E, Klein Z, et al. Replication pattern of the p53 and 21q22 loci in the premalignant and malignant stages of carcinoma of the cervix. Cancer. 1998;83:1966–1971. [PubMed] [Google Scholar]
- 64.Amiel A, Litmanovitch T, Lishner M, Mor A, Gaber E, et al. Temporal differences in replication timing of homologous loci in malignant cells derived from CML and lymphoma patients. Genes Chromosomes Cancer. 1998;22:225–231. doi: 10.1002/(sici)1098-2264(199807)22:3<225::aid-gcc8>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 65.Amiel A, Kirgner I, Gaber E, Manor Y, Fejgin M, et al. Replication pattern in cancer: asynchronous replication in multiple myeloma and in monoclonal gammopathy. Cancer Genet Cytogenet. 1999;108:32–37. doi: 10.1016/s0165-4608(98)00107-1. [DOI] [PubMed] [Google Scholar]
- 66.Cytron S, Stepnov E, Bounkin I, Mashevich M, Dotan A, et al. Epigenetic analyses in blood cells of men suspected of prostate cancer predict the outcome of biopsy better than serum PSA levels. Clin Epigenetics. 2011;2:383–388. doi: 10.1007/s13148-011-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reish O, Orlovski A, Mashevitz M, Sher C, Libman V, et al. Modified allelic replication in lymphocytes of patients with neurofibromatosis type 1. Cancer Genet Cytogenet. 2003;143:133–139. doi: 10.1016/s0165-4608(02)00858-0. [DOI] [PubMed] [Google Scholar]
- 68.Litmanovitch T, Altaras MM, Dotan A, Avivi L. Asynchronous replication of homologous alpha-satellite DNA loci in man is associated with nondisjunction. Cytogenet Cell Genet. 1998;81:26–35. doi: 10.1159/000015003. [DOI] [PubMed] [Google Scholar]
- 69.Dotan ZA, Dotan A, Ramon J, Avivi L. Altered mode of allelic replication accompanied by aneuploidy in peripheral blood lymphocytes of prostate cancer patients. Int J Cancer. 2004;111:60–66. doi: 10.1002/ijc.20237. [DOI] [PubMed] [Google Scholar]
- 70.Grinberg-Rashi H, Cytron S, Gelman-Kohan Z, Litmanovitch T, Avivi L. Replication timing aberrations and aneuploidy in peripheral blood lymphocytes of breast cancer patients. Neoplasia. 2010;12:668–674. doi: 10.1593/neo.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fritz AJ, Sinha S, Marella NV, Berezney R. Alterations in replication timing of cancer related genes in malignant human breast cancer cells. J Cell Biochem. 2012 doi: 10.1002/jcb.24447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bras A, Cotrim CZ, Vasconcelos I, Mexia J, Leonard A, et al. Asynchronous DNA replication detected by fluorescence in situ hybridisation as a possible indicator of genetic damage in human lymphocytes. Oncol Rep. 2008;19:369–375. doi: 10.3892/or.19.2.369. [DOI] [PubMed] [Google Scholar]
- 73.Dotan ZA, Dotan A, Ramon J, Avivi L. Aberrant allele-specific replication, independent of parental origin, in blood cells of cancer patients. BMC Cancer. 2008;8:390. doi: 10.1186/1471-2407-8-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagler A, Cytron S, Mashevich M, Korenstein-Ilan A, Avivi L. The aberrant asynchronous replication - characterizing lymphocytes of cancer patients - is erased following stem cell transplantation. BMC Cancer. 2010;10:230. doi: 10.1186/1471-2407-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adolph S, Hameister H, Schildkraut CL. Molecular analysis of the aberrant replication banding pattern on chromosome 15 in murine T-cell lymphomas. Chromosoma. 1992;101:388–398. doi: 10.1007/BF00582833. [DOI] [PubMed] [Google Scholar]
- 76.zur Hausen H. Chromosomal changes of similar nature in seven established cell lines derived from the peripheral blood of patients with leukemia. J Natl Cancer Inst. 1967;38:683–696. [PubMed] [Google Scholar]
- 77.Miles CP, O’Neill F. 3H labeling patterns of permanent cell line chromosomes showing pulverization or accentuated secondary constrictions. J Cell Biol. 1969;40:553–561. doi: 10.1083/jcb.40.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith L, Plug A, Thayer M. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc Natl Acad Sci U S A. 2001;98:13300–13305. doi: 10.1073/pnas.241355098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang BH, Smith L, Huang J, Thayer M. Chromosomes with delayed replication timing lead to checkpoint activation, delayed recruitment of Aurora B and chromosome instability. Oncogene. 2007;26:1852–1861. doi: 10.1038/sj.onc.1209995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breger KS, Smith L, Turker MS, Thayer MJ. Ionizing radiation induces frequent translocations with delayed replication and condensation. Cancer Res. 2004;64:8231–8238. doi: 10.1158/0008-5472.CAN-04-0879. [DOI] [PubMed] [Google Scholar]
- 81.Breger KS, Smith L, Thayer MJ. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum Mol Genet. 2005;14:2813–2827. doi: 10.1093/hmg/ddi314. [DOI] [PubMed] [Google Scholar]
- 82.Farkash-Amar S, David Y, Polten A, Hezroni H, Eldar YC, et al. Systematic Determination of Replication Activity Type Highlights Interconnections between Replication, Chromatin Structure and Nuclear Localization. PLoS One. 2012;7:e48986. doi: 10.1371/journal.pone.0048986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gondor A, Ohlsson R. Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat Rev Genet. 2009;10:269–276. doi: 10.1038/nrg2555. [DOI] [PubMed] [Google Scholar]
- 84.Loupart ML, Krause SA, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 85.Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, et al. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 86.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 87.Zhou J, Snyder AR, Lieberman PM. Epstein-Barr virus episome stability is coupled to a delay in replication timing. J Virol. 2009;83:2154–2162. doi: 10.1128/JVI.02115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cosgrove AJ, Nieduszynski CA, Donaldson AD. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 2002;16:2485–2490. doi: 10.1101/gad.231602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watanabe Y, Shibata K, Sugimura H, Maekawa M. p53-dependent change in replication timing of the human genome. Biochem Biophys Res Commun. 2007;364:289–293. doi: 10.1016/j.bbrc.2007.09.136. [DOI] [PubMed] [Google Scholar]
- 90.Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- 91.Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Mol Cell. 2010;40:736–748. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 92.Jorgensen HF, Azuara V, Amoils S, Spivakov M, Terry A, et al. The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol. 2007;8:R169. doi: 10.1186/gb-2007-8-8-r169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwaiger M, Kohler H, Oakeley EJ, Stadler MB, Schubeler D. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 2010;20:771–780. doi: 10.1101/gr.101790.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu R, Singh PB, Gilbert DM. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol. 2006;174:185–194. doi: 10.1083/jcb.200601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hansen RS, Stoger R, Wijmenga C, Stanek AM, Canfield TK, et al. Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum Mol Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- 97.Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol Cell Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cimbora DM, Schubeler D, Reik A, Hamilton J, Francastel C, et al. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol Cell Biol. 2000;20:5581–5591. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goren A, Tabib A, Hecht M, Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 102.Zappulla DC, Sternglanz R, Leatherwood J. Control of replication timing by a transcriptional silencer. Curr Biol. 2002;12:869–875. doi: 10.1016/s0960-9822(02)00871-0. [DOI] [PubMed] [Google Scholar]
- 103.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 104.Zhou J, Chau CM, Deng Z, Shiekhattar R, Spindler MP, et al. Cell cycle regulation of chromatin at an origin of DNA replication. EMBO J. 2005;24:1406–1417. doi: 10.1038/sj.emboj.7600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jablonka E, Goitein R, Marcus M, Cedar H. DNA hypomethylation causes an increase in DNase-I sensitivity and an advance in the time of replication of the entire inactive X chromosome. Chromosoma. 1985;93:152–156. doi: 10.1007/BF00293162. [DOI] [PubMed] [Google Scholar]
- 106.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 107.Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, et al. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 108.Aladjem MI, Groudine M, Brody LL, Dieken ES, Fournier RE, et al. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 109.Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- 110.Lande-Diner L, Zhang J, Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell. 2009;34:767–774. doi: 10.1016/j.molcel.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhong Q, Chen Y, Jones D, Lee WH. Perturbation of TSG101 protein affects cell cycle progression. Cancer Res. 1998;58:2699–2702. [PubMed] [Google Scholar]
- 112.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 113.Tatematsu KI, Yamazaki T, Ishikawa F. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells. 2000;5:677–688. doi: 10.1046/j.1365-2443.2000.00359.x. [DOI] [PubMed] [Google Scholar]
- 114.Gribnau J, Hochedlinger K, Hata K, Li E, Jaenisch R. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 2003;17:759–773. doi: 10.1101/gad.1059603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lock LF, Takagi N, Martin GR. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 116.Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10:149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- 117.Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci. 2004;117:5353–5365. doi: 10.1242/jcs.01412. [DOI] [PubMed] [Google Scholar]
- 118.Korenstein-Ilan A, Barbul A, Hasin P, Eliran A, Gover A, et al. Terahertz radiation increases genomic instability in human lymphocytes. Radiat Res. 2008;170:224–234. doi: 10.1667/RR0944.1. [DOI] [PubMed] [Google Scholar]
- 119.Bergstrom R, Whitehead J, Kurukuti S, Ohlsson R. CTCF regulates asynchronous replication of the imprinted H19/Igf2 domain. Cell Cycle. 2007;6:450–454. doi: 10.4161/cc.6.4.3854. [DOI] [PubMed] [Google Scholar]
- 120.Bianchi A, Shore D. Early replication of short telomeres in budding yeast. Cell. 2007;128:1051–1062. doi: 10.1016/j.cell.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 121.Lin CM, Fu H, Martinovsky M, Bouhassira E, Aladjem MI. Dynamic alterations of replication timing in mammalian cells. Curr Biol. 2003;13:1019–1028. doi: 10.1016/s0960-9822(03)00382-8. [DOI] [PubMed] [Google Scholar]
- 122.Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, et al. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 2007;26:1327–1339. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ofir R, Wong AC, McDermid HE, Skorecki KL, Selig S. Position effect of human telomeric repeats on replication timing. Proc Natl Acad Sci U S A. 1999;96:11434–11439. doi: 10.1073/pnas.96.20.11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Calza RE, Eckhardt LA, DelGiudice T, Schildkraut CL. Changes in gene position are accompanied by a change in time of replication. Cell. 1984;36:689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 125.Amiel A, Levi E, Reish O, Sharony R, Fejgin MD. Replication status as a possible marker for genomic instability in cells originating from genotypes with balanced rearrangements. Chromosome Res. 2001;9:611–616. doi: 10.1023/a:1012966221692. [DOI] [PubMed] [Google Scholar]
- 126.Sun Y, Wyatt RT, Bigley A, Krontiris TG. Expression and replication timing patterns of wildtype and translocated BCL2 genes. Genomics. 2001;73:161–170. doi: 10.1006/geno.2000.6479. [DOI] [PubMed] [Google Scholar]
- 127.Stoffregen EP, Donley N, Stauffer D, Smith L, Thayer MJ. An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum Mol Genet. 2011;20:2366–2378. doi: 10.1093/hmg/ddr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Diaz-Perez S, Ouyang Y, Perez V, Cisneros R, Regelson M, et al. The element(s) at the nontranscribed Xist locus of the active X chromosome controls chromosomal replication timing in the mouse. Genetics. 2005;171:663–672. doi: 10.1534/genetics.105.043026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Diaz-Perez SV, Ferguson DO, Wang C, Csankovszki G, Tsai SC, et al. A deletion at the mouse Xist gene exposes trans-effects that alter the heterochromatin of the inactive X chromosome and the replication time and DNA stability of both X chromosomes. Genetics. 2006;174:1115–1133. doi: 10.1534/genetics.105.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 131.Thayer MJ. Mammalian chromosomes contain cis-acting elements that control replication timing, mitotic condensation, and stability of entire chromosomes. Bioessays. 2012;34:760–770. doi: 10.1002/bies.201200035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 133.Boumil RM, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet. 2001;10:2225–2232. doi: 10.1093/hmg/10.20.2225. [DOI] [PubMed] [Google Scholar]
- 134.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]