Abstract
Objectives
Brain molecular aging, the pervasive and consistent transcriptome changes associated with normal brain aging, appears to overlap with disease pathways and may be anticipated in neurodegenerative and neuropsychiatric diseases, including major depressive disorder (MDD). Here we characterize the global interaction of MDD-related gene changes with age, starting from our previous report of downregulated brain-derived neurotrophic factor (BDNF) and BDNF-dependent genes in the amygdala of women with MDD.
Methods
A large scale gene expression dataset in the amygdala from a postmortem cohort of 21 women with MDD and 21 age-matched controls (age range: 16–74 years old) was analyzed for correlations of gene transcript changes with age, in the presence or absence of a diagnosis of MDD.
Results
(1) The age-related decrease in BDNF transcripts observed in control subjects corresponds with further age-related decreases in BDNF and BDNF-dependent gene expression in MDD subjects; (2) most MDD-related genes are frequently age-regulated in both MDD and in control subjects; (3) the effects of MDD and age are positively correlated; (4) most genes that are age-dependent in control subjects display greater age effects in MDD subjects, and (5) the increased prevalence of age effects in MDD corresponds to similar trends in controls, rather than representing de novo age effects.
Conclusions
MDD strongly associates with robust and anticipated gene expression changes that occur during normal aging of the brain, suggesting that an older molecular age of the brain represents an early biological event and/or a marker of risk for subsequent onset of MDD symptoms.
Keywords: Aging, Brain, Human, Postmortem, Neuropsychiatric, Major Depression, Microarray, Amygdala, BDNF
INTRODUCTION
Along with increased lifespan comes an increased risk for neuropsychiatric and neurological disorders.1 Studies of the functional correlates of aging consistently report increased incidence of low mood, motor deficits and declines in certain cognitive functions.2–4 Mood disorders per se do not necessarily increase with age, but studies report increasing numbers of depressive symptoms in older individuals; hence the under-diagnosing of depression may potentially reflect diagnostic structure and procedural issues, since older individuals tend to under-report psychiatric symptoms, and psychiatric symptoms can be perceived as part of normal aging (See4 and McKinney et al5 in this issue). Low mood also occurs in pre-symptomatic phases of neurodegenerative disorders. Together, these findings suggest that aspects of mood regulatory mechanisms may be selectively vulnerable during normal and pathological aging, or that depressive symptoms may represent a common output for various underlying age-related brain declines.4, 6 Although it is difficult to untangle causal links, evidence suggests that mood symptoms may represent an early marker for subsequent spiraling functional declines.3
On the other hand, depression has been compared to a state of “accelerated aging”, since subjects with depression display an increased incidence of various diseases of aging, such as cardiovascular and cerebrovascular diseases, metabolic syndrome, and dementia.7–8 However, the mechanisms underlying normal aging processes and disorders of old age are still being characterized.9–10 Specifically, recent progress in investigating the molecular, cellular and neural bases of depression in adult subjects, and the observation of a continuum of changes between normal aging and depression-related changes, have suggested a framework where molecular aging of the brain may promote biological changes that may in turn place the system at higher vulnerability for several age-related diseases.5, 9–10 Specifically, age-related gene expression changes have been described in the human brain and affect approximately 10% of all genes.11–18 These changes in gene transcript levels are continuous throughout adult life, include both upregulated and downregulated genes, affect specific cellular functions, and overlap both in terms of affected genes and direction of change with those observed in the context of neuropsychiatric disorders.11, 18 For instance, gene transcript levels for brain-derived neurotrophic factor (BDNF) decrease with age in the human brain,11, 18–19 but BDNF levels have also been reported to be low in the context of neuropsychiatric disorders.20–23 Note that these are cross-sectional measures; hence since the actual longitudinal molecular trajectories are not known, we use here the term “anticipated” rather than “accelerated” aging.
We recently reported robust large-scale gene expression changes in the amygdala in the post-mortem brains of women affected with major depressive disorder (MDD) compared to control subjects (See details and discussion in Guilloux et al22). MDD-related changes included a significant downregulation of BDNF, which also correlated with similar changes in a set of BDNF-related genes. Using analyses of covariance, age was identified as a significant covariate for BDNF and BDNF-related gene signals, while multiple additional clinical and demographic parameters did not affect the findings.22 Here, in the broader context of an age-by-disease interaction hypothesis,5, 10 we investigated these prior findings in more detail in the same cohort. Specifically, we hypothesized that MDD-related gene changes, in addition to those related to BDNF, i) would correlate with chronological age across subjects, ii) would correspond to normal age effects in control subjects, and iii) that the magnitude of age-related changes would be greater in MDD subjects.
METHODS
Human postmortem subjects
After consent was obtained from the next of kin, brain specimens were obtained during autopsies conducted at the Allegheny County Medical Examiner’s Office (Pittsburgh, PA, USA). For all subjects, consensus DSM-IV diagnoses of MDD were made by an independent committee of experienced clinical research scientists at a case conference utilizing information obtained from clinical records, toxicology exam and a standardized psychological autopsy.24 This latter incorporates a structured interview, conducted by a licensed clinical psychologist with family members of the index subject, to assess diagnosis, psychopathology, medical, social and family histories, as well as history of substance abuse. Rates of death by suicide, disease recurrence, evidence for antidepressant treatment at time of death, and alcohol dependence in MDD subjects were recorded. Toxicological screens on peripheral fluids were used to identify the presence of antidepressant medication. Twenty-one pairs of subjects were analyzed, each pair consisting of one female subject with MDD and a one control subject matched for sex, race (88% white Caucasian), and as closely as possible for age, postmortem interval (PMI) and brain pH (for complete details of the cohort, see Table S1 and Guilloux et al22). Brains were analyzed for adequate brain pH (>6.4) and RNA integrity by optical density (OD≥1.6) and Agilent bioanalyzer analysis (Agilent Technologies, Palo Alto, CA; RIN expert scoring system ≥ 7) as described.25 Accordingly, subject groups did not differ in mean age, PMI, brain pH, RNA integrity number (RIN), or RNA ratio (p>0.05).
Microarray Samples
The sample collection and generation of gene microarray data was described in Guilloux et al.22 In brief, rostral amygdala samples enriched in lateral, basolateral, and basomedian nuclei were dissected from frozen coronal blocks ~2cm caudal to the temporal pole. Total RNA was isolated and assessed using the Bioanalyzer 2100 (Agilent Technologies, Walbronn, Germany). To reduce the influence of technical variability, paired samples were processed together, but different pairs were randomly distributed at each experimental step. 1μg of total RNA was reverse-transcribed and converted into double-stranded cDNA. A biotinylated cRNA was transcribed in vitro, using an RNA polymerase T7 promoter site introduced during the reverse-transcription step. Fragmented labeled cRNA sample were hybridized onto Human Whole-Genome Expression Beadchips (Human HT12 v4.0, Illumina, Inc., San Diego, CA), assessing around 31,000 annotated genes. Probeset signals (i.e., transcript levels) were extracted and batch-normalized with the Beadarray (Illumina, Inc., SanDiego, CA) software following the default quality control parameters.
Real-time quantitative PCR (qPCR)
qPCR was performed as previously described.25 Small PCR products were amplified in quadruplets on a Mastercycler real-time PCR machine (Eppendorf, Hamburg, Germany), Results were averaged across the four replicates and transformed into relative expression levels (2−ΔΔCt) as the geometric mean of relative intensities compared to three internal controls (actin, glyceraldehyde-3-phosphate dehydrogenase and cyclophilin) with comparable expression in both subject groups.
Statistical Analyses
Statistical analysis of disease effect
The selection of MDD-related genes was made using a random-intercept statistical model (RIM) to adjust for clinical, demographic and technical variables (Antidepressant treatment, Suicide, Severity, pH, PMI and RIN), as described in Guilloux et al22 and reproduced below. Alternatively, parametric (paired T-test) or non-parametric (Wilcoxon test) statistical tests were also applied. Their respective use is described in the “Generation of gene lists” section below.
RIM was applied to account for paired design (MDD samples paired with corresponding controls with no disease) and existence of clinical, demographic and technical variables. For a given gene g, we fit the model where Ygik is the gene expression value of gene g (1≤g≤G) and sample i (i=1 for controls and i=2 for MDD) in pair k (1≤k≤K), X0ik takes one if the sample is MDD and zero if the sample is a control subject, Xlik represents values for clinical variables (e.g. 0–1 binary for respective parameters, i.e. taking alcohol or not, antidepressant exposure, or numerical for brain pH or PMI values) and αkis the random intercept, which are the deviations of average of all expression values at kth pair from the average in the whole population and is assumed to follow normal distribution with mean zero and variance t2g. Finally, ∈gik are independent random noises with normal distribution with mean zero and variance s2g. Under this model, βg0 is the disease effect of gene g of interest to be estimated. To obtain a MDD-associated biomarker list, likelihood ratio test (LRT) is used to assess the p-values of testing H0:βg = 0 and p-values are then corrected by Benjamini-Hochberg procedure for multiple comparison,26 with a false discovery rate (FDR) set at 5%.
Generation of gene lists
- List #1 (exploratory) was obtained using uncorrected parametric and non-parametric statistical tests (RIM, paired T-test and Wilcoxon test), with no threshold on effect size for MDD effect, resulting in an exploratory list of 4,131 genes with uncorrected statistical differences between MDD and control subjects.
- List #2 (intermediate) contained 307 genes obtained using uncorrected parametric and non-parametric statistical tests (RIM, paired T-test and Wilcoxon test), moderate statistical stringencies for gene selection (p<0.05 in at least one statistical test), and MDD effect size greater than 20%.
- List #3 (stringent and FDR-corrected) was composed of gene selection made using RIM to adjust for clinical, demographic and technical variables, followed by correction for multiple testing by the Benjamini-Hochberg method,26 resulting in a stringent selection of 116 differentially expressed genes (Adjusted p-value, i.e. q-value<0.05; mean change>20%).
Statistical analysis of age effects on gene transcripts
For each individual gene, we calculated the individual ratio of gene expression to the average expression of this single gene. For age effect, a Pearson correlation was performed using individual gene expression values and age, in the control population and in the MDD population. Based on a cohort of 21 subjects (control or MDD), a Pearson correlation factor >0.369 or <−0.369 gives a p-value p<0.05. Changes in slopes of combined age-dependent gene changes were calculated by Student’s t-tests, using slope values for the same individual genes between the control and MDD subjects. The relative proportions of age-related genes were compared between groups using the Chi-square test.
RESULTS
Reduced BDNF expression with age and MDD associates with greater age-effects on BDNF-related genes in MDD subjects
We previously reported that disease-associated genes, including BDNF, are robustly modulated by age in control subjects11, 18 and we also described a correlation with age for several genes that are under the control of BDNF in a cohort of postmortem female subjects with MDD (See Supplementary Figure S3 in Guilloux et al22). Here, we further investigated the relationships between age, MDD and altered transcript levels, starting with changes in BDNF-related genes in subjects with MDD compared to control subjects.
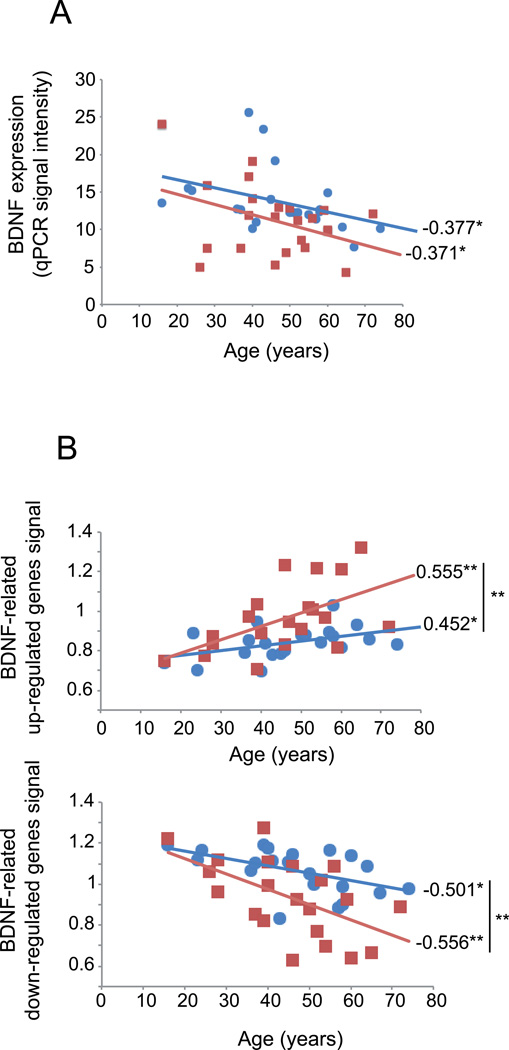
Displaying qPCR measures as a function of age, we confirm that BDNF mRNA expression decreases with age, both in the control and MDD groups, with a greater mean reduction in MDD subjects (Figure 1A; −22%, p<0.05, respective to age-matched control subjects), but no difference in slope values (p=0.133). Since BDNF is a neurotrophic factor that regulates the expression of many downstream genes, we tested whether MDD-related genes that are also BDNF-related may display greater change with age than do control subjects. From an exploratory gene list for MDD effect in the amygdala-based microarray dataset in a cohort of 21 female MDD compared to 21 matched controls (See List #1 in Methods), 52 genes were identified as BDNF-related through the presence of literature-based links in the Ingenuity database. BDNF-dependent genes were then split based on direction of MDD effect on gene expression (33 upregulated, 19 downregulated; See Supplementary Table S2). Individually, 11 of these 52 BDNF-related genes displayed significant age-effects in controls (4 up-regulated and 7 down-regulated genes with age), compared to 26 in the MDD cohort (8 up-regulated and 18 down-regulated genes with age). The combined age effects for these MDD up- or down-regulated genes within each subject are displayed in Figure 1B, as a function of age. As expected from our prior combined studies,22 these average values displayed significant positive correlation with age for MDD-upregulated genes and significant negative correlation with age for MDD-downregulated genes. Notably, although age-dependent changes display parallel trajectories for BDNF (Figure 1A), cross-subjects trajectories of combined changes for BDNF-dependent genes displayed increasing effect sizes with age in MDD subjects (Figure 1B; MDD versus control slope differences: upregulated genes, p=0.006; downregulated genes, p<0.0001).
FIGURE 1. BDNF and BDNF-related genes expression decrease with age.
(A) BDNF expression, measured by qPCR, is significantly and inversely correlated with chronological age in control and MDD subjects. Values are in arbitrary units of qPCR signal intensity. Respective to age-matched control subjects, MDD subjects display greater BDNF downregulation (−22%; p<0.05, Guilloux et al22). (B) Age-regulation of BDNF- and MDD-related genes. The average relative age effects for each individual subject are shown for the set of BDNF-related genes that display significant MDD effects. Microarray-based gene expression values were normalized to the group means of each gene and averaged per subject. The BDNF-related gene set was split based on the effect of age on those genes in control subjects (top panel, age-upregulated; bottom panel, age-downregulated). The results show that age-effects are systematically in the same direction, and of greater effect sizes in MDD subjects (red squares) compared to controls (blue circles). Values are Pearson correlation factors. *, p<0.05; **, p<0.01.
These results demonstrates that gene expression changes for BDNF and BDNF-related genes in MDD display similar age-correlation, although to a higher extent and age-dependent rate than control subjects; together suggesting that normal age-related changes may contribute to MDD-related biological changes.
Greater numbers of genes affected by age in MDD subjects and over-representation of age-effects on MDD-related genes in control subjects
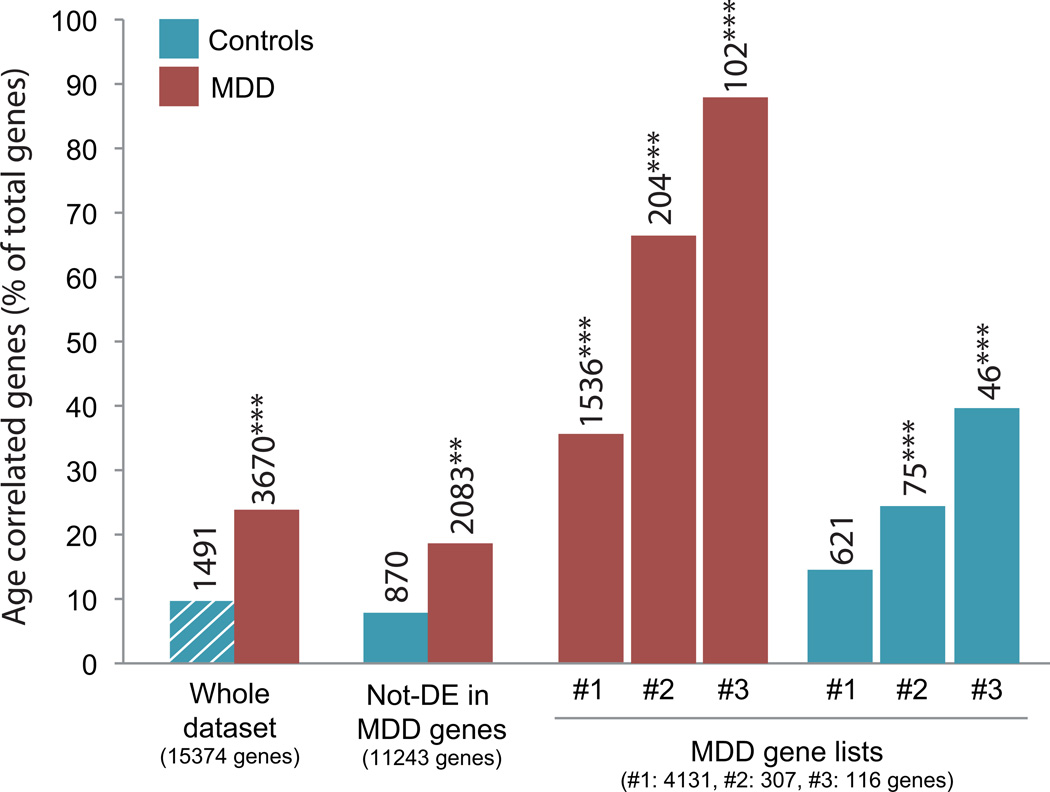
To test whether the increased age effect on MDD-related genes in MDD subjects was limited to BDNF-related genes, we next assessed the relative frequencies of genes displaying significant correlations between transcript levels and chronological age (i.e., age effect) for several sets of genes, based on increasing statistical stringency for MDD effects. The baseline for age effect was assessed in control subjects, where 9.7% of the genes (n=1491) displayed significant age effects (Pearson correlation with age, p<0.05; See methods) (Figure 2, hashed bar column), consistent with prior findings.18 In comparison, 3670 genes were correlated with age in MDD subjects (23.9%; Χ2=22.9 p<0.0001) (Figure 2). Restricting the pool of genes to only those affected in MDD progressively increased the relative proportion of genes significantly correlated with age (Figure 2; List #1: 35.3%, Χ2=74.9 p<0.0001; List #2: 66.4%, Χ2=367.7 p<0.0001), reaching a remarkable 87.9% in the list with the most stringent criteria for MDD effects (Figure 2; List #3: Χ2=698.7 p<0.0001). Notably, a similar trend of increased relative proportion of age effects was observed for the same gene lists in the control subjects (Figure 2; List #1: 14.3%, Χ2=2.42, p=0.11; List #2, 24.4%, Χ2=24.7 p<0.0001; List #3: 39.6%, Χ2=102.5 p<0.0001).
FIGURE 2. Increased numbers of age-correlated genes in MDD subjects.
The percentages of age-related genes (Y-axis) in MDD (red bars) and control subjects (blue bars) are displayed for various lists of genes with increasing stringengy for MDD effects (List #3 is the most stringent; see Methods). The numbers on top of bars represent the number of age-related genes. The numbers in parentheses at the bottom of the columns represent the number of genes in the corresponding gene lists. **p<0.01, ***p<0.0001 using Chi-square analyses, comparing the frequency of age-dependent genes in the different groups to the expected frequency, i.e. the frequency of age correlated genes observed in the control group (blue bar with white dashes). DE: differentially expressed.
Increased age effects in MDD subjects were also observed for genes that did not display MDD-related changes compared to the expected frequency, i.e., the frequency of age correlated genes observed in the control group (Figure 2, blue bar with white dashes), as demonstrated by a relative increase in age effect in that gene list (Figure 2; “Not-differentially-expressed (DE) in MDD genes”; red bar, 18.5%, Χ2=10.7 p=0.0011). In comparison, the same gene list (not-DE in MDD) did not show an increased proportion of age effect in control subjects (Figure 2: “not-DE in MDD”; blue bar, 7.7%, Χ2=0.44, p=0.5).
Together, these results demonstrate that the overlap between the MDD-related pathology and the gene expression correlates of aging extends well beyond the indirect effects of decreased BDNF on gene transcript levels, and in fact suggests that the presence of MDD accentuates the propensity of a gene to be age-regulated.
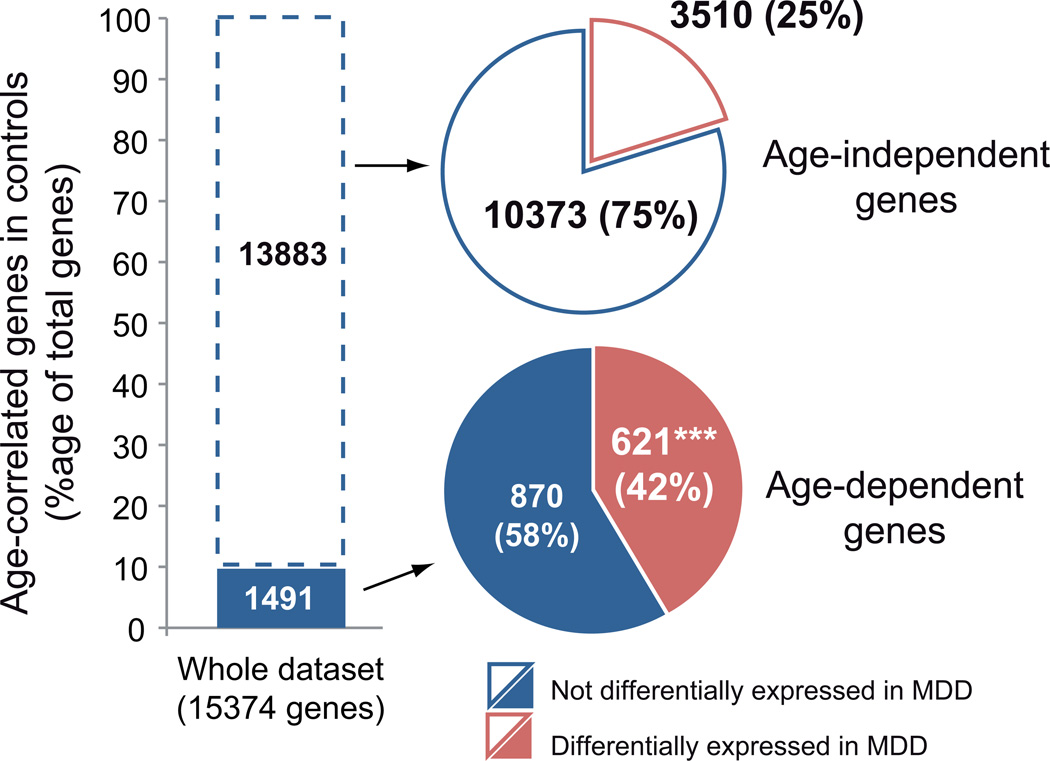
Age-correlation in control subjects predicts greater risk of association with MDD
Conversely the increased probability for any gene that is affected in MDD to also display age-related changes in control subjects, combined with the absence of increased age-effects in the list of genes not affected in MDD (Figure 2, blue bar compared to blue bar with white dashes), suggest that age-dependency in control subjects may predict putative involvement in MDD pathology. Accordingly, Figure 3 shows that 42% of the genes significantly correlated with age in control subjects were also differentially expressed in MDD subjects (Figure 3; lower pie-chart). This proportion is significantly greater than the 26.9% of genes displaying significant MDD correlation for the whole dataset (Χ2=11.11, p<0.001). On the other hand, age-independent genes showed a non-significant difference in rate of association with MDD compared to the whole dataset (Figure 3; 25.3%; Χ2=0.13 p=0.72; upper pie-chart).
FIGURE 3. Over-representation of MDD effects in gene set displaying age-dependent changes in controls.
The histogram shows the relative proportion of genes showing age-effects in the amygdala of control subjects (Age-dependent, Dark blue, ~10%). The lower pie-chart indicates that 42% of the age-dependent genes in controls are also differentially expressed in MDD subjects (Based on List # 1, See methods). ***p<0.0001; Chi-square analysis comparing frequency of genes differentially expressed in MDD in the age-dependent genes selection compared to their frequency in the whole dataset. The upper pie-chart indicates that the frequency of MDD effect in the set of genes that are age-independent was nominally greater, but not significant different, from the control population.
Together, these results demonstrate that the set of genes displaying age-dependent changes in control subjects largely overlaps with the set of genes that is affected in MDD, and suggest that genes which are age-dependent in control subjects may be more vulnerable to the pathophysiological processes of MDD.
The increased age-effect on MDD-related gene changes corresponds to an increase in effect size of subthreshold age-related changes in control subjects, rather than a true de novo age-effect
Our results show that genes differentially-expressed in MDD are for the most part affected by age in control subjects. But the lists of MDD-related genes also contain genes that do not display significant age effects. Therefore, we next investigated whether this increase in age-dependency for MDD-related genes represented an increase in sub-threshold age effect in control subjects (i.e., trend-like), or a true de novo age-dependency effect (i.e., unique to MDD subjects).
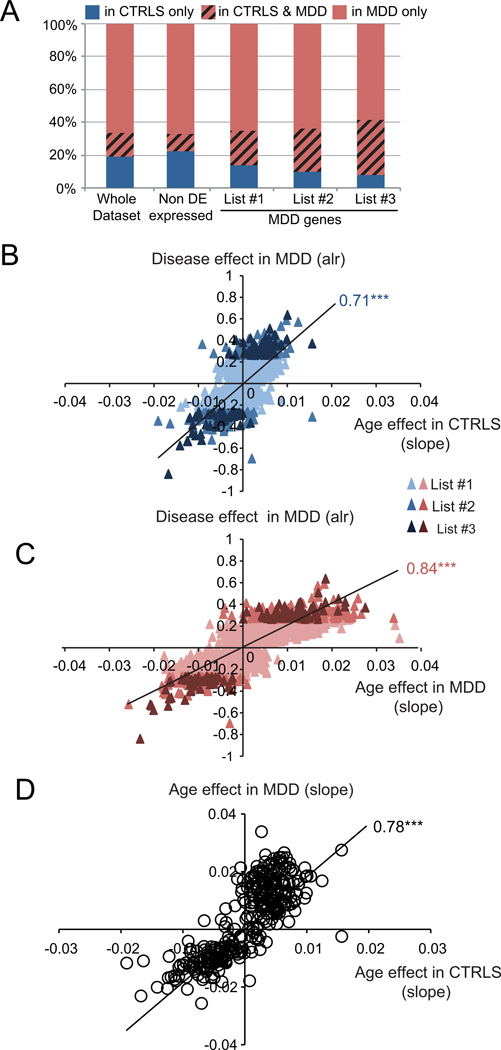
We first addressed this question by showing that the relative proportion of MDD-related genes that display significant age effects solely in control subjects greatly decreases as the stringency for MDD effect increases (Figure 4A, from list #1 to #3). This demonstrates that any core MDD gene that is affected by age in controls will almost systematically be affected by age in the context of MDD.
FIGURE 4. MDD-related gene changes are globally and positively correlated with age in MDD subjects, and are positively correlated with sub-threshold effects in control subjects.
(A) The relative proportion of genes correlated with age solely in controls (blue) decreases in gene lists with increasing stringency for statistical association with MDD. Red indicates proportion of genes with age effects solely in MDD subjects. The red blocks with black hashed lines indicate the proportion of genes displaying age-effects in both subject groups. (B, C) Correlation between the effect of MDD (Average log-ratio (alr) value of the MDD effect) and the age-related effect on gene expression ratio in controls (CTRLS) (B) and MDD subjects (C). Age effects were determined here using the slope of the linear regression of gene expression ratio as a function of age. Note that the unit of slope values (x-axis) correspond to changes per year in expression values that are normalized to the mean expression of the respective genes, as in Figure 1. So a value of 1 corresponds to the mean expression value, and slope values ranging from 0.01 (in controls) to 0.03 or 0.04 (in MDD) correspond to ~1% or 3–4% of change per chronological year. (D) Correlation of effect of age in controls with effect of age in MDD population. The values in graphs B–D are Pearson Correlation factors. ***, p<0.0001.
We next investigated correlations between significant MDD effects and age-related changes in gene expression in control subjects, regardless of age-significance in the control group. For this, we speculated that in control subjects, the slope values of the cross-subjects trajectory of changes in expression with chronological age may capture small trends in gene changes that would otherwise not reach statistical significance for age correlation. Accordingly, we show that the extent of MDD effect on gene transcript levels (average log-ratio of MDD versus control signal) is positively correlated with the slope values of age-related trajectories in control subjects (Figure 4B). We also show that the spread of the age-related slope values increases in MDD subjects (Figure 4C; see x-axis spread compared to Figure 4B). Finally, Figure 4D directly compares the age-related slope values for MDD-related genes between MDD and control subjects. The tight correlation (R=0.77, p<0.0001) and elevated slope (>1.8) of this latter comparison demonstrate that the age-related changes of MDD-related genes correspond to an almost doubling of already-existing age-related trends in control subjects, consistent with the differences in slope values observed in Figure 1 for the restricted set of BDNF-related genes (Figure 1[B]). This effect is illustrated here for genes corresponding to MDD list #2, but is observed for all selections (Figure S1). Notably, the tight scatter of the data points along the trend-lines in figures 4B–D indicate that these effects are not driven by outlier values.
Together, these results are consistent with the notion that the gene expression correlates of MDD are globally and positively correlated with age, and that these effects represent increased effect sizes of small or subthreshold age-effect in control subjects rather than de novo changes of age-related trajectories in MDD subjects.
DISCUSSION
Expanding from our previous findings of low BDNF expression in MDD and during normal aging in the amygdala of adult women affected with MDD compared to control subjects,22 we now show that most MDD-related genes are age-regulated both in MDD and in control subjects, and that gene expression changes in MDD and aging are in the same direction. We also show that most genes that are age-dependent in control subjects display greater age effects in MDD subjects and that age-correlation in control subjects predicts a greater likelihood of association with MDD. Finally, we report that the overall increased prevalence of age effects on genes in MDD subjects corresponds with similar changes in control subjects, although of reduced effect size, rather than to de novo age effects. Together these results demonstrate that MDD is strongly associated with robust and anticipated gene expression changes that occur during normal aging of the brain, and suggest that normal aging of the brain, as measured by changes in gene transcript levels, may represent an early biological event in the disorder, which potentially accelerates with older age. Indeed, since many age-related gene changes are affected in MDD in the same direction and to a greater extent, we speculate that normal age-related events can contribute to the pathophysiological processes engaged in MDD, and that the impact of MDD on gene expression becomes greater as subjects get older.
Arguments in support of an “older brain” in MDD have been reviewed in details by Wolkowitz et al,7–8 with an emphasis on stress-related co-morbid conditions, such as vascular diseases, metabolic syndrome and neurosteroid dysregulation, contributing to the biological profile that is observed in MDD subjects and that suggests a state of accelerated aging. The reviewed evidence suggests that disruptions in multiple cellular pathways, which occur downstream from stress and other metabolic disturbances, may lead to systemic and brain diseases, including depression, and overall accelerated cell aging.7–8 The data presented here overall fit with the presence of an “accelerated” or “anticipated” state of older biological age. That is, our results demonstrate that age- and MDD-related gene changes greatly overlap, and specifically that expression changes that occur in the context of MDD for a specific subset of genes are actually very similar in nature, but greater in effect size and rate of change, to what happens in the normal brain during aging. An alternative hypothesis is that an older brain is less able to tolerate the effects of MDD, although it is less likely as our assay measures age-trajectories in subjects covering a large age-span, including subjects in early adulthood. It is also important to note that when considering all genes, although 42% of age-dependent gene were also affected in MDD, a large proportion of age-dependent genes in control subjects (58%) were unchanged in MDD (Figure 3), arguing against a global increase in age effect in MDD, and rather suggesting a selectivity in anticipated age changes in MDD. On the other hand, our data also show that the majority of MDD-related gene changes correspond to similar age-dependent changes in controls, even if of small magnitude. It is also possible that small effects that correspond to few percentage points may remain biologically silent in control subjects, and that the moderate increases in effect size observed in MDD subjects may be sufficient to impact the function of associated biological systems. So in short, the anticipated brain molecular aging profile that is observed in MDD is robust, not global, and does not represent de novo age effects. This observation of MDD affecting a subset of age-dependent genes is consistent with our prior finding showing that the global “molecular age” of individual subjects, i.e., the predicted chronological age of a subject based on linear regression for all age-affected genes, did not differ between MDD and control cohorts.11 Our current results suggest instead that predictions of molecular ages based on gene changes within more restricted biological pathways may more accurately identify relevant biological changes at the intersection of age and diseases. We are currently pursuing these lines of research in this and other larger cohorts.
These latter considerations may also provide an answer to the basic but valid question: Why are most old people not suffering from depression, if normal brain aging promotes biological changes that are similar to those observed in depressed subjects? Selective versus global effects may be one answer (discussed above), but we also need to consider differences in terms of biological context and trajectory over time. First, in terms of biological context, normal molecular aging affects ~10% of all genes and has been hypothesized to represent the combined output of maturation processes, deleterious events (oxidative damage, neurosteroid dysregulation, etc), and compensatory mechanisms that together contribute to the maintenance of homeostasis in the face of a changing biological and biochemical environment, and in the context of the genetic and lifetime history of the individual.9–10, 18 So it is likely that molecular aging has a strong adaptive component, and that disruptions of subsets of age-dependent genes may offset this homeostatic state, without necessarily affecting global molecular aging. Second, in terms of trajectory of changes, MDD correlates with numerous changes in gene expression that have at best very small age-related effects on gene transcripts in control subjects, which on their own may never reach threshold for disrupted functions leading to MDD (Figures 2, 4). Finally, a progressive normal loss of ~40–50% of BDNF expression in 50 years may have a very different impact on neurobiological processes, compared to a more drastic loss of 50%, which is reported to occur in depressive episodes, as measured by circulating blood levels.27 Although the origin and trajectory of BDNF loss in MDD is not known, peripheral measures suggest that BDNF changes in MDD correlate with depressive episodes and recovery,27 demonstrating a plasticity of BDNF changes in MDD that is most likely very different from that of aging. Here since the majority of subjects were depressed at time of death (<25% in partial remission), we could not test the prediction of milder age-related deviations in recovered MDD subjects. So together, the emerging picture is that of a distorted age profile in MDD, with some age-related genes displaying normal trajectories, and others displaying much greater changes or amplified sub-threshold effects.
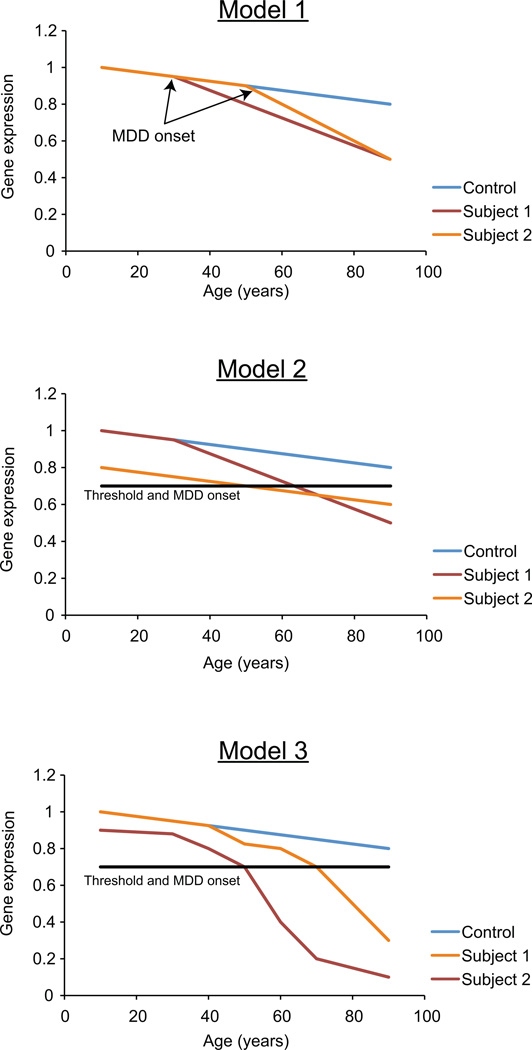
Thus, if anticipated molecular aging is observed in MDD subjects, the next set of unresolved questions is as follows: what is the actual trajectory of these changes and what could be the precipitating factors? Here for heuristic purposes, and since we cannot currently follow actual brain molecular trajectories within individuals, we speculate on putative trajectories to illustrate possible relationships between aging and depressed states. The first possibility is that the level of MDD-related gene expression is at control level prior to the onset of MDD (Figure 5; Model 1). The onset of the pathology causes drastic changes in gene expression and accelerates molecular aging. Second (Figure 5, Model 2), parallel and/or independent trajectories of altered gene expression eventually reach a threshold of disease onset, and genes may reside on “vulnerability trajectories” from variable length of times. Precipitating factors could be stress, cardio-vascular accidents, early-life events, pregnancy, etc. A combination of the two models could also be hypothesized, where early changes are accelerated by precipitating factors (Figure 5, Model 3). Conversely, protecting factors may place molecular changes on decelerated or “resilient” trajectories, hence forming the basis for individual variability in disease onset and presentation (not shown; See McKinney et al5 in this issue).
FIGURE 5. Putative trajectories of gene expression changes underlying aging and onset of MDD.
See Discussion for details. Model 1: the onset of the pathology causes drastic changes in gene expression and accelerates molecular aging. Model 2: independent trajectories of altered gene expression reach a threshold of disease onset after variable length of times. Model 3: in a combination of the first two models, early changes are accelerated by precipitating factors and reach threshold of disease onsets at variable times.
There are numerous limitations to this study. The first is that all conclusions are drawn from cross-sectional observations, since longitudinal studies of molecular changes are not feasible in the human brain. Moreover we cannot extrapolate on expression level of MDD-related genes before MDD episodes, or prior to the beginning of the pathology. Second, the expression levels of genes were measured using combined gray matter tissue samples, so small changes in expression levels or in the magnitude of changes over time may represent either true small changes, which may remain biologically-silent in control subjects (as discussed earlier), or may represent changes of greater magnitude in subsets of cells for which the effect size would be diluted in the combined samples. This latter point will need to be addressed by single cell studies, using laser-capture microscopy for instance. The third limitation is that we have not tested for a causality relationship, and specifically investigated whether the overlap of age-related changes and aging actually cause MDD-related symptoms. Genetic studies in rodent systems may provide appropriate models for testing cause and effect. Finally, these results were obtained in a cohort of women with relatively severe MDD profiles (33% death by suicide; 71% recurrent MDD) and will need to be confirmed in independent and larger cohorts, where the potential moderating effects of multiple factors, such as gender, disease severity and other clinical variables (i.e., death by suicide, age of onset and comorbid condition) can be fully evaluated.
Supplementary Material
Acknowledgement
This work was supported by National Institute of Mental Health (NIMH) MH084060 and MH093723 grants to ES, and MH043784 and MH084053 to DAL. The funding agency had no role in the study design, data collection and analysis, decision to publish and preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIMH or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest David A. Lewis currently receives investigator-initiated research support from Bristol-Myers Squibb, Curridium Ltd and Pfizer and in 2009–2011 served as a consultant in the areas of target identification and validation and new compound development to BioLine RX, Bristol-Myers Squibb, Merck and SK Life Science. The other authors declare no conflicts of interest.
REFERENCES
- 1.Reynolds CF, 3rd, Kupfer DJ. Depression and aging: a look to the future. Psychiatr Serv. 1999;50:1167–1172. doi: 10.1176/ps.50.9.1167. [DOI] [PubMed] [Google Scholar]
- 2.Steffens DC, Otey E, Alexopoulos GS, et al. Perspectives on depression, mild cognitive impairment, and cognitive decline. Arch Gen Psychiatry. 2006;63:130–138. doi: 10.1001/archpsyc.63.2.130. [DOI] [PubMed] [Google Scholar]
- 3.Lenze EJ, Schulz R, Martire LM, et al. The course of functional decline in older people with persistently elevated depressive symptoms: longitudinal findings from the Cardiovascular Health Study. J Am Geriatr Soc. 2005;53:569–575. doi: 10.1111/j.1532-5415.2005.53202.x. [DOI] [PubMed] [Google Scholar]
- 4.Fiske A, Wetherell JL, Gatz M. Depression in older adults. Annu Rev Clin Psychol. 2009;5:363–389. doi: 10.1146/annurev.clinpsy.032408.153621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKinney BCS. E.: The Age-by-Disease Hypothesis of Late-Life Depression. American Journal of Geriatric Psychiatry. 2012 doi: 10.1016/j.jagp.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexopoulos GS, Schultz SK, Lebowitz BD. Late-life depression. a model for medical classification. Biol Psychiatry. 2005;58:283–289. doi: 10.1016/j.biopsych.2005.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolkowitz OM, Reus VI, Mellon SH. Of sound mind and body: depression, disease, and accelerated aging. Dialogues Clin Neurosci. 2011;13:25–39. doi: 10.31887/DCNS.2011.13.1/owolkowitz. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolkowitz OM, Epel ES, Reus VI, et al. Depression gets old fast. do stress and depression accelerate cell aging? Depress Anxiety. 2010;27:327–338. doi: 10.1002/da.20686. [DOI] [PubMed] [Google Scholar]
- 9.Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 10.Glorioso C, Sibille E. Between destiny and disease: genetics and molecular pathways of human central nervous system aging. Prog Neurobiol. 2011;93:165–181. doi: 10.1016/j.pneurobio.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erraji-Benchekroun L, Underwood MD, Arango V, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 13.Blalock EM, Chen KC, Sharrow K, et al. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CK, Klopp RG, Weindruch R, et al. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 15.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 16.Blalock EM, Geddes JW, Chen KC, et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glorioso C, Oh S, Douillard GG, et al. Brain molecular aging, promotion of neurological disease and modulation by Sirtuin5 longevity gene polymorphism. Neurobiol Dis. 2011;41:279–290. doi: 10.1016/j.nbd.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webster MJ, Weickert CS, Herman MM, et al. BDNF mRNA expression during postnatal development, maturation and aging of the human prefrontal cortex. Brain Res.Dev.Brain Res. 2002;139:139–150. doi: 10.1016/s0165-3806(02)00540-0. [DOI] [PubMed] [Google Scholar]
- 20.Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 21.Weickert CS, Hyde TM, Lipska BK, et al. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 22.Guilloux JP, Douillard-Guilloux G, Kota R, et al. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Mol Psychiatry. 2011 Sep 13; doi: 10.1038/mp.2011.113. doi: 10.1038/mp.2011.113. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto T, Bergen SE, Nguyen QL, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glantz LA, Lewis DA. Reduction of synaptophysin immunoreactivity in the prefrontal cortex of subjects with schizophrenia. Regional and diagnostic specificity. Arch Gen.Psychiatry. 1997;54:660–669. doi: 10.1001/archpsyc.1997.01830190088009. [DOI] [PubMed] [Google Scholar]
- 25.Sibille E, Wang Y, Joeyen-Waldorf J, et al. A molecular signature of depression in the amygdala. Am J Psychiatry. 2009;166:1011–1024. doi: 10.1176/appi.ajp.2009.08121760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B, Methodological. 1995;57:289–300. [Google Scholar]
- 27.Sen S, Duman R, Sanacora G. Serum brain-derived neurotrophic factor, depression, and antidepressant medications: meta-analyses and implications. Biol Psychiatry. 2008;64:527–532. doi: 10.1016/j.biopsych.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.