Abstract
Background
Historically, VHL was the only frequently mutated gene in clear cell renal cell carcinoma (ccRCC), with conflicting clinical relevance. Excitingly, recent sequencing efforts identified several novel, frequent mutations of histone modifying and chromatin remodeling genes in ccRCC, including PBRM1, SETD2, BAP1 and KDM5C. Intriguingly, PBRM1, SETD2 and BAP1 are located in close proximity to VHL within a commonly lost (~90%) 3p locus. To date the clinical and pathologic significance of mutations in these novel candidate tumor suppressors is unknown.
Objective
To determine the frequency of and render the first clinical and pathologic outcome associated with mutations of these novel candidate tumor suppressors in ccRCC.
Design, Setting, and Participants
Targeted sequencing was performed in 185 ccRCC and matched normal tissues from a single institute. Pathologic features, baseline patient characteristics and follow-up data were recorded.
Statistical Analysis
The linkage between mutations and clinical and pathologic outcomes was interrogated with Fisher’s exact test (for stage and Fuhrman nuclear grade) and the permutation log-rank test (for cancer specific survival).
Results and Limitations
PBRM1, BAP1, SETD2 and KDM5C are mutated at 29%, 6%, 8% and 8%, respectively. Tumors with mutations in PBRM1 or any of BAP1, SETD2 or KDM5C (19%) are more likely to present with stage 3+ diseases, p=0.01 and p=0.001, respectively. Small tumors (<4cm) with PBRM1 mutations are more likely to exhibit stage 3 pathologic features (OR 6.4, p=0.001). BAP1 mutations tend to occur in Fuhrman Grade 3–4 tumors (p=0.052) and associate with worse cancer specific survival (p=0.01). Clinical outcome data is limited by the number of events.
Conclusion
Most mutations of chromatin modulators discovered in ccRCC are loss-of-function, which associate with advanced stage, grade, and possibly worsened cancer specific survival. Further studies validating the clinical impact of these novel mutations and future development of therapeutics remedying these tumor suppressors are warranted.
Keywords: Chromatin, Histone, Mutation, Outcome, Renal Cell Carcinoma
Introduction
The mutational landscape of clear cell renal cell carcinoma (ccRCC), the most common and aggressive form of renal cell carcinoma, has been radically transformed in the last two years due to several kidney cancer genomics projects. Long considered to be a disease dominated by the mutation of a single gene, VHL, recent studies identified several frequently mutated genes, including PBRM1 (1), SETD2, KDM5C (2, 3) and BAP1(4, 5).
Dalgliesh et al. performed selected exon sequencing of candidate cancer genes on 101 ccRCCs and discovered several novel recurrent mutations in chromatin remodeling genes, including SETD2 and KDM5C (2). In a follow-up study, these authors identified truncating mutations of PBRM1 in 41% of ccRCCs, nominating PBRM1 as the second most common mutated gene behind VHL (1). A subsequent report by Guo et al. confirmed the frequent mutations of VHL (27%), PBRM1 (21%), KDM5C (9%) and SETD2 (4%) in ccRCC, and identified BAP1 mutations in 8% cases (4). Interestingly, a recent study suggested a strong association between BAP1 mutations and advanced ccRCC tumor grade (5). Although whether and how mutations of these chromatin modulating genes contribute to the pathogenesis of ccRCC is unknown, disruption of chromatin biology has become an emerging theme constituting a new pathobiology underlying oncogenesis.
These findings are particularly intriguing for several reasons. First, these four genes all function in chromatin biology. Although the significance of this chromatin connection is currently unclear, histones and chromatin are essential building blocks of genomic architecture and dysregulation leads to transcriptional disruption of gene expression (6, 7). Second, PBRM1, SETD2 and BAP1 are located in close proximity at the 3p21 locus, right next to the 3p25 locus where VHL resides (Supplemental figure 1). Hence, the signature single copy loss of 3p (>90%) in ccRCC (8) would simultaneously impair four tumor suppressors that might be functionally linked. Third, PBRM1, BAP1 and SETD2 have recently been implicated in the pathogenesis of cancers other than ccRCC (9–13).
The clinical relevance of the VHL mutation in ccRCC has thus far been conflicting in both mutation frequency and its relationship to adverse tumor features and clinical outcomes. (14) Plausible causes underlying the wide range of VHL loss include tumor purity, tumor and patient heterogeneity, promoter methylation, etc. Nevertheless, many contemporary series estimate that VHL is impaired through mutations and promoter silencing in upwards of 90% of ccRCC (14, 15). Hence, the mutation status of VHL alone is quite unlikely to be useful as a biomarker for disease aggressiveness. On the other hand, a recent phylogenetic assessment of tumor heterogeneity in ccRCC suggested that mutations of chromatin modulators are secondary events and contribute to invasive and metastatic phenotypes (16). Hence, the mutation status of chromatin modulators in ccRCC might offer important prognostic insights and thus render novel therapeutic strategies. Accordingly, we set out to investigate the prevalence and the pathologic and clinical significance of these mutations in ccRCC patients from a single institute.
Methods
Patient Samples
Tissue samples from 185 treatment naive patients, undergoing either radical or partial nephrectomy for sporadic ccRCC from December of 2001 to December of 2011 were collected based on tissue availability and quality of samples with 85% of tumors collected after 2007 All patients had signed informed consents for tissue utilization and the study had been approval from our Institutional Review Board. All tumor staging was based on the AJCC/UICC TNM 7th Edition. All tumors were reviewed by a group of dedicated uropathologists to confirm the histopathologic diagnosis. Fresh frozen tumors and paired normal tissue blocks were identified and macro-dissected from areas marked by a uropathologist for maximal tumor density. DNA was extracted from tissue samples using DNEasy (Qiagen) and was quantified using a Nanodrop spectrophotometer (Invitrogen).
Integrated Mutation Analysis
Mutation analysis of entire coding regions of VHL, PBRM1, SETD2, BAP1 and KMD5C was performed using PCR amplification and bidirectional Sanger sequencing. Details of mutation analysis can be seen in Supplemental Methods.
Statistical Analysis
The association between the clinical and pathologic outcome and individual mutations was evaluated using Fisher’s exact test with two-tailed p values (for stage and Fuhrman nuclear grade) or log-rank permutation test (for cancer specific survival) (17). Odds ratios were used to estimate the strength of association for co-expression or mutual exclusivity among mutations as well as between mutations and pathologic features with 95% confidence intervals estimated using a logarithmic transformation. Multiplicity adjustment is not considered and, as a result, our conclusions are presented in suggestive rather than confirmatory language.
Results
Patients
The demographics, clinical, and pathologic characteristics of the 185 ccRCC patients are presented in Table 1. Patients were grouped into low risk (AJCC 1–2) and high risk (AJCC 3–4) for an analysis of association with gene mutations. Pathologic Fuhrman nuclear grades were grouped into low grade (1–2) and high grade (3–4) for mutation associations. Eighteen percent of patients developed metastasis: 21 patients presented with metastatic disease (cytoreductive nephrectomy) and 12 patients developed de novo distant metastasis. Seventeen patients (9%) died at last follow-up with 10 (5.4%) dying from ccRCC.
Table 1.
Clinical and pathologic characteristics of the cohort.
| Number of Patients | 185 |
|
| |
| Age (Median, IQR) | 61.4 (53.4,68.4) |
|
| |
| Gender | Female–29% |
| Male–71% | |
|
| |
| Race | White–90% |
| Black–4% | |
| Other–6% | |
|
| |
| BMI | 29.5 (25.6,34.1) |
|
| |
| Labs | Hb–13.8 (12.5,14.7) |
| Ca –9.3 (9,9.6) | |
|
| |
| Presentation | Incidental–77% |
| Local–18% | |
| Systemic–5% | |
|
| |
| Pathologic Stage | T1a–25.9% |
| T1b–13.5% | |
| T2a/b–7% | |
| T3–51.4% | |
| T4–2.2% | |
|
| |
| AJCC Clinical Stage | 1–38.4% |
| 2–5.9% | |
| 3–43.8% | |
| 4–11.9% | |
|
| |
| Fuhrman Nuclear Grade | 1–0.5% |
| 2–40% | |
| 3–48% | |
| 4–11.5% | |
|
| |
| Tumor Size (cm) (median, IQR) | 5 (3.3,8.2) |
|
| |
| Metastasis | 18% (33) |
| Metastasis at Presentation | 11.5% (21) |
| De Novo Metastasis | 6.5%(12) |
| Contralateral Recurrence | 1%(2) |
|
| |
| Followup Survivors (months) – average | 31 (1–123) |
|
| |
| Death | 17 (9%) |
| Death from RCC | 10 (5.4%) |
Frequencies and Types of Mutations
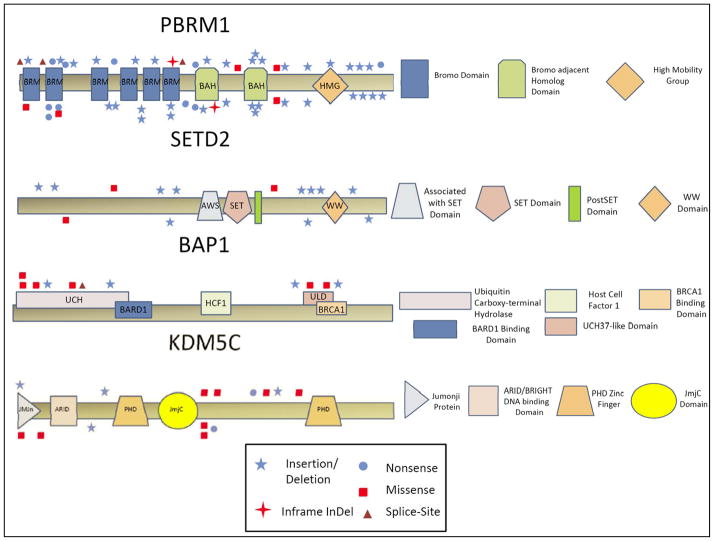
Overall, 119 of the 185 (65%) tumors harbored genetic mutations in at least one of the five target genes. VHL mutations were found in 49.2% of tumors, PBRM1 in 29.2%, SETD2 in 7.6%, KDM5C in 7.6%, and BAP1 in 5.9% (Figure 1 heat map). Supplemental Table 2 lists clinical and pathologic information of these 119 patients. In total, truncating mutations, i.e., frameshift and nonsense, represent 67%, 82.5%, 78.5%, 35.5% and 36% of mutations in VHL, PBRM1, SETD2, KDM5C and BAP1, respectively (Supplemental Tables 3 and 4, and Supplemental Figures 2 and 3). Individual gene maps with notations of mutation types and their positional relationships to the indicated functional domains are depicted in Figure 2.
Figure 1.

Heat map of mutations in affected samples (65% – 119/185).
Figure 2.
Gene maps with mutation types, locations, and gene domains.
Mutations and Clinical Outcomes
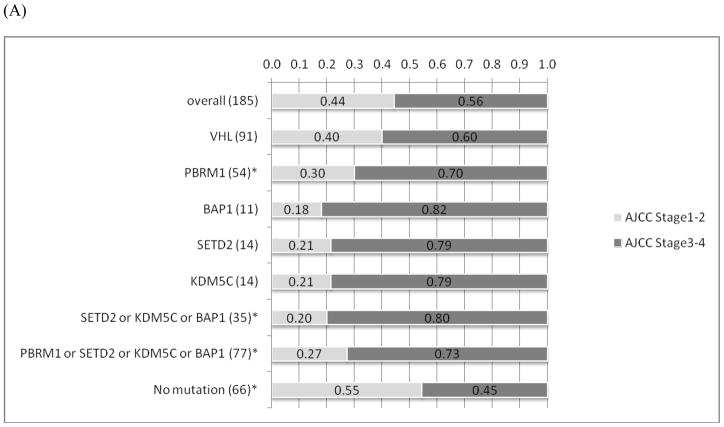
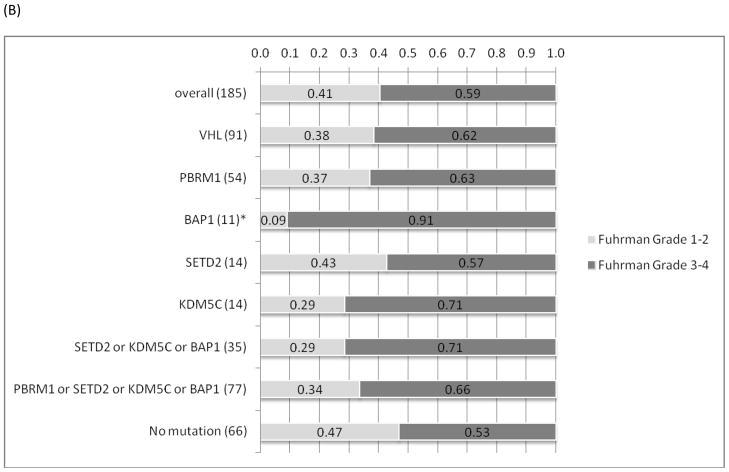
There is a tendency of mutual exclusivity between PBRM1 and BAP1 mutations, which is supported by a recent report (5), but it did not reach statistical significance (p=0.18) (Figure 1). Interestingly, tumors with PBRM1 mutations (n=54) were more likely to present with advanced stages (p=0.01) (Figure 3). Although there was no association between tumors with mutations in BAP1, SETD2 or KDM5C alone and advanced stages at presentation, there was a statistically significant connection (p=0.001) between those with any of the three mutations (n=35) and advanced stages, likely reflecting the relatively low prevalence of individual mutations (<10%). A subset analysis of tumor by mutation type was not suggestive of any clinical or pathologic trends with the exception of SETD2 mutations. All SETD2 mutations were frameshift (n=11) with the exception of 3 missense mutations that score as either neutral or low by functional impact (18). When excluding missense mutations, SETD2 mutations alone were associated with advanced tumor stages (p=0.02). Furthermore, tumors with BAP1 mutations were more likely to have higher Fuhrman nuclear grades (3–4), but this was not quite statistically significant (p=0.052), whereas none of the remaining gene mutations was associated with Fuhrman grade (Figure 3). There were 12 tumors that underwent sarcomatoid de-differentiation; however, no associations between specific individual mutations with sarcomatoid changes were detected. (data not shown).
Figure 3.
(A) The associations of between individual mutations and mutation combinations and AJCC stages. (B) The associations of individual mutations and mutation combinations with Furhman nuclear grades. * Indicates statistical significance (p<0.05; Fischer’s exact test).
Mutation Impacts on Small Tumors
In terms of tumor sizes, there was no statistically significant association between mutation and size (Data not shown). Since small kidney tumors (≤4cm), incidentally found by imaging studies, are generally considered as less aggressive and might be reasonably managed with close monitoring, we wished to determine if there is a connection between underlying genetic abnormalities and pathologic stages in such tumors. Primary kidney tumors, less than 4cm, are classified as pathologic T1a when no aggressive pathologic features are observed. On the other hand, pathologic T3a tumors are those that grossly extend into the renal vein or muscle containing branches, or invade into sinus or perirenal fat. Overall, 69 tumors were smaller than 4cm, comprising 48 (pT1a) and 21 (pT3a) tumors. Remarkably, we discovered that small tumors with PBRM1 mutations alone (n=12) were greater than 6 times more likely to be at pathologic T3a than those (n=9) with wild type PBRM1 (OR 6.44 [1.8–24.7], p=0.001). Similarly, small tumors with mutations in BAP1, SETD2 and/or KDM5C also exhibited a higher propensity to be at pT3a (OR 5.3 [1.2–28.8], p=0.03). In total, tumors with any of the PBRM1, BAP1, SETD2 and/or KDM5C mutations were likely to be at higher stage (OR 10.3 [2.8–44.7], p<0.001), supporting the notion that chromatin dysfunction participates in disease progression.
Mutations and Metastasis
Based on our cohort and targeted genetic studies, there was no statistically significant association between specific mutations of chromatin modulators and the presence or the subsequent development of metastasis. Nevertheless, 27% and 29% of patients with either BAP1 or SETD2 mutations presented with or developed metastasis, compared to 18% of metastasis in the entire cohort (p value not statistically significant, Supplemental Table 5).
Mutations and Survival
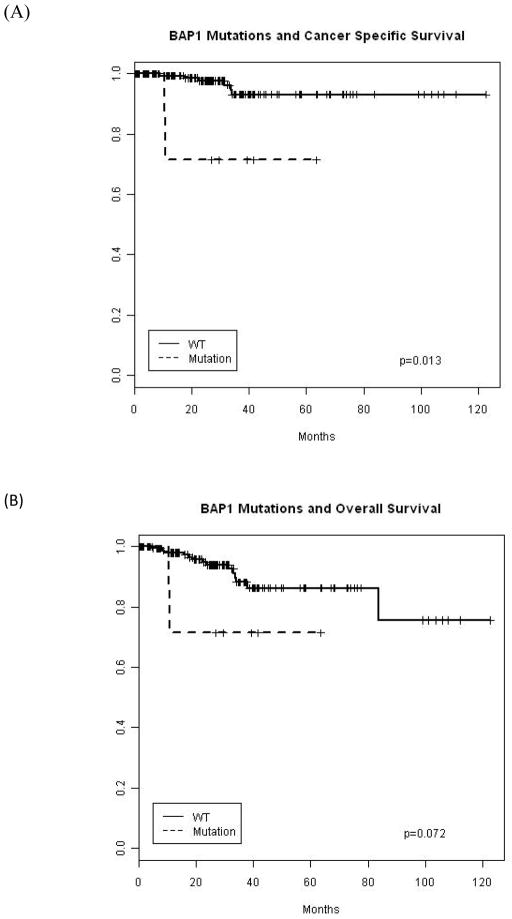
Survival analysis was limited by the number of events at the time of analysis. However, BAP1 mutations were significantly associated with worse survival outcomes on Kaplan Meier analysis (Fig 4; p=0.013 permutation log rank). On the contrary, there was no statistically significant association between other mutations and survival.
Figure 4.
Kaplan Meier survival plot for BAP1 mutation and (A) cancer specific and (B) overall survival.
Discussion
Although inactivation of the VHL gene is the most prevalent (upwards of 90%) genetic alteration based on certain reports and likely represents the primary initiating event for the pathogenesis of ccRCC, numerous lines of evidence indicate that the loss of VHL alone is insufficient to cause ccRCC and provides neither prognostic nor therapeutic prediction values (19, 20). Consistent with these notions, our study identified VHL mutations in nearly half of our cohort and detected no prognostic significance. Genetic loss of heterozygosity studies of ccRCC have implicated the chromosome 3p21 locus as a region harboring additional tumor suppressors besides VHL (21). Three large-scale genomic studies have identified several recurrently mutated genes in ccRCC, including PBRM1, BAP1, SETD2 and KDM5C. Remarkably, all have important roles in the epigenetic control of gene expression, and three of them (PBRM1, BAP1 and SETD2) are closely situated in the frequently lost 3p21 locus. Hence, our current study focused on addressing the clinical and/or pathologic significance of mutations in this newly identified class of candidate tumor suppressors.
In agreement with the recent reports, our data validate prevalent mutations of PBRM1, positioning it as the second most commonly mutated gene in ccRCC, right behind VHL. The PBRM1 gene encodes the Polybromo 1 (BAF180) protein, the chromatin targeting subunit of the Polybromo SWI/SNF complex (PBAF) (22). SWI/SNF complexes are large ATP-dependent chromatin-remodeling machines that mobilize nucleosomes along the DNA (23). Genetic data on PBRM1 from both ours and others’ indicate loss-of-function as the leading feature of tumor-derived PBRM1 mutations. However, how dysfunction of these SWI/SNF genes contributes to tumorigenesis is largely unknown. Nevertheless, our clinical and pathologic interrogation provides an invaluable hint. We found that small tumors with PBRM1 mutations are 6 fold more likely to be categorized as pT3a instead of pT1a, suggesting a role of PBRM1 in bridling cell invasiveness. SETD2 is a histone H3 lysine 36 (H3K36) methyltransferase that regulates mRNA splicing and transcription elongation (24, 25); BAP1 interacts with and deubiquitinates host cell factor-1 (HCF-1), a transcription co-activator, that regulates cell proliferation (26); and KDM5C a histone 3 trimethyl-lysine 4 (H3K4Me3) demethylase that erase active transcription marks (27). Mutation rates of these three genes occur in 5–10% of ccRCC and most mutations are loss-of-function, supporting their roles as tumor suppressors. Despite the relatively low mutation incidences and short clinical follow up, we were able to deduce certain clinical/pathologic features associated with individual mutations. In terms of SETD2, we identified 11 loss-of-function and 3 low impact missense mutations(18). Among the 11 tumors bearing SETD2 loss-of-function mutations, 10 occurred in stage 3+, of which 2 presented with and 2 subsequently developed metastatic diseases. In total, the overall metastatic rate of patients with SETD2 mutated primary tumors is 36% (4/11), implicating a functional connection between SETD2 and cancer metastasis. With respect to BAP1 mutations, we demonstrated associations with high Fuhrman nuclear grades and worse cancer specific survivals, underscoring its tumor suppressor role in both mesothelioma and uveal melanoma. In regard to the KDM5C mutation alone, we were unable to detect significant associations with specific clinical/pathologic states, indicating that a larger cohort and a longer follow up may be required to unveil its role in the pathogenesis of ccRCC.
Limitations of this study include the lack of a significant number of cancer specific outcomes and gene expression. Nevertheless, the majority of mutations in VHL, PBRM1 and SETD2 are nonsense/frameshift truncating mutants and invariably cause loss of the protein function/product, as do the essential splice site mutations we identified in BAP1. While there is a fair amount of missense mutations in BAP1 and KMD5C, we use the validated MUTATIONASSESSOR computational algorithm to predict the functional impact of these mutations (Supplemental table 2). This analytic filter suggests that the mutations are functionally deleterious.. Furthermore, we were unable to address the issue of epigenetic gene silencing through mechanisms such as methylation or polyadenylation. However, publically available, unpublished data from the Cancer Genome Atlas project (TCGA) does not implicate any of these genes as frequently methylated other than VHL.
Rationales underlying mechanism-based therapeutics in advanced and metastatic ccRCC have evolved predominantly around the VHL/HIF angiogenic axis. The subsequently proven clinical benefit of administering anti-angiogenic agents to ccRCC patients has led to the approved first-line use of such agents, including Sunitinib and Pazopanib. However, despite these mark strides against this deadly disease, most advanced-stage patients eventually succumb to their illness, urging for the development of new targeting strategies. Data thus far demonstrated that the involvement of this new class of tumor suppressors, i.e. chromatin modulators, in cancers is clearly beyond ccRCC. Thus, further clinical, pathologic and mechanistic interrogations likely yield novel therapeutic insights that impact diverse cancers.
Conclusions
To our knowledge, our data presents the first clinical/pathologic assessment of a novel class of tumor suppressors, consisting of chromatin modulating factors, in ccRCC. Our study indicates that mutations of PBRM1, SETD2, BAP1 and/or KDM5C in kidney cancers are associated with advanced stage, grade and tumor invasiveness. Remarkably, small (<4cm) tumors with PBRM1 mutations, the second most commonly mutated gene in ccRCC, are significantly linked to a higher tumor stage. Furthermore, BAP1 mutations appear to associate with worse cancer specific survival. Further studies to validate clinical impacts and future therapies to target this new class of cancer contributory chromatin-modulating genes in ccRCC are warranted.
Supplementary Material
Table 2.
Pathologic stages (pT1a to pT3a) in tumors ≤4cm. Statistically significant findings are bolded (p<0.05; Fischer’s exact test).
| Mutation | VHL | PBRM1 | BAP1 | SETD2 | KDM5C | BAP1 or SETD2 or KDM5C | PBRM1 or BAP1 or SETD2 or KDM5C |
|---|---|---|---|---|---|---|---|
| Overall Frequency (n=185) | 49% | 29% | 6% | 8% | 8% | 19% | 42% |
| Frequency in Tumors ≤4cm (n=69) | 49% | 29% | 3% | 9% | 6% | 16% | 39% |
| Pathologic T1a (n=48) | 21 (44%) | 8 (17%) | 1 (2%) | 3 (6%) | 1 (2%) | 4 (8%) | 11 (23%) |
| Pathologic T3a(n=21) | 13 (62%) | 12 (57%) | 1 (5%) | 3 (15%) | 3 (15%) | 7 (33%) | 16 (76%) |
| Odds Ratio (CI) | 2.1 (0.7 –6.9) | 6.4 (1.8 –24.7) | 2.3 (0.03 –188) | 2.5 (0.3 –20.2) | 7.6 (0.6 –418.6) | 5.3 (1.2 –28.8) | 10.3 (2.83–44.7) |
| p=0.20 | p=0.001 | p=0.52 | p=0.36 | p=0.08 | p=0.03 | p<0.001 |
Acknowledgments
Financial Support:
This work has been supported by the Paula Moss Trust for research into the cure and treatment of kidney cancer (Hsieh). The National Cancer Institute T32 CA082088-12 and the Stephen P Hanson Family Fund Fellowship in Kidney Cancer (Hakimi).
Footnotes
Conflict of Interest:
None to report for any author
References
- 1.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–42. doi: 10.1038/nature09639. Epub 2011/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–3. doi: 10.1038/nature08672. Epub 2010/01/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duns G, van den Berg E, van Duivenbode I, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer research. 2010;70(11):4287–91. doi: 10.1158/0008-5472.CAN-10-0120. Epub 2010/05/27. [DOI] [PubMed] [Google Scholar]
- 4.Guo G, Gui Y, Gao S, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature genetics. 2012;44(1):17–9. doi: 10.1038/ng.1014. Epub 2011/12/06. [DOI] [PubMed] [Google Scholar]
- 5.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44(7):751–9. doi: 10.1038/ng.2323. Epub 2012/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–52. doi: 10.1038/38664. Epub 1997/10/06. [DOI] [PubMed] [Google Scholar]
- 7.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding--wrapping up transcription. Science. 2002;297(5588):1824–7. doi: 10.1126/science.1074200. Epub 2002/09/14. [DOI] [PubMed] [Google Scholar]
- 8.Toma MI, Grosser M, Herr A, et al. Loss of heterozygosity and copy number abnormality in clear cell renal cell carcinoma discovered by high-density affymetrix 10K single nucleotide polymorphism mapping array. Neoplasia. 2008;10(7):634–42. doi: 10.1593/neo.08160. Epub 2008/07/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shain AH, Giacomini CP, Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(5):E252–9. doi: 10.1073/pnas.1114817109. Epub 2012/01/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–3. doi: 10.1126/science.1194472. Epub 2010/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nature genetics. 2011;43(7):668–72. doi: 10.1038/ng.855. Epub 2011/06/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–63. doi: 10.1038/nature10725. Epub 2012/01/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Sarakbi W, Sasi W, Jiang WG, Roberts T, Newbold RF, Mokbel K. The mRNA expression of SETD2 in human breast cancer: correlation with clinico-pathological parameters. BMC cancer. 2009;9:290. doi: 10.1186/1471-2407-9-290. Epub 2009/08/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossage L, Eisen T. Alterations in VHL as potential biomarkers in renal-cell carcinoma. Nature reviews Clinical oncology. 2010;7(5):277–88. doi: 10.1038/nrclinonc.2010.42. Epub 2010/04/07. [DOI] [PubMed] [Google Scholar]
- 15.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(15):4726–34. doi: 10.1158/1078-0432.CCR-07-4921. Epub 2008/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. Epub 2012/03/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heller G, Venkatraman ES. Resampling procedures to compare two survival distributions in the presence of right censored data. Biometrics. 1996;52:1204–13. [Google Scholar]
- 18.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39(17):e118. doi: 10.1093/nar/gkr407. Epub 2011/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandriota SJ, Turner KJ, Davies DR, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer cell. 2002;1(5):459–68. doi: 10.1016/s1535-6108(02)00071-5. Epub 2002/07/19. [DOI] [PubMed] [Google Scholar]
- 20.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer research. 2006;66(5):2576–83. doi: 10.1158/0008-5472.CAN-05-3241. Epub 2006/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER. Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes, chromosomes & cancer. 1998;22(3):200–9. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. Epub 1998/06/13. [DOI] [PubMed] [Google Scholar]
- 22.Thompson M. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91(3):309–19. doi: 10.1016/j.biochi.2008.10.019. Epub 2008/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400(6746):784–7. doi: 10.1038/23506. Epub 1999/08/31. [DOI] [PubMed] [Google Scholar]
- 24.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes & development. 2008;22(24):3422–34. doi: 10.1101/gad.1720008. Epub 2009/01/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exonsby H3K36me3. Nature genetics. 2009;41(3):376–81. doi: 10.1038/ng.322. Epub 2009/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. The Journal of biological chemistry. 2009;284(49):34179–88. doi: 10.1074/jbc.M109.046755. Epub 2009/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. Epub 2007/05/22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.