SUMMARY
Superfamily ATPases in Type IV pili (T4P), Type 2 secretion (T2S), and archaella (formerly archaeal flagella) employ similar sequences for distinct biological processes. Here we structurally and functionally characterize prototypical superfamily ATPase FlaI from Sulfolobus acidocaldarius showing FlaI activities in archaeal swimming organelle assembly and movement. FlaI solution X-ray scattering and crystal structures with and without nucleotide reveal a hexameric crown assembly with key cross-subunit interactions: rigid building blocks form between N-terminal domains (points) and neighboring subunit C-terminal domains (crown ring). Upon nucleotide binding, these six cross-subunit blocks move with respect to each other distinctly from secretion and pilus ATPases. Crown interactions and conformations regulate assembly, motility and force direction by a basic-clamp switching mechanism driving conformational changes between stable, backbone-interconnected moving blocks. Collective structural and mutational results identify in vivo functional components for assembly and motility, phosphate triggered rearrangements by ATP-hydrolysis, and molecular predictors for distinct ATPase superfamily functions.
INTRODUCTION
The assembly and movement of archaella (formerly termed archaeal flagella), bacterial Type IV pili (T4P), plus Type 2 and 4 secretion (T2S and T4S) systems are driven by superfamily ATPases. Both archaella and bacterial flagella function as rotating swimming organelles. Yet, archaella lack the hollow core of bacterial flagella and are assembled at the base (not the tip) by ATPase motors that are not homologous to bacterial flagella. Archaella are thus a unique motility structure with functional similarity to bacterial flagella, but structural similarity to T4P (Ghosh and Albers, 2011; Pohlschroder et al., 2011; Trachtenberg and Cohen-Krausz, 2006). The archaellum (Jarrell and Albers, 2012) and T4P assembly machinery components (Bardy et al., 2004; Ng et al., 2006) are also homologous. Both bacterial prepilins and prearchaellins contain class III signal peptides processed by homologous T4P peptidases, PilD in bacteria and FlaK/PibD in archaea (Albers et al., 2003; Bardy et al., 2002). Moreover, secretion system ATPase superfamily member FlaI and FlaJ membrane protein are homologous to PilT/B ATPases and the main transmembrane protein PilC in bacterial T4P, and likely function as a core platform for assembly and rotation of the archaellum (Planet et al., 2001; Thomas et al., 2002). However, due to limited biochemical and structural information for archaellum components we lack a mechanistic understanding of archaella assembly and motility.
From genetic manipulation on the two major archaeal kingdoms, euryarchaeota and crenarchaeota, all archaella assembly components encoded in the genomes of archaellated archaea are essential for assembly (Chaban et al., 2007; Lassak et al., 2011). As in haloarchaea, based on conserved archaella operons, archaellum movement is likely ATP dependent and not proton motive force driven (Streif et al., 2008).
Archaella, bacterial T4P, T2S, and T4S assembly systems all use homologous ATPase motors (Craig and Li, 2008; Hansen and Forest, 2006) (Figure S1). Those ATPases - PilE/PilF/TcpT/BfpD/CofH/LngH (T4P) (Chakraborty et al., 2008; Crowther et al., 2004; Gomez-Duarte et al., 2007; Tripathi and Taylor, 2007), GspE/XcpR/PulE/EpsE/ExeE/XpsE/OutE (T2S) (Filloux et al., 1998; Johnson et al., 2006; Russel, 1998; Shiue et al., 2006), VirB11 (T4S) (Savvides et al., 2003) or FlaI (archaellum) (Ghosh and Albers, 2011; Thomas et al., 2002) - are PilT/FtsK secretory ATPases, a subgroup of RecA/Rad51-like motors (Iyer et al., 2004; Planet et al., 2001; Shin et al., 2003). In T4P systems, these ATPases act in assembly (e.g. assembly ATPase PilB) of the respective filaments and in pilus disassembly or retraction of the pilus (e.g. retraction ATPase PilT) (Burrows, 2005). Structures of T4P filaments from Pseudomonas aeruginosa (Craig et al., 2003) and Neissera gonorrhoeae (Craig et al., 2006) suggest a model for T4P assembly-disassembly, which merits testing.
An added puzzle is how these sequence-similar T4P, T2S and archaellum ATPases use ATP binding and hydrolysis to drive distinct processes: T4P for assembly/disassembly, T2S for the secretion of proteins, and archaella for assembly and filament motion. Informative crystal structures and models of T2S assembly ATPases (Abendroth et al., 2005; Chen et al., 2005; Robien et al., 2003; Yamagata and Tainer, 2007) and the T4P retraction ATPase (Misic et al., 2010; Satyshur et al., 2007) allowed few functional tests due to challenges of working with pathogens for genetic in vivo studies.
Fortunately, the crenarchaeon Sulfolobus acidocaldarius provides a prototypic system for investigating T4P/T2S/archaellum ATPases, as genetic tools and non-pathogenicity aid genetic in vivo studies. Recent genetic tools to generate in-frame deletion mutants and functional in vivo experiments with proteins expressed in trans aid more thorough and detailed analysis of the archaellum system (Albers et al., 2006; Wagner et al., 2009). Furthermore, FlaI, which shows ATP-dependent oligomerization and stimulation by archaeal tetraether lipids (Ghosh et al., 2011), provides an ATPase for examining both archaella assembly and rotation.
To examine the molecular basis for FlaI ATPase dual activities and the functional diversity of this ATPase superfamily, we employed structural analyses by crystallography and small angle X-ray scattering (SAXS) in combination with in vitro and in vivo analyses. Collective results identify distinct residues and components essential for archaella assembly and motility, a striking cross-subunit interface for transmission of ATPase force, inter-subunit interactions to the active site, and a novel ADP-PO4 release intermediate that together support a unified mechanistic understanding of filament assembly and motility functions.
RESULTS
FlaI is the Assembly ATPase for S. acidocaldarius Archaella
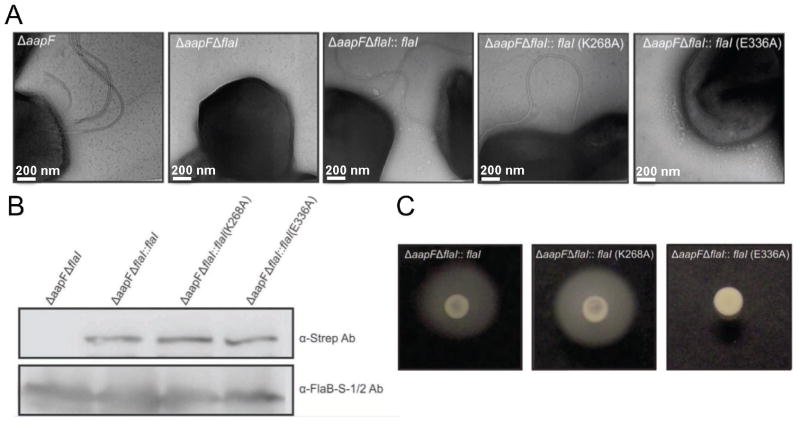
The S. acidocaldarius ΔaapF strain lacks the membrane protein of the archaeal attachment-association pili (aap) assembly system, resulting in a hyper-archaellated and hyper-motile phenotype (Henche et al., 2012). Deletion of FlaI in this background (ΔaapFΔflaI) eliminates surface archaellation, implicating FlaI as the assembly ATPase (Lassak et al., 2011). To test and extend these observations and further test FlaI roles in archaellum assembly and filament movement, we complemented the ΔaapFΔflaI strain in trans, using maltose inducible expression vectors, with wild-type flaI, or the Walker AK268A and Walker BE336A flaI mutants (Figure 1). All strains were examined for archaella by electron microscopy (EM) (Figure 1A), for FlaI expression levels by Western blot (Figure 1B) and for swimming in soft gelrite medium (Figure 1C) after inducing expression of complemented genes. While the ΔaapFΔflaI cell is non-motile in swimming assays (Movie S1 and S2), EM revealed that wild-type flaI and the flaI Walker AK268A mutant could assemble archaea, and these complemented strains were motile in swimming assays (Figure 1). Importantly, complementation with the flaI Walker BE336A mutation, which severely limits FlaI ATPase activity (Ghosh et al 2011), does not support archaella assembly or swimming. This essential role for FlaI ATPase activity in archaella assembly motivated our detailed structural characterizations of this ATPase as the basis for archaella assembly and swimming.
Figure 1. FlaI ATPase activity is essential for archaellum formation.
(A) EM images of the Sulfolobus cell surface (from left to right): The background is a hyper-archaellated strain (ΔaapF) that lacks pili. Deletion of the flaI gene results in no surface archaella. FlaI function is restored by introducing wild-type flaI or the Walker A mutant. The Walker B mutant strain is incapable of complementation, indicating the need of enzymatic ATPase activity for archaellum assembly.
(B) Wild-type FlaI, FlaIK268A Walker A and FlaIE336A Walker B mutants are expressed in the ΔaapFΔflaI mutant strain. Expression was detected by western blot, with the archaellin protein (FlaB) used as an endogenous control.
(C) Motility assays showing the ability of wild-type and FlaI variants to complement the motility phenotype in the ΔaapFΔflaI strain. From left to right: Swimming behavior of the in trans complemented ΔaapFΔflaI with wild-type FlaI, FlaI Walker AK268A mutant and FlaI Walker B mutant. See also Figure S1, Movie S1 and S2).
Domain Folds of FlaI ATPase
To examine FlaI structures and conformations for insights into archaellum assembly, full-length FlaI Walker B mutant (FlaIE336A) was purified and crystallized with nucleotide (ATP/ADP). The crystal structure was solved in space group C2 by phasing with seleno-methionine, allowing building of contiguous polypeptide chains (minus the N-terminal methionines) for the three FlaI molecules in the asymmetric unit and refinement to 2.0 Å resolution (Table 1). Generation of the crystallographic hexamer using the 2-fold axis provided a detailed FlaI structure with bound nucleotide for comparisons to other superfamily ATPases (Figures 2A and 2B).
Table 1.
X-ray Diffraction Data and Refinement Statistics
| Data set | FlaIE336A-ADP (native) | FlaIE336A-ADP (SeMet) peak/remote | FlaI-Apo |
|---|---|---|---|
| Diffraction Data Statistics | |||
| Synchrotron Beamline | ALS 8.3.1 | ALS 8.3.1 | ALS SIBYLS |
| Space group | C 2 | C 2 | P 63 |
| Cell dimensions | a=169.5Å; b=148.1Å; c=123.6Å á=ã=90°; â=131.6° |
a=169.6Å; b=148.0Å; c=124.4Å á=ã=90°; â=131.7° |
a=b=130.0Å; c=311.8 Å á=â=90°; ã=120° |
| Wavelength (Å) | 1.09 | 0.98/0.96 | 1.07 |
| Resolution range (Å)1 | 46.0 – 2.0 (2.2-2.0) | 57.9 – 2.5 (2.7-2.5) | 49.9 – 3.6 (3.8-3.6) |
| Completeness (%)1 | 99.7 (95.6) | 99.9 (97.6) | 99.5 (98.3) |
| Observed reflections | 5,453,245 | 2,088,354/2,170,286 | 1,135,996 |
| Unique reflections | 178,574 | 80,054 | 34,746 |
| Rsym (%)1 | 7.1 (49.7) | 5.4 (43.7) | 9.8 (86.0) |
| I/ó1 | 25.9 (2.5) | 36.4 (1.8) | 17.7 (3.0) |
| Refinement Statistics | refined as rigid body only | ||
|
| |||
| Resolution range (Å) | 46 - 2 | 50 – 3.6 | |
| Rwork/Rfree (%)1 | 18.5 (25)/21.4 (29) | 29.6 (37)/31.8 (40) | |
| No. atoms | |||
| Protein | 12,565 | 16,448 | |
| Ligand/ion | 98 | - | |
| Water | 766 | - | |
| Average B-factor | 57.4 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | ||
| Bond angles (°) | 1.18 | ||
| PDB ID | 4IHQ.pdb | 4II7.pdb | |
Values in parentheses are for the highest resolution shell.
Rsym is the unweighted R value on I between symmetry mates.
Rfree is the cross-validation R factor for 10% of reflections against which the model was not refined.
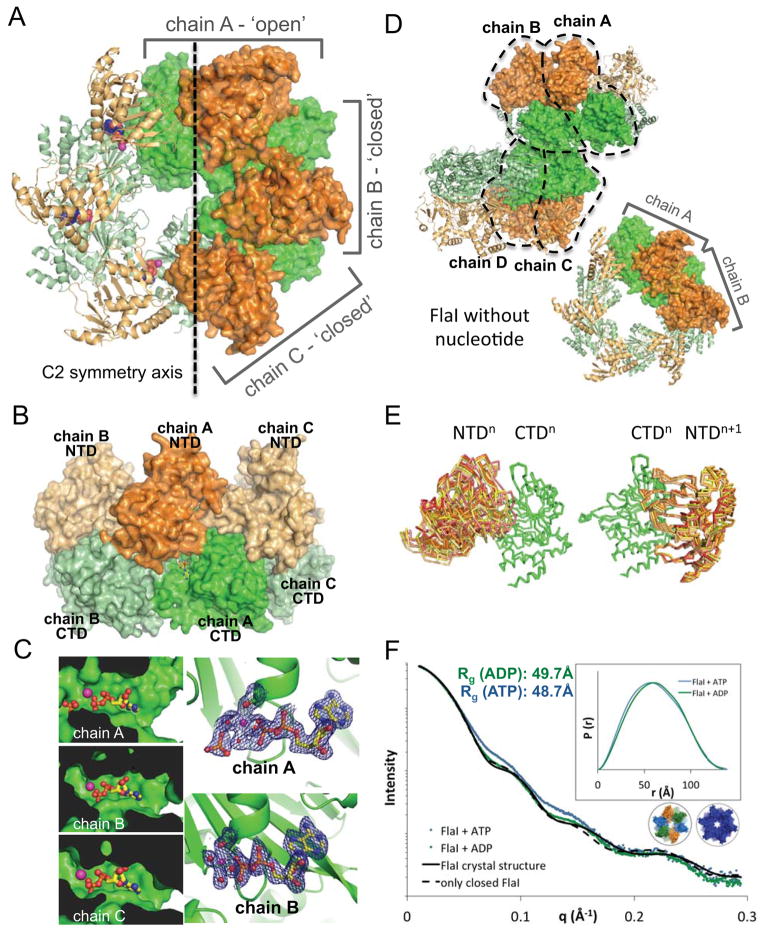
Figure 2. FlaI crystal structures show different subunit conformations.
(A) Crystal structure of the Walker B mutant FlaIE336A with NTDs (orange) and CTDs (green). A C2 symmetry axis divides the hexameric ring into 2 halves, three chains form the asymmetric unit (surface representation): chain A adopts a much more open conformation than chains B and C.
(B) The FlaI ring forms a crown-like structure. Side view of the FlaIE336A hexameric ring shows the crown-like structure with the CTDs forming the base, the NTDs the points, with nucleotide shown as yellow sticks.
(C) The three active sites are not identical. The active site in chain A is more open and solvent accessible than in chains B and C. Electron density (2|Fo|-|Fc| map at 1.5σ) for released phosphate is seen in chain A but not for chains B and C, which have similar electron densities (chain B shown as an example).
(D) Apo-FlaI without nucleotide also crystallized as a hexameric ring. Two subunits from two different hexameric rings form the asymmetric unit (surface representation), resulting in four unique subunit conformations. Colored as in A.
(E) Comparison of the seven observed FlaI conformations reveals FlaI building blocks (three from ADP-FlaI and four from apo-FlaI). Left, alignment of the CTDs shows many different conformations of the corresponding NTDs. Right, Building blocks of a CTDn with NTDn+1 superimpose very well for all seven conformations.
(F) ATP-bound FlaI is more compact than ADP-FlaI. SAXS data of FlaI with ATP (blue) and ADP (green) shows a more compact structure for ATP-FlaI. Theoretical scattering curves for the crystal structure (black line) and an all closed model (dashed black line) are shown. Inset: P(r) distribution of the same data. See also Figure S2.
Each FlaI monomer is a bi-lobed structure consisting of a C-terminal ATPase domain (CTD) conserved among homologous ATPases, and a much more variable N-terminal domain (NTD) (Figures 5A and S1). Both NTD and CTD form similar-sized globular structures with a diameter of 45–55 Å, and a flexible linker connects the two domains (Figures 2A and 2B). The NTD forms two regions; NTD-1 (residues 1–126) and NTD-2 (residues 127–225). NTD-1 consists of a 29-residue α-β-α-structure, followed by a three-stranded β-sheet and a three α-helix bundle. The NTD-1 electron density, and especially the three α-helix bundle, is less well-defined, suggesting high flexibility. NTD-2 has well-defined electron density, consisting of two α-helices and a large six-stranded β-sheet.
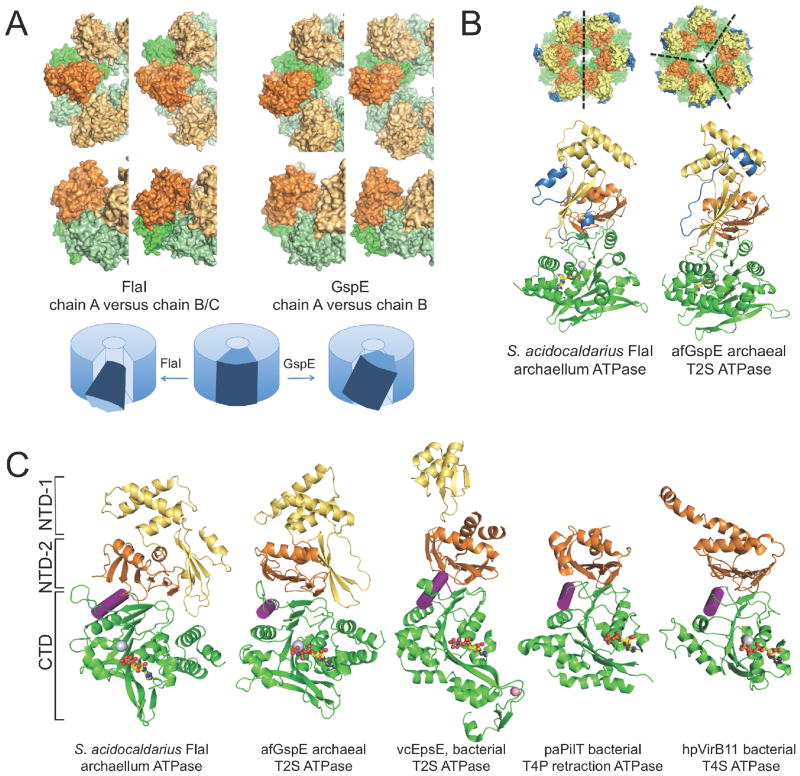
Figure 5. FlaI compared to other secretion family ATPases.
(A) Movements induced by ATP hydrolysis differ between FlaI and GspE. The two building blocks to compare are shown in the same orientation within the hexameric ring. For FlaI NTDchainA + CTDchainB is compared to NTDchainB + CTDchainC; for GspE the only two building blocks present in the structure are shown. The different building block conformations in the FlaI and GspE rings allow us to propose domain movements during ATPase hydrolysis. The directionality of movement differs between FlaI and GspE (top, top view; bottom, side view).
(B) The largest structural difference between FlaI and GspE is in residues 1-29 (blue). This region forms the outer side surface of the crown-like structure (top). Crystal related symmetry axes are indicated as black dashed lines.
(C) Like FlaI, other secretion system ATPases form building blocks. CTDs (green) are shown with NTD-2 (orange) and when present the NTD-1 (yellow) of the neighboring subunit with nucleotides (sticks), Mg2+ (silver) and Zn2+ ions (pink spheres). The α-helix mediating the block movement described in Figure 3 is highlighted (purple). See also Figure S4, Movies S3 and S4.
The CTD also forms two subdomains. CTD-1 (residues 226–418) contains all the structural and functional features of a RecA-like ATPase with Walker A and B and His box motifs. As in other RecA-like ATPases, it has a central β-sheet core flanked on both sides by α-helices. Two of the β-strands are very long and extend towards the CTD of the neighboring subunit. CTD-2 (residues 419–C-terminus) has two β-strands and four α-helices.
FlaI Hexameric Rings are Built of Different Monomer Conformations
FlaI crystallized as a hexameric ring with intrinsic 2-fold symmetry resulting in three unique subunits. The overall hexamer shape resembles a crown: CTDs form the crown ring and NTDs form crown points (Figure 2B). The FlaI crown has a diameter of 140 Å and a height from base to tip of 75 Å, with small and large openings formed by the CTD (9.5 Å) and NTD (75 Å) rings. Besides the linker, the CTD and NTD rings are connected within each FlaI monomer by a relatively small interface (buried surface area; B.S.A of 365–400 Å2). The FlaI monomers form the hexamer through much larger inter-subunit interfaces with the six-stranded β-sheet of NTD-2 packing against the neighboring FlaI CTD, a CTDn:NTDn+1 interaction, resulting in a B.S.A of ~1,240 Å2.
Interestingly, the FlaI hexamer is asymmetric and superimposition of the three full-length monomers shows that while chains B and C superimpose relatively well (RMSDCα = 0.68 Å), chain A adopts a dramatically different conformation (RMSDCα = 2.4 Å to chain B and 2.8 Å to chain C). Superimposition of separated NTDs and CTDs shows that despite differences in full length FlaI, the individual NTD and CTD structures are all similar, with only small changes in the NTD three-helix bundle at the crown points (average RMSDCα of NTDs = 0.95 Å and CTDs = 0.75 Å). In contrast, the linker connecting the CTD and NTD is flexible allowing a range of different conformations (RMSDCα of individual subunits = 4.7 Å). Comparison shows that the FlaI hexamer bound to ADP contains two open active sites (from chain A) and four closed and buried active sites (from chains B and C) (Figures 2A–C and Movie S3).
To compare nucleotide-bound and -free FlaI structures we also crystallized wild-type FlaI without nucleotide, in space group P63, and solved the structure by molecular replacement using PHASER (McCoy, 2007). The CTD structure of ADP-bound FlaIE336A was used as a first search model, followed by a search with the NTD structure. The structure was refined to 3.6 Å resolution by rigid body refinement (Table 1). Although FlaI exists as a monomer in solution without nucleotide (Ghosh et al., 2011), it forms hexameric rings in our structure, likely due to the high protein concentration within the crystal. The asymmetric unit consists of four unique subunits as part of two different hexameric rings (Figure 2D). Similar to the ADP-bound structure, the flexible linker connecting the NTD and CTD adopts four different conformations, however the hexameric arrangement is different with alternating open and closed active sites. Together with the three conformations seen in the ADP-bound structure, this gives a total of seven different FlaI conformations.
Comparing the seven structurally different FlaI subunits by superposition of their CTDs shows that the NTDs adopt multiple conformations (Figure 2E, left). Similarly, the intra-subunit interface between NTD and CTD is small compared to that between one NTD and the neighboring subunit CTD. Therefore we compared building blocks consisting of a CTD plus the tightly bound NTD from the next subunit. Superposition of the CTDn’s of these building blocks shows that the NTDn+1’s all align with each other (Figure 2E, right).
An ADP-PO4 Release Intermediate is Visible in Two Hexamer Active Sites
Nucleotide binding occurs at the interface between two FlaI subunits (Figures 2A and S2). As FlaIE336A shows strongly reduced ATPase activity (Ghosh et al., 2011), we expected to trap an ATP-bound state when we crystallized it with ATP and MgCl2. However, the electron density in the active site showed that ADP and Mg2+ were bound in all subunits (Figure 2C), indicating ATP hydrolysis during crystallization. Interestingly, the major difference between FlaI subunits is the rotation of one molecule (chain A), resulting in an open active site compared to a closed conformation for chains B and C. Closer examination of the chain A conformation revealed extra electron density consistent with a phosphate moiety (Figure 2C and S3). The phosphate is 4 Å away from the active site Mg2+ and 4.2 Å from the ADP β-phosphate, suggesting this a release intermediate that has not previously been seen in this superfamily. ADP and Mg2+ binding residues are similar within the CTD of all three subunits, with the only major rearrangement being a dramatic conformational switch of Arg338, which moves to coordinate the released phosphate in chain A (Figure 3). Importantly, the most dramatic difference between chain A and chains B/C is the rotation of the NTD. The NTD of chain A rotates ~20°, and due to their extensive shared interface this conformational rearrangement shifts the chain B CTD towards the chain A active site (Figure 3A). The net result is the fitting of a chain B α-helix into a groove near the chain A active site, positioning two basic residues into hydrogen bonding distance of the released phosphate: Arg326 is 3.6 Å away and Lys322 is 3.4 Å away (Figure 3B). These residues are conserved through most of this ATPase superfamily (Figure 5C and S1).
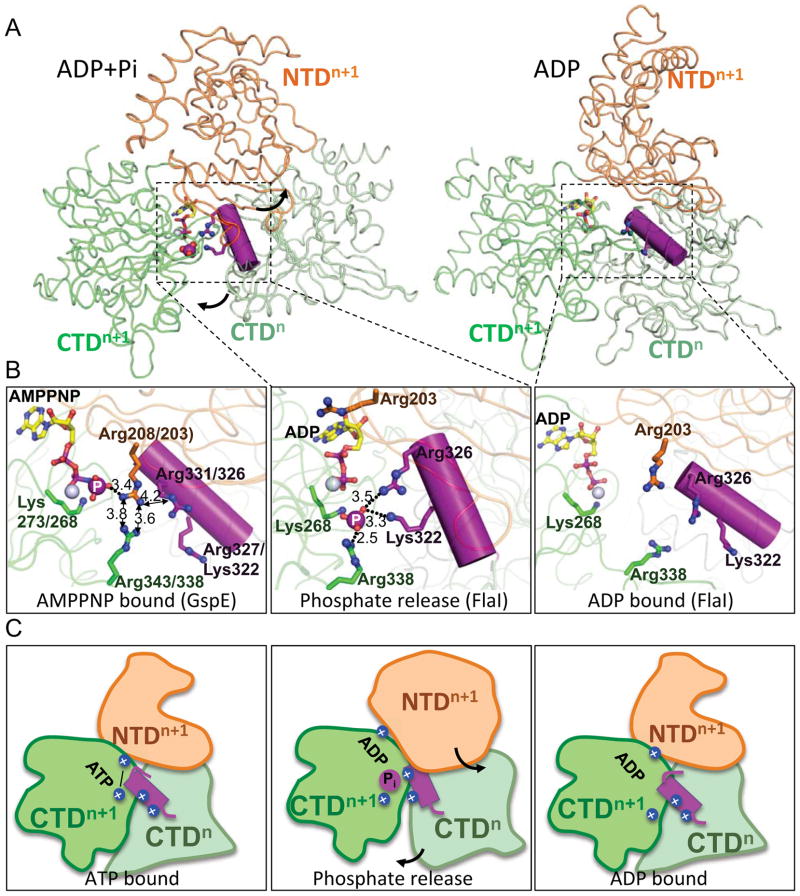
Figure 3. Basic-clamp switching drives secretion family ATPase conformational changes.
(A) Overview of conformational changes between FlaI ADP+Pi and ADP states. A CTDn helix containing a basic-clamp (purple) enters the neighboring active site to coordinate the released Pi in the ADP+Pi state (left). Phosphate (purple sphere) dissociation results in conformational rotation of CTDn towards CTDn+1, and NTDn+1 away from CTDn+1 (arrows) with the basic-clamp helix moving away from the active site (right). Boxed regions are shown in (B).
(B) Close up of ATP hydrolysis and Pi release-driven conformational changes. The GspE structure bound to AMPPNP (left) shows close proximity (distances are in Å) of intramolecular and intermolecular Arg-clamps, likely stabilized by Arg203 hydrogen bonding to the γ-phosphate. In this state the intermolecular basic-clamp (purple) on CTDn is outside the active site. Residues are numbered for GspE/FlaI. Upon ATP hydrolysis (middle) the basic-clamp helix rotates into the active site, with FlaI Arg326 and Lys322 of CTDn displacing Arg203 of NTDn+1 to make intersubunit interactions to the released phosphate (large purple sphere). Residues are numbered for FlaI. Upon phosphate release (right), the basic-clamps reset with the basic-clamp helix moving out of the active site and NTDn+1 Arg203 moving back into the active site, positions similar to those seen in the AMPPNP state, ready for subsequent rounds of ATP binding and hydrolysis. Residues are numbered for FlaI.
(C) Schematic showing the conformational changes observed between ATP bound (left), phosphate release intermediate (middle) and ADP bound (right) states for FlaI and superfamily members. Colors are as for (A) with basic-clamp residues highlighted with blue spheres. See also Figure S3.
Solution Analysis Reveals ATP Binding Induces a More Compact Form of FlaI
Experiments to soak ATP into the crystals to obtain ATP-bound FlaI failed as crystals dissolved, suggesting conformational differences between ATP and ADP-bound states. To test differences between ATP and ADP-bound states in solution, we did SAXS experiments with FlaIE336A with either nucleotide (Figure 2F). The Walker B mutation significantly slows the rate of ATP hydrolysis, allowing the ATP-bound state to be observed during the short time frame of the SAXS experiment. The experimental scattering curves for FlaIE336A-ADP and FlaIE336A-ATP are clearly different. The calculated theoretical scattering curve for the crystal structure of FlaIE336A-ADP (Figure 2F, solid black line) overlaps well with the equivalent experimental scattering curve, with an improved fit obtained by generating an all-closed 6-fold symmetric FlaI ring (dotted black line) with SymmDock, a geometry based docking program (Schneidman-Duhovny et al., 2005). The FlaI-ATP pair distribution (P(r)) function, which provides experimental electron pair distances to accurately compare structures by SAXS (Putnam et al., 2007)), is shifted to smaller distances compared to FlaI-ADP, particularly between 20–90 Å, indicating a more compact structure. Collectively, the solution and crystal structures suggest that ATP binding locks the FlaI hexamer into a more symmetrical and less dynamic conformation, which may promotes hexamer assembly by stabilizing interactions across adjacent subunits.
Archaellum Assembly Requires FlaI Hexameric Assembly
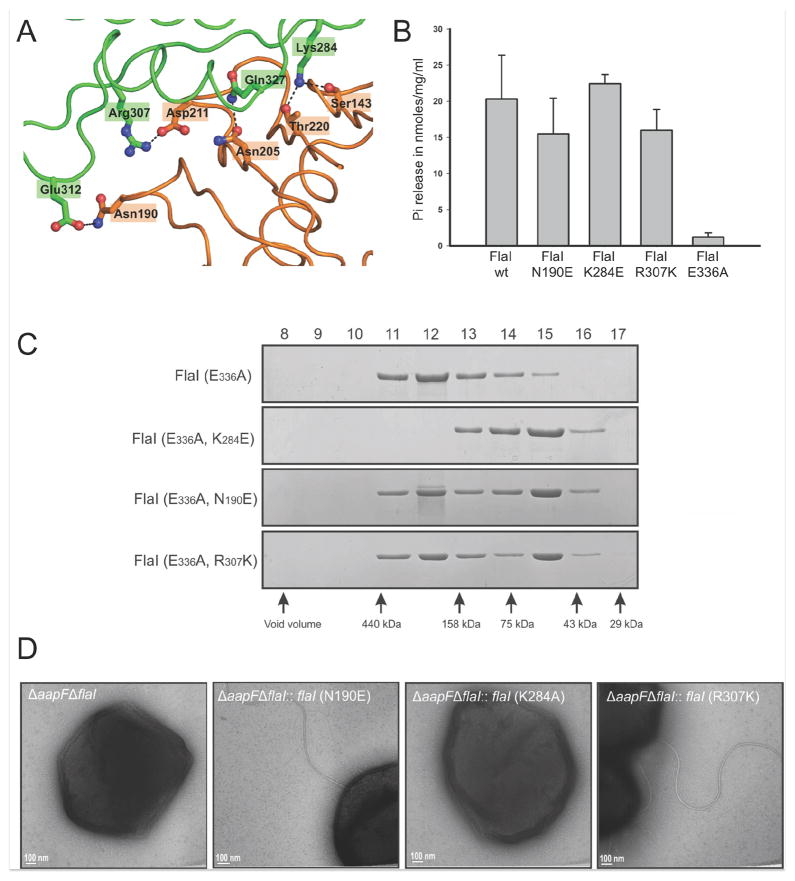
FlaI undergoes hexameric assembly in solution upon ATP binding (Ghosh et al., 2011), as do other secretion ATPase super-family members. Yet, a key question is if this hexameric assembly is indispensible for the association and functionality of their respective macromolecular machines. To test if ATP-dependent oligomerization is important for archaellum assembly in S. acidocaldarius, we made three FlaI mutations, designed to disrupt subunit interactions based on our structures. Residues in salt-bridges at the haxameric interface of two subunits were mutated: Asn190 to Glu; Lys284 to Glu and Arg307 to Lys (Figure 4A), and the FlaI variants were characterized in vitro and in vivo.
Figure 4. FlaI hexameric conformation is necessary for function.
(A) Residues involved in salt bridges between FlaI subunits were mutated.
(B) Interface mutants are active ATPases. ATPase activity was analyzed by measuring released phosphate (Pi) upon ATP-hydrolysis at 70°C (mean of three independent experiments ± standard deviation).
(C) Analytical gel filtration shows the effect of interface mutation on hexamerization. Analysis of interface mutations (FlaIK284E/E336A, FlaIN190E/E336A, FlaIR307K/E336A) shows that these interface mutants have reduced ability to form hexamers upon ATP binding compared to FlaIE336A.
(D) Complementation analysis of the ΔaapFΔflaI mutant strain with FlaI interface mutants. Expression of FlaI interface mutants in trans in a ΔaapFΔflaI strain revealed partial complementation phenotypes for FlaIN190E and FlaIR307K. However, FlaIK284E was unable to complement the mutant phenotype (ΔaapFΔflaI) indicating that FlaI needs to be in its hexameric form to be functional.
We made these mutations in wild-type flaI, to analyze the effect on ATPase activity, and also in the Walker BE336A flaI mutant, to test the impact on oligomerization. Purified interface mutants in the wild-type background were analyzed for ATP hydrolysis. All three point mutants were active ATPases (Figure 4B), showing that the ATP binding and hydrolysis motifs conserved within each FlaI monomer are sufficient for ATPase activity. The interface mutants in the WalkerBE336A background were tested for oligomerization by analytical gel filtration analysis. For oligomerization experiments, the WalkerBE336A background is necessary to disable the ATPase activity and trap the structure in the hexameric state. Self-assembly into hexameric rings for all mutants was decreased compared to FlaIE336A (Figure 4C). The most striking effect was evident for FlaIK284E/E336A, which was unable to form stable hexamers even with ATP.
To test the functional impact of disrupting FlaI oligomerization, we performed in trans complementation of the ΔaapFΔflaI S. acidocaldarius strain with each point mutant. The ΔaapF mutant is a hyper-archaellated strain that therefore gives stronger motility phenotypes (Henche et al., 2012). EM analysis of complemented strains revealed that FlaIN190E and FlaIR307K restored the wild-type phenotype, whereas FlaIK284E did not, with only 6% of cells over-expressing FlaIK284E assembling archaella (Figure 4D). These collective analyses indicate that FlaI must be able to assemble into its hexameric state to function as the archaella assembly ATPase.
Covalently Interlocked Blocks Move Within the Ring to Make FlaI a Stable Motor
How GspE and FlaI achieve their different physiological functions in protein secretion and movement has been a puzzle. Comparison of FlaI and GspE conformations (Figure 5) shows that the directionality of movement induced by ATP binding, hydrolysis and release differs. FlaI moves with a rotating translation parallel and across the plane of the hexameric ring (Movie S3), a movement suitable to drive both assembly and rotation of archaella. In contrast, GspE moves up and down perpendicular to the plane of the ring, with little rotary movement (Movie S4). The building block seen for FlaI and GspE is also conserved across other superfamily ATPases (Figures 5C and S4).
The FlaI NTD is Essential for Archaellum Formation but not ATPase Activity
The most variable region of the T2S/T4S/T4P ATPase superfamily is the NTD, with the T4P retraction ATPase PilT (Misic et al., 2010) and the T4S ATPase VirB11 (Savvides et al., 2003) lacking the NTD-1 of the T2S ATPases GspE (Yamagata and Tainer, 2007), EpsE (Abendroth et al., 2005; Robien et al., 2003) and FlaI (Figure 5C). Although the overall fold of FlaI NTD-1 resembles GspE NTD-1, it is unrelated to the fold of the EpsE NTD-1. Full-length EpsE has yet to be crystallized, but EspE1 NTD-1 has been crystallized in complex with inner membrane protein EpsL cytoplasmic domain (Abendroth et al., 2005; Robien et al., 2003). Collectively, these results suggest that the superfamily NTD domains link the conserved ATPase domain to the distinct biological systems.
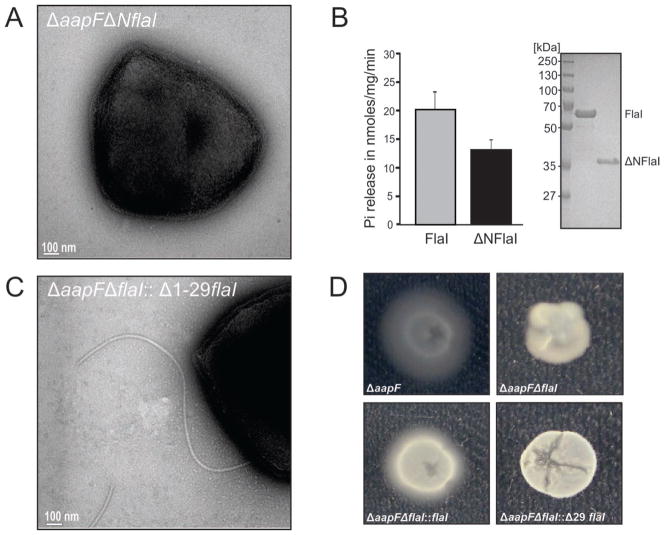
To investigate the structurally-implicated biological roles of the FlaI NTD, we generated a ΔaapFΔNflaI strain, which lacks FlaI amino acids 1-224 (yellow/orange in Figure 5C). EM analysis of the ΔaapFΔNflaI deletion mutant revealed that no archaellum was assembled (Figure 6A) and swimming motility was abolished (Movie S5), indicating a central role of the NTD in archaellum assembly. To test if this functional effect was due to a loss of FlaI ATPase activity from NTD deletion, we expressed and purified ΔNFlaI from E. coli. The ΔNFlaI protein still exhibits 75% of ATPase activity compared to the full-length FlaI, indicating that assembly failure in the ΔaapFΔNflaI deletion mutant was not due to the loss of ATPase activity (Figure 6B): the ΔaapFΔNflaI deletion provides a separation-of-function mutation for ATPase versus assembly and motility.
Figure 6. FlaI NTD is necessary for archaellum assembly, but not ATPase activity.
(A) Deletion of FlaI residues 1-224 (ΔaapFΔNflaI) results in a phenotype similar to flaI-deletion (ΔaapFΔflaI).
(B) The CTD alone is an active ATPase. ATPase assays comparing purified full-length FlaI in comparison to the CTD alone (ΔNFlaI). ΔNFlaI showed 75% activity compared to wild-type, indicating that the ΔaapFΔNflaI phenotype cannot be explained by a loss in enzymatic activity (mean of three independent experiments ± standard deviation). (C) FlaI residues 1-29 are not essential for archaellum assembly. Complementation analysis of ΔaapFΔNflaI mutant strain with FlaIΔ1-29 shows archaellum assembly in the complemented strain (10–20% cells showed flagella assembly).
(D) Motility assays showing inability of FlaIΔ1-29 to restore motility phenotype in the ΔaapFΔNflaI strain. From left to right: Swimming of the hyper-archaellated background strain (ΔaapF) was observed in gelrite plates while a deletion of FlaI resulted in non-motile cells (ΔaapFΔNflaI). Complementation of the strain with wild-type FlaI (flaI::ΔaapFΔNflaI) restored the wild-type phenotype whereas strains complemented with FlaIΔ1-29 had no motility. See also Movie S5.
The First 29 Residues of FlaI are Essential for Motility but not Assembly
From our structures and analyses, a striking difference between the FlaI and GspE crystal structures is in amino acids 1-29 (Figure 5B). In FlaI these residues form a surface exposed α-β-α-structure (Figures 5B and S1), located on the outside of the hexamer farthest away from the center. We hypothesized that this subdomain is a specific adaptor that links the ATPase to archaellum assembly or motility. To test this, we generated a construct of FlaI where amino acids 1-29 were deleted (FlaIΔ1-29), expressed this variant in a S. acidocaldarius ΔaapFΔflaI background, and assayed for archaellum assembly and motility. EM analysis of the strain over-expressing FlaIΔ1-29 revealed that 10–20% of the cell population can assemble archaella while complementation with wild-type FlaI leads to 40–50% cells with archaella (Figure 6C), indicating a non-essential role of the amino acids 1-29 in assembly. However, unlike hyper-archaellated ΔaapF and flaI-complemented ΔaapFΔflaI strains, which show motility under standard assay conditions, the FlaIΔ1-29 expressing ΔaapFΔflaI strain showed no motility similar to the non-motile ΔaapFΔflaI strain (Figure 6D). Together these analyses indicate a role of FlaI amino acids 1-29 in motility rather than assembly of the S. acidocaldarius archaellum: FlaIΔ1-29 provides a separation-of-function mutation for assembly versus motility.
The FlaI Three-Helix Bundle is Required for Membrane Localization and Archaellum Assembly
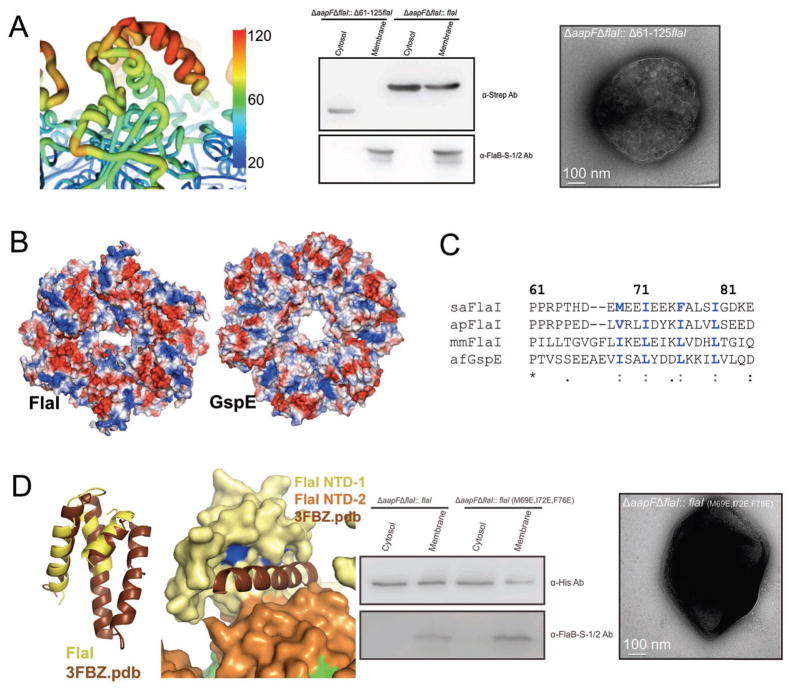
FlaI and the membrane protein FlaJ form the key platform on which the archaellum assembly takes place. Methanocaldococcus sp. FlaI was suggested to interact with the membrane and membrane anchored archaella assembly components (Thomas and Jarrell, 2001). Together with our findings that FlaI NTD is not responsible for enzymatic activity and the structure of the T2S ATPase EpsE-NTD in complex with membrane protein EpsL (Abendroth et al., 2005), we hypothesized that the FlaI ring top surface, formed by NTD-1, may constitute the membrane and/or membrane protein interaction surface. Mapping crystallographic temperature factors (B-values) onto the FlaI structure, as proven useful for observing differences in mobility with functional implications (Tainer et al., 1984), identifies a three-helix bundle (amino cids 61–125) that has higher B-factors than the rest of the protein (Figure 7A), suggesting increased local motion. This three-helix bundle forms the tip of the FlaI crown and is placed to interact with components of the membrane.
Figure 7. N-terminal helix bundle domain interaction site.
(A) A three-helix bundle is essential for archaellum formation and FlaI localization. Left: a strongly charged patch is the three-helix bundle with relatively high B-factors (red with thicker ribbon), indicative of intrinsic flexibility. Middle: FlaI missing the helix bundle domain (FlaIΔ61-125) does not localize to the membrane. Protein presence in membrane and cytoplasmic fractions is shown by Western blot. Right: the deletion of the helix bundle domain in vivo results in non-archaellated cells.
(B) Electrostatic surface (blue positive; red negative) shows differences between FlaI and GspE.
(C) Conserved, surface exposed hydrophobic residues in the helix bundle domain (blue) are good candidates for a protein-protein interaction surface.
(D) DALI search for the three-helix bundle identifies several structural domains that consist of an additional fourth helix. Left: superposition of the DALI result (3FBZ.pdb) positions the fourth helix directly in the groove between NTD-1 and NTD-2 and close to the conserved hydrophobic residues (blue). Mutation of these residues (Met79, Ile72 and Phe76) to glutamates results in both decreased FlaI association with the membrane, as compared to wild-type FlaI in our membrane association assays (middle), and in a non-archaellated phenotype (right). See also Movie S6.
We examined the effect of removing this three-helix bundle from FlaI on archaellum assembly by expressing the FlaIΔ61-125 internal deletion in trans in the ΔaapFΔflaI strain. EM analysis revealed that loss of the three-helix bundle abolishes archaella assembly (Figure 7A). Furthermore, cellular fractionation confirmed that the FlaIΔ61-125 protein was unable to localize to the membrane, indicating a role of the three-helix bundle in mediating protein-protein and/or protein-membrane interactions (Figure 7A).
To further examine interaction surfaces in the FlaI61-125 region, we did more detailed structural analysis. Comparison of the FlaI and GspE NTDs shows that the charge distribution differs greatly on the top surface where this three-helix bundle is located (Figure 7B). In FlaI this surface is positively charged, and we expect these regions to most likely interface with the membrane lipid negatively charged phosphates. In contrast, stable protein-protein interaction sites commonly involve surface-exposed, conserved hydrophobic residues. Examination of our FlaI NTD structures identifies three hydrophobic surface exposed residues (Met69, Ile72 and Phe76) that map to the three-helix bundle and are conserved among all the FlaIs in archaea (Figure 7C). Further, a DALI search for structural homologs (Holm and Rosenstrom, 2010) using only the three-helix bundle identified predominantly four-helix bundles, with the best hit shown in Figure 7D (3FBZ.pdb). Interestingly, superimposition of this four-helix bundle with FlaI positions the fourth helix precisely into a groove formed within the three-helix bundle that is lined with the conserved hydrophobic residues.
To test the structurally implicated FlaI-FlaJ interface, we constructed a FlaIM69E/I72E/F76E triple point mutant and expressed this in trans in the ΔaapFΔflaI deletion strain. EM analysis revealed no archaellum assembly and swimming assays showed no motility (Movie S5) in the complemented strain over-expressing the triple point mutant. Furthermore, membrane localization assays showed that FlaIM69E/I72E/F76E localization to the membrane was significantly reduced compared to wild-type (Figure 7D). Collectively, these results support an important role of these residues in archaellum assembly, most likely through mediating protein-protein interactions.
DISCUSSION
Combined structural and functional analyses of the prototypic archaeal motor ATPase FlaI suggest the basis for its dual function in archaella assembly and movement, which further informs superfamily functions. We found that S. acidocaldarius FlaI ATPase activity and hexameric assembly is essential for archaellum assembly, and identified N-terminal FlaI regions important for archaellum motility, assembly and FlaI localization. Moreover, our crystal structure of FlaI bound to nucleotide traps a previously unobserved intermediate that informs the mechanistic basis for conformational changes characteristic of this superfamily.
Arg-Clamp Switches Drive Building Block Movements
The 7 FlaI conformations in our crystal structures expand the number of conformations observed for this ATPase superfamily. GspE (Yamagata and Tainer, 2007), PilT (Misic et al., 2010; Satyshur et al., 2007) and VirB11 (Savvides et al., 2003) hexameric structures have two or more conformations, and symmetric hexamers of VirB11 and PilT have also been seen. Dynamic hexamer formation is therefore characteristic of this superfamily, and the propensity to form symmetric versus asymmetric states may reflect differences in synchronization of ATP binding and hydrolysis between subunits. Analyses of VirB11 and GspE led to suggestions they act as motors by linking a rigid basal ring formed by the ATP binding CTD with moving NTDs on top (Savvides et al., 2003; Yamagata and Tainer, 2007). Yet, we find that in all superfamily members with known structures neighboring NTD and CTD domains form a tight interface (Figure 2E) and act as a moving building block, supporting and extending a mechanism proposed for PilT (Satyshur et al., 2007). We propose this “flexible crown” structure is key to FlaI motor functions that drive archaellum assembly in an ATP-hydrolysis dependent manner. Further, differences in building block movements driven by ATP binding and hydrolysis may explain why similar proteins can provide the energy for distinct biological processes like protein secretion versus archaella assembly and movement.
Interestingly, in our crystal structure of FlaI bound to nucleotide, one subunit is trapped in a novel phosphate-release state. In this conformation, basic residues from the neighboring CTD subunit protrude into the active site cleft and coordinate with the released phosphate. Previous low-resolution PilT observations implicated analogous interactions, involving PilT Arg203 and Arg207, and led to suggesting that this could be similar to the F1F0ATPases Arg-finger (Satyshur et al., 2007). Our results thus test and extend these observations, providing the first direct evidence of intersubunit communication to the nucleotide in the active site, and suggest phosphate coordination immediately following ATP hydrolysis is a switch that triggers building block rearrangements. Closed conformations of GspE and PilT (Misic et al., 2010; Satyshur et al., 2007; Yamagata and Tainer, 2007) have identified an intrasubunit Arg-clamp that binds the ATP γ-phosphate. In our ADP-bound FlaI structures these clamp residues, Arg338 in the CTD and Arg203 in the NTD, are not in an active conformation. However, consistent with our comparison to GspE, where the Arg-clamp residues must move closer together to bind the ATP γ-phosphate, our SAXS data supports a model where the FlaI subunits compact further to adopt a closed conformation when bound to ATP. Surprisingly, in GspE these Arg residues are in close proximity (<4 Å), and without stabilization from the γphosphate would repel each other (Figure 3B). Further, Arg203 is close (4.2 Å) to Arg326 of the neighboring CTD. Collectively, this leads us to hypothesize a switch-model where hydrolysis of ATP to release the γ-phosphate would move the Arg-clamp residues into closer proximity, leading to their repulsion (Figure 3). As the NTD is no longer anchored to the nucleotide, this repulsion would drive the conformational rotation of the NTD and the concerted movement of the neighboring CTD to which it is interfaced, positioning the second conserved basic-clamp, Arg326 and Lys322 in FlaI, to intermolecularly bind to the released phosphate. Interestingly, this movement opens up the back face of the FlaI hexamer suitable for ADP dissociation (presumably high nucleotide concentration allows ADP to remain in our crystal structure). Due to the high concentration of positive charge around the phosphate this intermediate may be metastable (temperature-trapped for the thermophile), and repulsion from these residues would drive further subunit movements and subsequent phosphate release so that the cycle of ATP binding, hydrolysis and product release can continue efficiently for archaellum assembly and motility. Intriguingly, another insight from our structure is an intramolecular hydrophobic interface formed between the basic-clamp helix of the nucleotide binding subunit with secondary structure elements surrounding Arg338, which is in close contact with the leaving phosphate. This interface provides another potential mechanism by which ATP hydrolysis between subunits is coordinated: the basic-clamp helix may sense the phosphate release state through its connection to Arg338.
The NTD Links Building Block Movements to Archaellum Assembly and Motility
The FlaI NTDs, which form the crown points, are strategically located on the outside edge to readily form functional interactions (Figure 5B and 7A): striking differences between FlaI and GspE NTD’s indicate this is an area that interacts with the distinct membrane components in each system. Interestingly, we found that in FlaI the first 29 residues, on the crown tip sides, are required to drive archaellum motility; whereas the three-helix bundle, at the crown point, localizes FlaI to the membrane and is required for archaellum assembly and motility. Further, the three-helix bundle is a likely interaction surface for FlaJ, as at physiological pH (S. acidocaldarius intracellular pH is ~6.5) the three-helix bundle is negatively charged (pI 5.75) and the conserved cytoplasmic domains of FlaJ are positively charged (pI of 9.75 and 10.3). FlaI and FlaJ interactions may be further driven by hydrophobic regions between a groove in the FlaI three-helix bundle with an α-helix of FlaJ, with protruding helices for this interaction existing in the cytoplasmic domains of membrane proteins in the Eps (Abendroth et al., 2005) and Bfp systems (Yamagata et al., 2012).
Combining FlaI biochemical data with our structural and genetic data suggests three key steps for FlaI functions in both archaellum assembly and movement, which supports and extends ideas proposed from sequence homologous T2S and T4P systems (see Graphical Abstract). 1) Monomeric FlaI protein in the cytoplasm self-assembles into a hexameric ring upon ATP binding. 2) Specific interactions between the FlaI NTD and both membrane lipids and its binding partner (most likely FlaJ) move the FlaI hexamer to the membrane. 3) Basic clamp switching during successive rounds of ATP hydrolysis, product release and ATP binding results in conformational translation with rotation of the stable FlaI inter-subunit blocks, which drives both archaella assembly and rotation. Interestingly, the top surface of FlaI moves up (and down) 8–10 Å when ATP is hydrolyzed (Movie S3). This shift range matches the spacing between pilin proteins in T4P filaments (Craig et al., 2003; Craig et al., 2006; Hartung et al., 2011), with reconstructed pili from various organisms all showing a distance of 5–10 Å between pilin neighbors. Thus, the conformational change in FlaI may be transferred through the likely tight interaction with FlaJ to push the archaellum filament out of the membrane, allowing a new archaellin subunit to bind and grow the filament.
Our results elucidate how FlaI structures and conformations play key roles in archaellum assembly and motility functions with implications for superfamily evolution and functions: crown geometry determines sites of interaction, and tight inter-subunit moving blocks provide power for filament assembly and rotation, whereas loop connections provide interface elasticity by allowing functional movements without disrupting assembly. Overall, collective results reveal detailed and global FlaI activities in transducing nucleotide binding and hydrolysis into translational and rotatory motions suitable for assembly and motility without destabilizing the integrity of the hexameric crown assembly.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
S. acidocaldarius strain DSM639 was used for all in vivo experiments, with deletion experiments done in a ΔaapF background, see Supplemental Information.
ATPase Assay and Analytical Size Exclusion Chromatography
Cloning, expression and purification of recombinant FlaI and mutants is described in Supplemental Information. ATPase assays used the malachite green assay (Ghosh et al., 2011). For analytical size exclusion chromatography, the protein was incubated at 50°C for 10 min and then 4°C for 6 h with either 1 mM AMPPNP (A denylyl-imidodiphosphate) or ADP plus 10 mM MgCl2. The samples were applied to a Superdex 200 10/300 column and fractions analyzed by SDS-PAGE.
Small Angle X-ray Scattering
SAXS data were collected and processed (see Supplemental Information) at the Advanced Light Source (ALS) beamline 12.3.1 (SIBYLS) Lawrence Berkeley National Laboratory (Classen et al., 2010; Hura et al., 2009).
Crystallization and Structure Determination of FlaI
FlaI crystals were grown and data collected, processed and refined as described in the Supplemental Information. X-ray data were collected on beamlines 8.3.1 and 12.3.1 at the ALS. Refined models of FlaIE336A-ADP at 2.0 Å and FlaI-Apo at 3.6 Å have good statistics (Table 1).
In-frame Gene Deletion and Gene Complementation
Deletion mutant strains and recombinant plasmids were constructed as described in the Supplemental Information. All bacterial strains, primers and plasmids are listed in Table S1.
Cell Fractionation and Localization Analysis
To test roles of highly conserved and specific residues in FlaI, we used cellular fractionation of the complemented deletion mutant (ΔaapFΔflaI) expressing different FlaI variants (see Supplemental Information).
Electron Microscopy
For Transmission Electron Microscopy (TEM) cells were fixed with 2.5% glutaraldehyde for 15 min at room temperature. Cell suspensions were placed directly on glow-discharged Formvar-coated 200-mesh copper grids (Plano, Wetzlar, Germany). Samples were negatively stained for 20 s with 2% uranyl acetate, washed twice with water, and air-dried. TEM was performed using a JEOL JEM 3010 electron microscope (300 kV; LaB6 cathode).
Swimming Assays on Semi-Solid Plates and Thermomicroscopy
Analyses of motility and swimming behavior of S. acidocaldarius are described in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
Cross-subunit interface forms building block for transmission of ATPase force
Structures and mutants reveal functional components for assembly and motility
The directionality of movement explains FlaI activities and GspE differences
The phosphate released by ATP-hydrolysis triggers subunit rearrangements
Acknowledgments
We thank Albers and Tainer laboratory members for helpful comments, especially Susan Tsutakawa, Michal Hammel and ALS BL12.3.1 and BL8.3.1 staff. This study was supported by NIH grant AI22160 to J.A.T. The SIBYLS beamline (BL12.3.1) is supported by United States Department of Energy program IDAT and by National Institute of Health grant GM105404. A.G. received a Max Planck Postdoctoral fellowship and S.V.A. was supported by a VIDI grant of the Dutch Science Organization (NWO) and Max Planck Society intramural funds.
Footnotes
ACCESSION CODES
Coordinates and structure factors are in the Protein Data Bank under accession numbers 4IHQ and 4II7.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J Mol Biol. 2005;348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Albers SV, Jonuscheit M, Dinkelaker S, Urich T, Kletzin A, Tampe R, Driessen AJ, Schleper C. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Szabo Z, Driessen AJ. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol. 2003;185:3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy SL, Mori T, Komoriya K, Aizawa S, Jarrell KF. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae. J Bacteriol. 2002;184:5223–5233. doi: 10.1128/JB.184.19.5223-5233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy SL, Ng SY, Jarrell KF. Recent advances in the structure and assembly of the archaeal flagellum. J Mol Microbiol Biotechnol. 2004;7:41–51. doi: 10.1159/000077868. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Weapons of mass retraction. Mol Microbiol. 2005;57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa S, Jarrell KF. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Monfett M, Maier TM, Benach JL, Frank DW, Thanassi DG. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect Immun. 2008;76:2852–2861. doi: 10.1128/IAI.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shiue SJ, Huang CW, Chang JL, Chien YL, Hu NT, Chan NL. Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. J Biol Chem. 2005;280:42356–42363. doi: 10.1074/jbc.M506843200. [DOI] [PubMed] [Google Scholar]
- Classen S, Rodic I, Holton J, Hura GL, Hammel M, Tainer JA. Software for the high-throughput collection of SAXS data using an enhanced Blu-Ice/DCS control system. J Synchrotron Radiat. 2010;17:774–781. doi: 10.1107/S0909049510028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Li J. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Crowther LJ, Anantha RP, Donnenberg MS. The inner membrane subassembly of the enteropathogenic Escherichia coli bundle-forming pilus machine. Mol Microbiol. 2004;52:67–79. doi: 10.1111/j.1365-2958.2003.03963.x. [DOI] [PubMed] [Google Scholar]
- Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Albers SV. Assembly and function of the archaeal flagellum. Biochem Soc Trans. 2011;39:64–69. doi: 10.1042/BST0390064. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Hartung S, van der Does C, Tainer JA, Albers SV. Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem J. 2011;437:43–52. doi: 10.1042/BJ20110410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duarte OG, Chattopadhyay S, Weissman SJ, Giron JA, Kaper JB, Sokurenko EV. Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J Bacteriol. 2007;189:9145–9149. doi: 10.1128/JB.00722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JK, Forest KT. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J Mol Microbiol Biotechnol. 2006;11:192–207. doi: 10.1159/000094054. [DOI] [PubMed] [Google Scholar]
- Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L, Tainer JA. Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J Biol Chem. 2011;286:44254–44265. doi: 10.1074/jbc.M111.297242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henche AL, Koerdt A, Ghosh A, Albers SV. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ Microbiol. 2012;14:779–793. doi: 10.1111/j.1462-2920.2011.02638.x. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Albers SV. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- Lassak K, Neiner T, Ghosh A, Klingl A, Wirth R, Albers SV. Molecular analysis of the crenarchaeal flagellum. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07916.x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J Mol Biol. 2010;400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol. 2006;11:167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M, Ghosh A, Tripepi M, Albers SV. Archaeal type IV pilus-like structures--evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol. 2011;14:357–363. doi: 10.1016/j.mib.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J Mol Biol. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. Embo J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. Embo J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiue SJ, Kao KM, Leu WM, Chen LY, Chan NL, Hu NT. XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. Embo J. 2006;25:1426–1435. doi: 10.1038/sj.emboj.7601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streif S, Staudinger WF, Marwan W, Oesterhelt D. Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J Mol Biol. 2008;384:1–8. doi: 10.1016/j.jmb.2008.08.057. [DOI] [PubMed] [Google Scholar]
- Tainer JA, Getzoff ED, Alexander H, Houghten RA, Olson AJ, Lerner RA, Hendrickson WA. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature. 1984;312:127–134. doi: 10.1038/312127a0. [DOI] [PubMed] [Google Scholar]
- Thomas NA, Jarrell KF. Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. J Bacteriol. 2001;183:7154–7164. doi: 10.1128/JB.183.24.7154-7164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NA, Mueller S, Klein A, Jarrell KF. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol Microbiol. 2002;46:879–887. doi: 10.1046/j.1365-2958.2002.03220.x. [DOI] [PubMed] [Google Scholar]
- Trachtenberg S, Cohen-Krausz S. The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. J Mol Microbiol Biotechnol. 2006;11:208–220. doi: 10.1159/000094055. [DOI] [PubMed] [Google Scholar]
- Tripathi SA, Taylor RK. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J Bacteriol. 2007;189:4401–4409. doi: 10.1128/JB.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Berkner S, Ajon M, Driessen AJ, Lipps G, Albers SV. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem Soc Trans. 2009;37:97–101. doi: 10.1042/BST0370097. [DOI] [PubMed] [Google Scholar]
- Yamagata A, Milgotina E, Scanlon K, Craig L, Tainer JA, Donnenberg MS. Structure of an essential type IV pilus biogenesis protein provides insights into pilus and type II secretion systems. J Mol Biol. 2012;419:110–124. doi: 10.1016/j.jmb.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. Embo J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.