SUMMARY
Past studies have documented a cross-talk between H2B ubiquitylation (H2Bub) and H3K4 methylation, but little, if any, direct evidence exists explaining the mechanism underlying H2Bub-dependent H3K4 methylation on chromatin templates. Here, we took advantage of an in vitro histone methyltransferase assay employing a reconstituted yeast Set1 complex (ySet1C) and a recombinant chromatin template containing fully ubiquitylated H2B to gain valuable insights. Combined with genetic analyses, we demonstrate that the n-SET domain within Set1, but not Swd2, is essential for H2Bub-dependent H3K4 methylation. Spp1, a homolog of human CFP1, is conditionally involved in this cross-talk. Our findings extend to the human Set1 complex, underscoring the conserved nature of this disease-relevant, cross-talk pathway. As not all members of the H3K4 methyltransferase family contain n-SET domains, our studies call attention to the n-SET domain as being a predictor of H2B ubiquitylation ‘sensing’ in bringing about downstream H3K4 methylation.
INTRODUCTION
Methylation on lysine 4 of H3 (H3K4me) is one of the prominent histone modification marks that correlates strongly with active transcription in yeast (Santos-Rosa et al., 2002) and in metazoans (Schübeler et al., 2004). Accumulating studies in metazoans have implicated mis-regulation of H3K4 methylation in the pathogenesis of cancer and in developmental defects (Wang and Allis, 2009; Chi et al., 2010), further emphasizing the importance of understanding the regulation of H3K4 methylation.
Metazoans contain multiple H3K4 methyltransferase enzymes (SET1A, SET1B, MLL1-4) that reside in multisubunit complexes with several shared subunits. Despite a common histone methylation target site (H3K4), SET1/MLL complexes have non-redundant roles that complicate studies (Eissenberg and Shilatifard, 2010). In contrast, the budding yeast Saccharomyces cerevisiae contains a single H3K4 methyltransferase (Set1) that is found in a complex (ySet1C/COMPASS) with seven other subunits (Swd1, Swd3, Bre2, Sdc1, Spp1, Swd2 and Shg1) (reviewed in Dehé and Géli, 2006). Six of these subunits are conserved in the metazoan SET1 complexes, with four also conserved in the MLL complexes, thus making ySet1C a good model.
Interestingly, H3K4 methylation in yeast requires prior trans-tail ubiquitylation on histone H2B at lysine 123 (H2Bub) (Sun and Allis, 2002; Dover et al., 2002), at least for H3K4 di- and tri-methylation (Dehé et al., 2005; Shahbazian et al., 2005). Two groups reported that Swd2 plays a key role in this process, although by different mechanisms (Lee et al., 2007; Vitaliano-Prunier et al., 2008). One clear prediction of the above studies is that the Swd2-deficient ySet1C will not be able to di- and tri-methylate H3K4 efficiently on a chromatin template. However, due to a lethal phenotype related to the function of Swd2 as a component of the cleavage and polyadenylation factor (CPF) complex (Cheng et al., 2004; Dichtl et al., 2004; Nedea et al., 2008), this issue has yet to be thoroughly addressed.
Additional genetic analyses of ySet1C subunit deletions/mutations have implicated individual subunits in the regulation of H3K4 methylation (Nagy et al., 2002; Dehé et al., 2006; Nedea et al., 2008), but fail to rigorously rule out indirect effects of subunit mutations through other cellular processes. These considerations necessitate biochemical analyses with defined components of the ubiquitylation-methylation machinery and pathway under examination, notably the physiologically-relevant substrates and enzyme systems responsible for bringing about H3K4 methylation on a ubiquitylated H2B chromatin template.
Here, we have employed a biochemically defined system reconstituted with recombinant H2Bub-containing chromatin (McGinty et al., 2008; McGinty et al., 2009) and recombinant ySet1Cs, along with genetic analyses, to elucidate novel mechanistic insights underlying H2Bub-dependent H3K4 methylation. Our study identifies the n-SET domain of Set1, as well as a conditional role for Spp1, in mediating H2Bub-dependent H3K4 methylation, a medically-relevant pathway conserved between yeast and humans.
RESULTS
H2B Ubiquitylation Directly Stimulates Yeast Set1 Complex-Mediated H3K4 Methylation
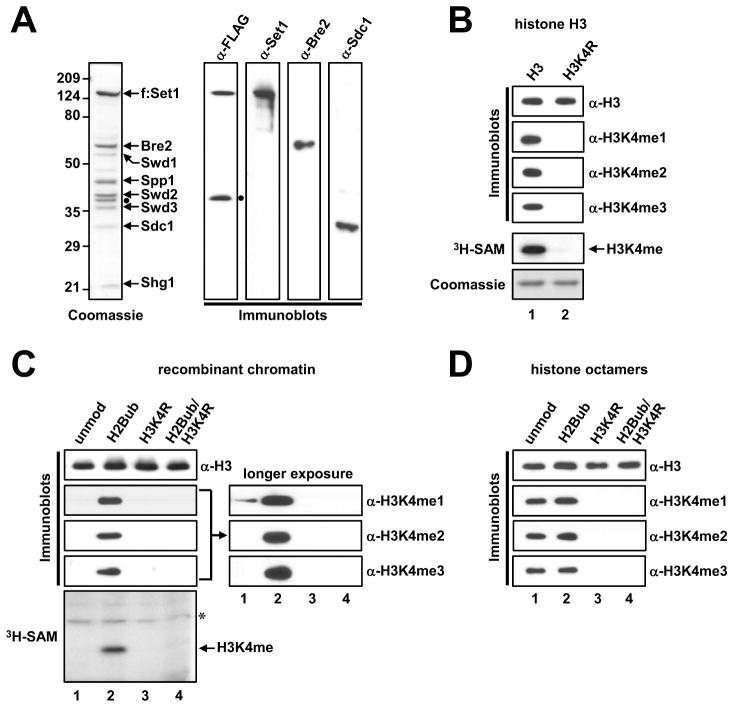
The yeast Set1/COMPASS complex (hereafter ySet1C) was reconstituted and affinity-purified from Sf9 cells coinfected with baculoviruses that express FLAG-Set1 and the other seven untagged ySet1C subunits (Figure 1A). Gel filtration confirmed that reconstituted ySet1C forms a stable complex containing all eight subunits (Figure S1A). Using purified ySet1C in an in vitro histone methyltransferase (HMT) assay with free histone H3 as substrate, H3K4me1, H3K4me2 and H3K4me3 were easily detected by immunoblots (Figure 1B). A parallel analysis in the presence of 3H-S-adenosylmethione (SAM) showed no detectable methylation of the H3K4R mutant substrates, indicating that H3K4 is the sole site methylated by purified ySet1C under these conditions.
Figure 1. Establishment of In Vitro H3K4 Methylation Assays with Purified Yeast Set1 Complex and Ubiquitylated H2B-Containing Recombinant Chromatin.
(A) SDS-PAGE/Coomassie Blue staining and immunoblot analyses of the purified, baculovirus-reconstituted ySet1C. Band identities were confirmed by LC/MS analysis and by immunoblot. The dot indicates an N-terminal degradation product of FLAG-tagged Set1.
(B, C and D) Ubiquitylated H2B directly stimulates H3K4 methylation. Free H3 or H3K4R (B), chromatin templates assembled with indicated pre-modified histones (C) and octamers with indicated pre-modified histones (D) were subjected to in vitro HMT assays with purified ySet1C. H3 methylation status was monitored by immunoblotting with indicated antibodies or by fluorography (in this and subsequent figures). The asterisk in (C) indicates an uncharacterized methylation signal from a component in the chromatin assembly factor or ySet1C preparations.
Note: unmod, unmodified histones; H2Bub, lysine(K)120-ubiquitylated H2B; H3K4R, H3 containing an arginine(R) substitution at lysine(K)4; H3K4me, lysine(K)4-methylated H3; H3K4me1, lysine(K)4-mono-methylated H3; H3K4me2, lysine(K)4-di-methylated H3; H3K4me3, lysine(K)4-tri-methylated H3.
See also Figure S1.
To assess a potential role for ubiquitylated H2B (H2Bub) in directly facilitating H3K4 methylation (H3K4me) by ySet1C, we assembled recombinant chromatin with Xenopus histone octamers containing either unmodified H2B or a semisynthetic H2B (McGinty et al., 2009; Figure S1B) fully ubiquitylated at lysine 120 (lysine 123 in yeast H2B) and performed an in vitro HMT assay. Consistent with prior studies using the human SET1 complex (Kim et al., 2009), ySet1C-generated H3K4me2/3 on the chromatin substrate exhibited a strict H2Bub-dependency (Figure 1C) that correlated with H2Bub levels (Figure S1C). A low basal level of H3K4me1 was observed on the unmodified chromatin substrate; this was also enhanced by H2Bub (Figure 1C, long exposure). Similar results were obtained with recombinant mononucleosomes (Figure S1D) and HeLa cell-derived natural oligonucleosomes that contain endogenous modifications (Figure S1E).
Interestingly, H2Bub also significantly stimulated H3K4 mono-methylation (Figures 1C, S1C and S1D), which contrasts with earlier reports of H2Bub being dispensable for H3K4 mono-methylation (Dehé et al., 2005; Shahbazian et al., 2005). However, we confirm reports of decreased levels of H3K4 mono-methylation in the rad6Δ (Dehé et al., 2005; Chandrasekharan et al., 2010) and H2BK123R (Shahbazian et al., 2005; Chandrasekharan et al., 2010) yeast strains (Figures S1F and S1G). Our results thus indicate that H2Bub directly stimulates all states of H3K4 methylation. H2Bub dependency was not observed when histone octamers were used (Figure 1D), indicating that ySet1C exhibits H2Bub-dependent H3K4 methylation activity only in a true nucleosomal context. Taken together, our in vitro HMT assays successfully recapitulate the H2Bub-dependent ySet1C H3K4 methylation activity reported by earlier studies, providing us with a means to directly assess the mechanism of H2Bub-H3K4me crosstalk, independent of indirect effects. Using this system, we sought to determine the minimal requirements for H2Bub-dependent H3K4 methylation in a nucleosomal context.
Set1, Swd1, Swd3, Bre2 and Sdc1 are the Minimal Yeast Set1 Complex Subunits Required for H2B Ubiquitylation-Dependent H3K4 Methylation
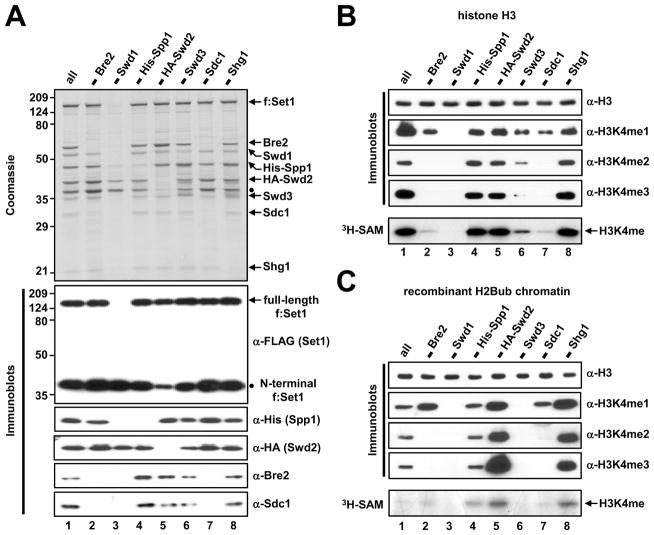
We first reconstituted and affinity-purified ySet1Cs lacking individual subunits (Figure 2A), leading us to make several conclusions regarding complex integrity: First, consistent with earlier reports (Dehé et al., 2006; Nedea et al., 2008; Halbach et al., 2009), we were unable to reconstitute an intact complex in the absence of Swd1 (lane 3), indicating a critical role of this subunit. In addition, lowered yields of complex were observed in the absence of Swd3, Bre2, Sdc1 or Swd2 (Figure S2A), suggesting additional contributions of these subunits to ySet1C stability. Second, in accordance with previous suggestions (Roguev et al., 2001; Dehé et al., 2006; South et al., 2010), a heterodimeric association of Bre2 and Sdc1 was required for their residence within ySet1C (anti-Bre2 and anti-Sdc1 immunoblots in Figure 2A, lanes 2 and 7). Third, reduced levels of the N-terminal Set1 degradation fragment (see dot) were observed in reconstitutions without Swd2 (compare lane 5 and other lanes in the staining and anti-FLAG immunoblot panels). Fourth, and importantly, the exclusion of either Spp1 or Swd2 did not affect the levels of other subunits (staining and anti-His and anti-HA immunoblots, lanes 4 and 5). These results suggest that Spp1 and Swd2 directly bind to a scaffold protein, likely Set1, in a manner independent of other subunits (detailed below).
Figure 2. Subunit Requirement for H3K4 Methylation Activity of the Yeast Set1 Complex.
(A) SDS-PAGE/Coomassie Blue staining and immunoblot analyses of purified ySet1Cs reconstituted with baculovirus vectors in the absence of the indicated subunits. Complex loading was normalized to FLAG-Set1 (except lane 3, see below).
(B and C) Free histone H3 (B) and H2Bub-chromatin templates (C) were subjected to in vitro HMT assays with indicated ySet1Cs.
Note that because of inefficient complex formation in the absence of Swd1, a volume of the Swd1-deficient sample (lanes 3) equal to that of the intact complex sample (lanes 1) was used for SDS-PAGE and in vitro HMT assays.
See also Figure S2.
Having ySet1 complexes lacking individual subunits, we then sought to assess the contribution of each subunit to the associated H3K4 methylation activity in an in vitro HMT assay, first using free H3 as substrate (Figure 2B). The residual complex reconstituted in the absence of Swd1 showed a complete loss of H3K4 methylation activity (lane 3). Omission of Swd3 resulted in moderately decreased H3K4me1 activity and markedly reduced H3K4me2/3 activities (lane 6 versus lane 1). Complexes lacking Bre2 or Sdc1 generated significant, albeit decreased, levels of H3K4me1, but failed to generate H3K4me2/3 (lanes 2 and 7 versus lane 1), indicating clear defects in “processivity” of the methylation reaction. Importantly, the Spp1, Swd2 and Shg1-deficient complexes exhibited H3K4 methylation activities comparable to that of the intact complex (lanes 4, 5, and 8 versus lane 1).
We next tested the contribution of each subunit to the H3K4 methylation activity of ySet1C on an H2Bub-containing chromatin template (Figure 2C). Consistent with previous reports (Nagy et al., 2002; Dehé et al., 2006; Nedea et al., 2008), complexes reconstituted in the absence of Swd1 or Swd3 were totally inactive for H3K4 methylation (lanes 3 and 6). Next, ySet1Cs lacking Bre2 or Sdc1 exhibited H3K4me1 activity but no H3K4me2/3 activities with the chromatin template (lanes 2 and 7). We further found that the Spp1 (lane 4)-, and Shg1 (lane 8)-deficient complexes showed modest and significant increased levels of H3K4 methylation activities, respectively. Thus, our in vitro system, employing minimal purified factors, reveals direct and varying contributions of given subunits to enzymatic activity.
Surprisingly, on recombinant chromatin templates, omission of Swd2 from the purified Set1 complex significantly increased the levels of H3K4me1, H3K4me2 and H3K4me3 relative to the intact complex (Figure 2C, lane 5 versus lane 1). This markedly enhanced activity remains dependent on H2Bub (Figures S2B–D), excluding the formal possibility that omission of Swd2 renders the H3K4 methylation activity of the complex H2Bub-independent. Moreover, this difference between intact and Swd2-deficient complexes does not reflect the presence of the HA-tag on Swd2 (Figures S2E and S2F). Rather, our findings suggest that Swd2 plays an intrinsic inhibitory role on H3K4 methylation activity within ySet1C that has previously been unrecognized. Finally, our results define Set1, Swd1, Swd3, Bre2 and Sdc1 as the minimal set of subunits directly required for H2Bub-dependent H3K4 methylation activity (Figures S2G–I) by ySet1C.
Structural Organization of the Yeast Set1 Complex
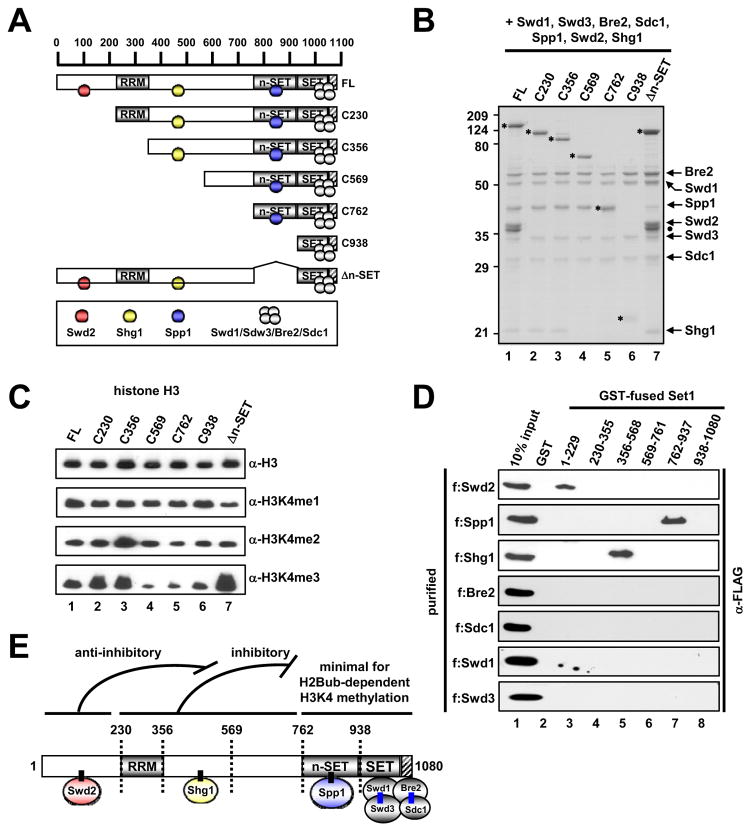
To determine regions in Set1 that are required for H3K4 methylation and for interactions with other ySet1C subunits, we generated a series of deletion mutants of Set1 (Figure 3A) before combining them (as FLAG-tagged Set1 or Set1 fragments) with the other seven (untagged) subunits. After reconstitution and purification, the wild type or mutant ySet1 complexes are referred to as full length (FL) or Δn-SET (internal deletion of residues 762–937) or by their N-terminal truncation site (C230, C356, C560, C762 and C938). Compositional analyses revealed (i) that the C230 and C356 complexes lack only Swd2 (Figure 3B, lanes 2 and 3), (ii) that the C569 and C762 complexes lack both Swd2 and Shg1 (lanes 4 and 5), (iii) that the C938 complex lacks Swd2, Shg1 and Spp1 (lane 6), (iv) that deletion of the n-SET domain (Δn-SET) leads to the selective loss of Spp1 (lane 7 and Figure S3A) and (v) that all complexes contain equal levels of Swd1, Swd3, Bre2 and Sdc1. These results indicate that C938, containing only the SET and post-Set domains of Set1, is sufficient for association of Swd1, Swd3, Bre2 and Sdc1 (see also Figure S3B). In functional assays, all complexes, including the C938-based complex, exhibited comparable levels of H3K4 mono-, di- and tri-methylation on free H3 (Figure 3C), whereas the C938 fragment alone was unable to methylate H3K4 (Figures S3C and S3D). These results indicate that the SET and post-SET domains, in conjunction with the other associated four subunits, have an intrinsic ability to methylate H3K4.
Figure 3. Subunit Interactions within the Yeast Set1 Complex.
(A) A schematic diagram of Set1 and derived fragments, with predicted RRM, n-SET, SET and post-SET domains, and subunit associations deduced from interaction studies. Hatched box, the post-SET domain in this and other figures.
(B) SDS-PAGE/Coomassie Blue staining of purified ySet1Cs reconstituted with baculoviruses expressing FLAG-Set1 or FLAG-Set1 fragments and all seven (untagged) ySet1C subunits. FLAG-Set1 polypeptides are marked by asterisks. Complex loading was normalized to Swd1, Swd3, Bre2 and Sdc1. Note that the C762 Set1 fragment co-migrates with Spp1.
(C) In vitro HMT assays with free histone H3 and indicated purified ySet1Cs.
(D) Direct binding of purified FLAG-tagged ySet1C subunits (Figure S3G) to GST-Set1 fragments (Figure S3F) relative to GST.
(E) Schematic model of subunit interactions with Set1. Direct interactions established in this study are depicted by short, black connecting lines. Direct Bre2-Sdc1 and Swd1-Swd3 interactions established in earlier studies (Roguev et al., 2001; Dehé et al., 2006; Halbach et al., 2009) are indicated by blue lines. Apparent functions of Set1 domains in methylation activity, indicated at the top, are deduced from Figure 4. Numbers indicate amino acid residues. Although not depicted, an interaction between Swd1-Swd3 and a region that lies N-terminal to the SET domain (probably the n-SET domain associated with Spp1: see Figure 6) is suggested by the observation that a Set1 fragment encoding residues 1–900 is co-immunoprecipitated with Swd1-Swd3 (Dehé et al., 2006).
See also Figure S3.
Given the above observations of subunit associations, we next examined direct interactions between purified Set1 fragments (Figures S3E and S3F) and purified subunits (Figure S3G). These analyses revealed direct binding of Swd2 to the N-terminal part (residues 1–229) of Set1 (Figure 3D, top panel), direct binding of Spp1 to the n-SET domain (residues 762–937) within Set1 (second panel) and direct binding of Shg1 to Set1 residues 356–568 (third panel). The specific interactions were also confirmed by other in vitro and in vivo approaches (Figures S3H–K). Despite efficient co-purification of Swd1, Swd3, Bre2 and Sdc1 by FLAG-tagged C938 (Figures 3B and S3B), none of these proteins alone were shown to interact directly with any Set1 fragments (Figure 3D, lower four panels), potentially reflecting the necessity of prior subunit interactions (e.g. Bre2-Sdc1 or Swd1–Swd3 heterodimer formation) for their association with Set1. Taken together, these results establish an organization of ySet1C shown in Figure 3E (also depicted in Figure 3A), and are generally consistent with earlier cell-based interaction studies (Dehé et al., 2006; Halbach et al., 2009).
The n-SET Domain within Set1 Is Essential for H3K4 Methylation on Ubiquitylated H2B-Containing Nucleosomes
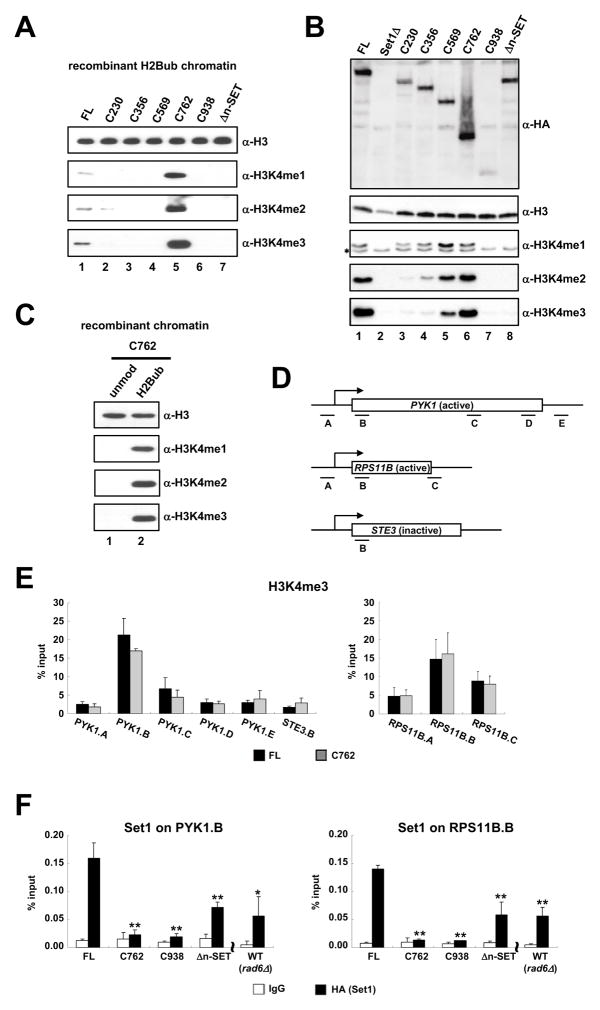
To rigorously examine the H2Bub-dependent nature of purified ySet1 complexes containing either FL Set1 or Set1 fragments (Figure 3B), all complexes depicted in Figure 3A were tested in an in vitro HMT assay with the H2Bub-containing chromatin substrate. Unlike an in vitro HMT assay with free H3 as substrate (Figure 3C), complexes containing Set1 N-terminal deletion mutants C230, C356 and C569 showed no H3K4 methylation activity (Figure 4A, lanes 2–4 versus lane 1). However, and interestingly, the C762 complex, a complex that contains a Set1 fragment encompassing the n-SET, SET and post-SET domains (along with the associated five subunits as depicted in Figure 3A), exhibited an H3K4 methylation activity that was markedly higher than that of the intact Set1 complex (Figure 4A, lane 5 versus lane 1). Furthermore, the C938 complex, which contains only the SET and post-SET domains along with the associated four subunits, showed no H3K4 methylation activity in this assay (lane 6) despite its intrinsic ability to methylate free H3 (Figure 3C). Hence, these data strongly suggest that the n-SET domain contains a previously unrecognized motif that is critical for H2Bub-dependent H3K4 methylation. In support, the complex containing only an internal n-SET deletion within Set1 exhibited no H3K4 methylation activity (Figure 4A, lane 7). These results indicate the presence of an inhibitory domain, N-terminal to the n-SET domain, that is counteracted by the N-terminus (residues 1–229) of Set1 (schematized in Figure 3E).
Figure 4. The n-SET Domain Is Required for the H2B Ubiquitylation-Dependent H3K4 Methylation Activity of the Yeast Set1 Complex.
(A) In vitro HMT assays with purified ySet1Cs (indicated in Figure 3B) and H2Bub-chromatin templates.
(B) Immunoblots with indicated antibodies of whole cell extracts from different set1 mutant strains carrying chromosomal genes expressing the indicated HA-tagged FL Set1 or Set1 fragments. Asterisks denote a faster-migrating, nonspecific band detected in all reactions with the specific batch of anti-H3K4me1 antibody used for this analysis.
(C) In vitro HMT assay with the purified C762 complex on chromatin templates assembled with unmodified or H2Bub octamers.
(D) Schematic representation of transcriptionally active (PYK1 and RPS11B) and inactive (STE3) loci and amplicons used for quantitative PCR.
(E) H3K4me3 ChIP analyses on the indicated genes/amplicons in yeast strains carrying chromosomal HA-tagged FL or C762 Set1 genes. Error bars denote standard deviations from three independent biological replicates both here and in Figures 4F and 5E.
(F) ChIP analyses with anti-HA antibody to determine chromatin association of HA-tagged FL, C762, C938 and Δn-SET Set1 proteins at 5′-transcribing regions (marked ‘B’ in Figure 4D) of PYK1 (left) and RPS11B (right) genes. A yeast strain that harbors an HA-tagged WT Set1 gene was also tested in a rad6Δ background. Anti-rabbit IgG was used as a control in this figure (and in Figure 5E). The significance of the differences in the ChIP signals was evaluated using the Student’s t-test (* denotes p-value 0.01–0.05 and ** denotes p-value < 0.01).
See also Figure S4.
To confirm a critical role for the n-SET domain in H3K4 methylation in vivo, we generated yeast strains that harbor chromosomal copies of HA-tagged Set1 and Set1 fragments, and monitored H3K4me levels (Figure 4B). Consistent with the in vitro results, as well as earlier in vivo studies (Briggs et al., 2001; Fingerman et al., 2005; Schlichter and Cairns 2005), the C762 strain exhibited a normal level of H3K4 methylation (lane 6 versus lane 1) whereas the C938 and n-SET deletion strains showed no detectable activity (lanes 7 and 8). However, in contrast to the in vitro HMT assay, the C569 strain, and to a lesser extent, the C356 strain, exhibited significant levels of H3K4 methylation (lanes 4 and 5). These more modest H3K4 methylation activities may reflect a more complicated H3K4 methylation process that is modulated in vivo by other factors that are missing in our minimal in vitro system.
Next, we undertook a more detailed characterization of the C762 complex, as this complex, which contains the n-SET domain, gives strong H3K4 methylation with our H2Bub-containing chromatin template. Like the intact ySet1 complex, the C762 complex exhibited H3K4 methylation activity only in the presence of H2Bub both in vitro (Figure 4C) and in vivo (Figure S4A), thus ruling out the possibility that markedly increased H3K4 methylation by the C762 complex in vitro is generated in an H2Bub-independent manner.
Beyond analyses of global H3K4 methylation levels in the SET1 serial deletion strains, transcriptionally active (PYK1 and RPS11B) and inactive (STE3) genes were chosen for analysis of the region-specific accumulation of H3K4 methylation and Set1 by chromatin immunoprecipitation (ChIP). As expected, H3K4me3 was highly enriched at the 5′ early transcribing regions (“B” regions in Figure 4D) relative to other regions (Figure 4E). Importantly, the C762 strain exhibited patterns of H3K4me3 accumulation on active and inactive genes almost identical to those of the wild-type (WT) strain with FL Set1. This suggests that the normal levels of global H3K4 methylation in the C762 strain (Figures 4B and S4A) are not due to aberrant increases of H3K4me at non-transcribing genes. In addition, the observation of decreased H3K4me3 levels on the active genes in the C569 strain (Figure S4B) is consistent with the global decrease of H3K4me3 in this strain (Figure 4B). Interestingly, a ChIP analysis of Set1 localization on active genes revealed that C762 occupancy is markedly decreased, relative to FL Set1 occupancy, at the 5′ early transcribing regions (Figure 4F), suggesting that some region N-terminal to the n-SET domain is important for proper recruitment and/or maintenance of Set1 on chromatin (Figure S4C). As to the comparable H3K4 methylation levels in the C762 and WT strains (Figures 4B and S4A), we reason that the increased intrinsic activity of the C762 complex relative to the intact complex (Figure 4A) compensates for its decreased chromatin association in cells (Figure 4F) such that the C762 strain shows a comparable level of H3K4 methylation compared to that of WT strain (Figures 4B and 4E). Lastly, Δn-SET Set1, albeit at a lower level than WT Set1, showed a clear accumulation on active genes, excluding the possibility that the absence of H3K4 methylation in the Δn-SET strain is caused by defect in the recruitment of Δn-SET Set1 to chromatin (Figure 4F and S4C).
To further verify that components (excluding Swd2) of the C762 complex are sufficient for H3K4 methylation in vivo, we constructed FL, C762 and C938 Set1 strains that were derived from the viable swd2 deletion background overexpressing the C-terminal fragment of Sen1, a transcription termination and processing factor (Nedea et al., 2008). In the FL Set1 derivative of the swd2 deletion strain, we observed a lower, but nonetheless significant, level of H3K4me2/3 compared to that in the strain containing WT Swd2 (Figure S4D). Consistent with the above results, the C762 derivative of the swd2 deletion strain exhibited a level of H3K4me2/3 comparable to that of the FL Set1 strain containing WT Swd2, whereas the C938 derivative of the swd2 deletion strain showed no H3K4 methylation. Taken together, these findings indicate a clear requirement of the n-SET domain for cellular H3K4 methylation that is persistent even without Swd2 -- thus calling attention to this domain of Set1 for the first time.
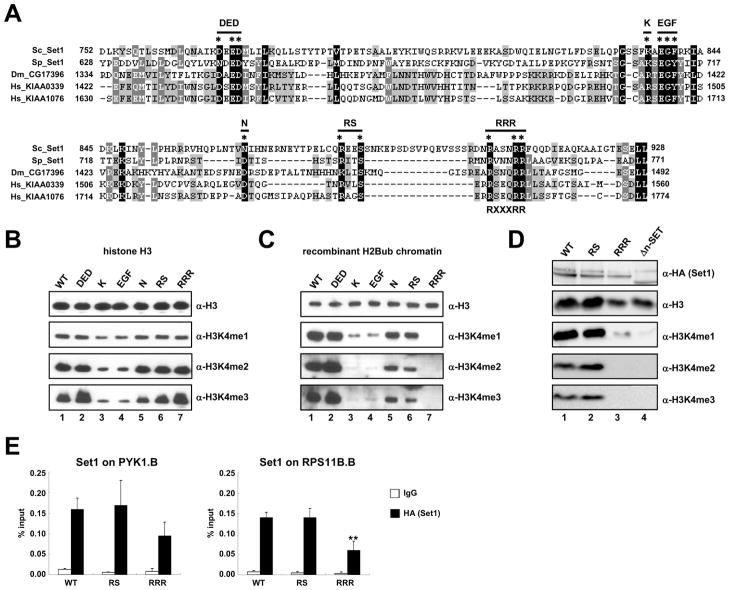
The RXXXRR Motif within the n-SET Domain Is Critical for H2B Ubiquitylation-Dependent H3K4 Methylation
The n-SET domain is highly conserved among various Set1 family members (Figure 5A; Roguev et al., 2001). Given the involvement of the n-SET domain in mediating H2Bub-dependent H3K4 methylation, we generated several point mutations at conserved amino acids within the n-SET domain to establish critical amino acid residue(s) required for H2Bub-dependent H3K4 methylation activity (Figure 5A). We then reconstituted and purified ySet1Cs expressing FLAG-tagged WT or mutant Set1 as in previous experiments (Figure S5A). All complexes displayed equal levels of ySet1C subunits, including Spp1 that directly associates with the n-SET domain, indicating that none of these mutations dramatically affect ySet1C subunit associations.
Figure 5. Identification of Amino Acid Residues within the n-SET Domain Responsible for H2B Ubiquitylation-Dependent H3K4 Methylation Activity of the Yeast Set1 Complex.
(A) ClustalW2 multiple sequence alignments of n-SET domains from Set1 family members: Saccharomyces cerevisiae (Sc_Set1, GeneBank Accession number: NP_011987), Schizosaccharomyces pombe (Sp_Set1, NP_587812), Drosophila melanogaster (Dm_CG17396, NP_001015221) and Homo sapiens (Hs_KIAA0339, BAA20797; Hs_KIAA1076, Q9UPS6). Encoded amino acid numbers are indicated. Conserved amino acids that were changed to alanine are marked by asterisks, and labels for derived Set1 mutants are indicated at the top. The consensus for the RXXXRR motif is indicated at the bottom.
(B and C) In vitro HMT assays with purified ySet1Cs bearing the indicated Set1 mutations and free histone H3 (B) or H2Bub-chromatin templates (C).
(D) Immunoblot analyses with indicated antibodies of whole cell extracts from yeast strains that carry chromosomal genes expressing the indicated HA-tagged WT or mutant Set1 proteins.
(E) ChIP analyses with anti-HA antibody to determine chromatin association of HA-tagged WT, RS and RRR Set1 proteins at 5′-transcribed regions (marked ‘B’ in Figure 4D) of PYK1 (left) and RPS11B (right) genes. The significance of the differences in the ChIP signals was evaluated using the Student’s t-test (** denotes p-value < 0.01).
See also Figure S5.
In an in vitro HMT assay with free H3 as substrate, the DED, N, RS and RRR mutants exhibited H3K4 methylation activities comparable to that of WT Set1 (Figure 5B, lanes 2 and 5–7 versus 1). However, the K and EGF mutants exhibited similar levels of H3K4me1 but decreased levels of H3K4me2/3 relative to those observed with WT (lanes 3 and 4 versus 1). Notably, the latter two mutations are located at the region considered to have homology to the Win motif within the pre-SET domain of the mammalian MLL family members. The Win motif is known to affect H3K4 methylation activity of MLL1 through an interaction with WDR5 (human homologue of yeast Swd3) (Patel et al., 2008; Song and Kingston, 2008). These results suggest a conserved role for this region in modulating H3K4 methylation activity of H3K4 methyltransferase members.
Next, these complexes were subjected to an in vitro HMT assay with the H2Bub-containing chromatin substrate (Figure 5C). The DED, N, RS mutants showed robust H3K4 methylation activities (lanes 2, 5 and 6) whereas the K and EGF mutants exhibited significantly decreased levels of H3K4 methylation activity relative to WT (lanes 3 and 4 versus 1). The decreased H3K4 methylation mediated by the latter two complexes is likely due to their diminished intrinsic abilities to methylate H3K4. Importantly, despite its intrinsic ability to methylate free H3, the RRR mutant is completely unable to carry out H3K4 di- and tri-methylation on either H2Bub-containing (lane 7) or unmodified (Figure S5B) chromatin. Notably, however, the RRR mutant, like the EGF mutant, effects a low level of H3K4 mono-methylation equivalent to that observed with the WT Set1 complex (Figure S5B). Thus, the selective n-SET function in nucleosomal versus free H3 methylation relates to its cooperative function with H2Bub rather than some H2Bub-independent function related to nucleosomal structure per se. In support, yeast cells containing a chromosomal RRR mutation showed complete loss of H3K4me2/3 and a significant decrease in H3K4me1 (Figure 5D, lane 3 versus lane 1). ChIP analysis demonstrated that the RRR Set1 mutant can be recruited to active genes (Figure 5E), indicating that the H3K4 methylation defect of this mutant is not due to its inability to associate with chromatin. By identifying a specific “RXXXRR” motif whose mutation causes complete loss of H2Bub-dependent H3K4 di-/tri-methylation, our results strengthen the argument for a critical role of the n-SET domain in H2Bub-dependent H3K4 methylation.
The n-SET Domain Bound to Spp1 Communicates with the ‘Catalytic Core’
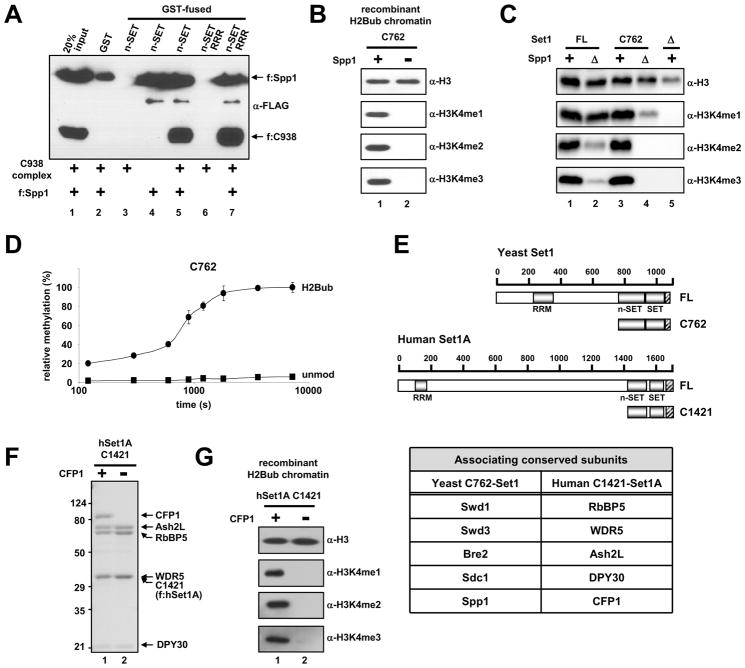
Based on the intrinsic methylation activity of the ‘catalytic core’ [equivalent to the C938 complex that contains the SET and post-SET domains of Set1 and the minimal subunits (Swd1, Swd3, Bre2 and Sdc1), Figures 3 and S3] and the specific requirement of the n-SET domain for methylation of H2B-ubiquitylated chromatin (Figures 4 and 5), we postulated that H2Bub-dependent H3K4 methylation activity of ySet1C might reflect a direct binding of ubiquitin to ySet1C via the n-SET domain. However, to date, our best attempts have failed to provide convincing evidence for a direct physical interaction of free ubiquitin, H2Bub or H2Bub-chromatin with any ySet1C subunit or derived Set1 fragment (data not shown; see Discussion). As an alternative, we considered the possibility that direct communication between the n-SET domain and the catalytic core might be involved in H2Bub-dependent H3K4 methylation. Binding of the C938 complex (containing FLAG-C938, Swd1, Swd3, Bre2 and Sdc1) to GST-immobilized n-SET fragment (Figure 6A, lane 3) was not detected. However, addition of purified Spp1 to the binding reaction resulted in a strong association of the C938 complex with n-SET (lane 5), whereas Spp1 alone does not bind to C938 complex (lane 2, see Figure legend). Thus, these results suggest an interaction between the catalytic core and the n-SET domain and/or between the catalytic core and Spp1 when Spp1 binds to n-SET -- which we refer to as an ‘internal domain interaction’.
Figure 6. Direct Internal Domain Interaction and Conformational Change in the Active Site of the Set1 Complex.
(A) The purified C938 complex (Figure 3B, lane 6) was tested for binding to GST versus GST-n-SET (Set1 residues 762–937, Figure S3F) in the presence and absence of purified FLAG-Spp1 (Figure S3G). Bound proteins were scored by immunoblotting with anti-FLAG antibody. n-SET RRR, the n-SET fragment that contains RRR mutation, is described in Figure 5. Note that the Spp1 that is nonspecifically bound to GST under these binding conditions (150 mM KCl, 0.05 % NP40 and 0.2 mg/ml BSA, lane 2) does not coimmunoprecipitate the C938 complex, indicating that Spp1 alone does not interact with the C938 complex.
(B) H2Bub-chromatin templates were subjected to in vitro HMT assays with purified C762 complexes containing (Figure 3B, lane 5) and lacking (Figure S3B, lane 5) Spp1.
(C) Whole cell extracts from yeast cells that carry the indicated chromosomal genes expressing HA-tagged FL and C762 Set1 and their isogenic spp1Δ derivatives were subjected to immunoblot analyses with indicated antibodies.
(D) Kinetic analysis of methylation of H2Bub- and unmodified chromatin by the C762 complex. The highest methylation level observed at 2h is arbitrarily set as 100 %. X-axis is shown in a log10 scale to emphasize early time points. Error bars denote standard deviations from three independent assays.
(E) A schematic diagram of full-length and C1421 (encompassing n-SET, SET and post-SET domains) human Set1A fragments along with a table of minimal components required for H3K4 methylation in the yeast and human Set1 complexes.
(F) SDS-PAGE/Coomassie Blue staining of purified human Set1A complexes reconstituted with baculoviruses expressing FLAG-tagged C1421 fragment, RbBP5, WDR5, Ash2L, DPY30 and with or without CFP1.
(G) H2Bub-chromatin templates were subjected to in vitro HMT assays with purified human C1421 complexes containing and lacking CFP1.
See also Figure S6.
In assessing the involvement of Spp1 in n-SET-mediated H2Bub-dependent H3K4 methylation, the C762-based complex lacking Spp1, unlike the complete C762 complex that contains Spp1, was unable to methylate H3K4 on H2Bub-chromatin (Figure 6B). Consistent with this observation, Spp1 deletion in the C762 strain resulted in both the complete disappearance of H3K4me2/3 and a significant decrease of H3K4me1 (Figure 6C, lane 4 versus 3). Taken together, these results indicate that, in the absence of N-terminal Set1 sequences, the n-SET domain and Spp1 act in conjunction with the catalytic core to mediate H2Bub-dependent H3K4 methylation.
Importantly, the above-described internal domain interaction (e.g. between Spp1-bound n-SET and the catalytic core) raises the possibility of a conformational change in the active site of the Set1 complex in the presence of H2Bub as its chromatin substrate. To explore this possibility, we performed a kinetic analysis with an excess amount (saturating level) of enzyme in relation to the substrate. The C762 complex exhibits more efficient methylation of H2Bub-chromatin as compared to unmodified chromatin even at early reaction time points, which likely mimic a single turnover paradigm (Figure 6D). This result suggests that H2Bub and the internal domain interaction somehow combine to enhance the chemical step of H3K4 methylation by the SET catalytic core. This kinetic enhancement could reflect changes in the structure of the enzyme active site, the nucleosome or both.
We next sought to determine if the above findings in yeast extend to human; and, in as much as humans have multiple Set1-like family members along with conserved complex subunits, we explored whether a conserved role existed for the Spp1 homolog in H2Bub-mediated H3K4 methylation. Our findings show that CFP1, the human homolog of yeast Spp1, is also necessary for the activity of a partial human Set1A complex that contains an hSet1A fragment encompassing the n-SET, SET and post-SET domains (Figures 6E-G). These findings lend strong support of a conserved role(s) of the n-SET domain and its associated Spp1 homologs in mediating an H2Bub “sensing” mechanism for from yeast to human (see Discussion).
DISCUSSION
To elucidate an H2Bub-dependent H3K4 methylation mechanism that involves an interplay between domains within Set1 and subunits of ySet1C, we have used a reconstituted, biochemically-defined system as well as yeast genetic analyses. Our findings permit valuable mechanistic insights into this cross-talk pathway at several levels: (i) H2Bub directly stimulates mono-, di-, and tri-methylation of H3K4 by ySet1C, (ii) the conserved Swd1, Swd3, Bre2, and Sdc1 subunits are sufficient for H2Bub-dependent methylation of nucleosomal H3K4 by a reconstituted ySet1C that contains full-length Set1, (iii) the Swd2 subunit, while able to modulate the activity of ySet1C, is not directly required for the H2Bub-dependent H3K4 methylation activity of the complex, (iv) the n-SET domain within Set1 plays a critical regulatory role in H2Bub-dependent H3K4 methylation and (v) Spp1 is also involved in n-SET-mediated H2Bub-dependent H3K4 methylation -- but that its function may be conditional and redundant with that of an N-terminal region of Set1. Altogether, these results provide fundamentally new mechanistic insights into H2Bub-mediated H3K4 methylation by ySet1C.
H2B Ubiquitylation Directly Stimulates H3K4 Mono-, Di- and Tri-methylation by the Yeast Set1 Complex
The participation of many factors that directly or indirectly affect H3K4 methylation underscore a remarkably complex picture of the mechanism of H2Bub-dependent H3K4 methylation (Ruthenburg et al., 2007). In this study, using a semi-synthetic H2B that is fully ubiquitylated at lysine 120 and purified ySet1C, we demonstrate that ubiquitylated H2Bub, in the absence of other histone modifications and other factors, directly stimulates H3K4 methylation by ySet1C -- as previously shown for human DOT1L (McGinty et al., 2008) and human (Kim et al., 2009) and fission yeast (Racine et al., 2012) Set1 complexes.
Our in vitro assays demonstrate that H2Bub directly stimulates not only di- and tri- but also mono-methylation of nucleosomal H3K4 by ySet1C (Figure 1), which is supported by our own (Figure S1) and other (Chandrasekharan et al., 2010) in vivo studies showing reduced H3K4 mono-methylation in the absence of H2B ubiquitylation. Thus, our biochemical analyses indicate that H2Bub directly stimulates all three H3K4 methylation states and, given the additional observation that H2Bub is essential for H3K4 methylation on a nucleosomal substrate, but not on a histone octamer without DNA, point to the necessity for using more physiologically-relevant nucleosomal substrates for mechanistic studies of histone cross-talk.
The SET and Post-SET Domains of Set1, Along with Swd1, Swd3, Bre2 and Sdc1, Form the ‘Catalytic Core’ for H3K4 Methylation
The essential Swd1, Swd3, Bre2 and Sdc1 subunits of ySet1C were found to associate directly with the C-terminal SET and post-SET domains of Set1; and the C938 complex that is minimally composed of these components was shown to have an H3K4 methyltransferase activity toward free H3. Thus, the intrinsic H3K4 methyltransferase activity of ySet1C is minimally contributed by the Set1 SET and post-SET domains and the associated four subunits (designated the ‘catalytic core’). Of note, the Swd1, Swd3, Bre2 and Sdc1 subunits are homologs, respectively, of the RbBP5, WDR5, Ash2L and DPY30 proteins that are common subunits in the mammalian SET1/MLL family of H3K4 methyltransferases and that, with SET1/MLL SET domains, were shown to form catalytically active core methyltransferase complexes (reviewed in Cosgrove and Patel, 2010). Therefore, we postulate that formation/function of the catalytic pocket of ySet1C might similarly be regulated by association of the Set1 SET domain with the interacting core subunits and potentially by additional interactions of other subunits or Set1 domains (see below) with the catalytic core.
Swd2 Is Not Directly Required for H2B Ubiquitylation-Dependent H3K4 Methylation by the Yeast Set1 Complex
We also uncovered some surprising results in relation to the Swd2 subunit. In contrast to the conclusions of two previous reports (Lee et al., 2007; Vitaliano-Prunier et al., 2008), the current study provides several lines of biochemical and genetic evidence indicating that Swd2 does not necessarily participate directly in H2Bub-dependent H3K4 methylation by ySet1C (Figures 2, 4, 6, S2, S4, and S6). Most compelling is our demonstration that exclusion of Swd2 from the reconstituted Set1 complex markedly enhances, rather than compromises, the H2Bub- dependent H3K4 mono-, di- and tri-methylation activity on a chromatin substrate. We postulate that the apparent discrepancies in results or interpretations of the various studies are potentially due to an indirect role(s) of Swd2 in regulating the structure and function of Set1C through factors present in vivo but not in our in vitro system. In this regard, the intracellular stability of Set1, and thus the integrity of the Set1 complex, is critically dependent upon the presence of Swd2 (Dichtl et al., 2004; Nedea et al., 2008) and the Swd2-containing Set1 and APT complexes show antagonistic functions (Soares and Buratowski, 2012). Thus, in light of the new data presented here, as well as previous observations, the exact role of Swd2 in ySet1C-mediated histone methylation remains unclear and needs to be reevaluated.
Role of the Set1 n-SET Domain and Spp1 in Mediating H2B Ubiquitylation-Dependent H3K4 Methylation and Potentially Redundant Functions of Other Domains within Set1
Unlike the competent Set1 C762 complex (containing only the n-SET, SET and post-SET domains), the C938 complex that lacks the n-SET domain has H3K4 methylation activity toward free H3 but not toward H2Bub-chromatin. Therefore, we conclude that a region necessary for H2Bub-dependent H3K4 methylation resides within the n-SET domain. This conclusion is further supported by our additional finding that an internal deletion of the n-SET domain almost completely abolishes H2Bub-dependent H3K4 methylation both in vitro and in vivo.
The n-SET domain, located immediately N-terminal to the SET domain, is more divergent than the SET and post-SET domains, but is still evolutionarily conserved among Set1 family members (Roguev et al., 2001). Interestingly, MLL family H3K4 methyltransferases also have SET and post-SET domains at their C-termini, but the MLL ‘pre-SET’ domain that lies immediately in front of the SET domain is different from the Set1 n-SET domain. In light of our finding that the n-SET RXXXRR motif plays a critical role in H2Bub-dependent H3K4 methylation, we note that the MLL pre-SET domain does not contain the RXXXRR motif. These observations raise the intriguing possibility that Set1 and MLL family members employ somewhat different mechanisms for H3K4 methylation. In support, H3K4 methylation in the ciliated protozoan Tetrahymena, which contains a single MLL-like H3K4 methyltransferase (with a pre-SET domain but no n-SET domain), is H2Bub-independent (Wang et al., 2009).
In addition to the n-SET domain, the interacting Spp1 subunit was also found to be critical for n-SET-mediated H2Bub-dependent H3K4 methylation in the absence of the Set1 N-terminal domain; and this function is conserved in the human complex (Figure 6). These results suggest that some region(s) within the first 761 residues of Set1 may act in a redundant manner with Spp1, raising the possibility of both a normal Spp1-independent pathway and a conditional Spp1-dependent pathway that may depend upon the modulation of Set1 N-terminal functions by other physiologically variable factors. Importantly, the n-SET RXXXRR motif, while critical for H2Bub-dependent H3K4 methylation, is not responsible either for association of n-SET with Spp1 (Figure S5) or for association of the catalytic core with the n-SET domain (Figure 6). These results imply that the n-SET RXXXRR motif contributes to H2Bub-dependent H3K4 methylation activity in an Spp1-independent manner. Interestingly, Set1-mediated H3K4 methylation requires Spp1 in fission yeast (Roguev et al., 2003). Our findings re-emphasize the importance of Spp1 for H2Bub-dependent H3K4 methylation and further suggest that our mechanistic studies on Spp1 may be relevant to the function of Spp1 homologues (CFP1/CXXC) in higher organisms.
Models for H2B Ubiquitylation-Dependent H3K4 Methylation by the Yeast Set1 Complex
The fundamental question remains as to the exact role of the n-SET domain in “sensing” H2Bub on chromatin templates to bring about H3K4 methylation. Despite extensive efforts, no convincing evidence was obtained for interactions of either the n-SET domain alone or the Spp1-assocated n-SET domain with free ubiquitin, free H2Bub or H2Bub chromatin under a variety of binding conditions. Thus, there either is no direct physical interaction between the n-SET domain and H2Bub chromatin or such an interaction is too weak or too transient to be detected in vitro. Consistent with this view, it is unlikely that H2Bub alone accounts for the entire mechanism of ySet1C recruitment to chromatin in vivo since RAD6 deletion only partially impairs Set1 accumulation on chromatin (Figure 4F; Ng et al., 2003; Dehé et al., 2005). Possibilities for H2Bub-independent recruitment include reported Set1C interactions with the RNA polymerase II-associated Paf1 elongation complex and the phosphorylated RNA polymerase II CTD (reviewed in Dehé and Géli, 2006).
In our proposed model (Figure 7), we envision that H3K4, either on free H3 or in the context of the histone octamer, is freely accessible to the yeast Set1C “catalytic core’” that is composed of the SET–post-SET domain and associated Swd1, Swd3, Bre2 and Sdc1 subunits (Figure 7A). In contrast, and despite Set1C recruitment to chromatin in the absence of H2B ubiquitylation (above), we propose that the unmodified nucleosomal substrate structure occludes free access of the catalytic core to H3K4 (Figure 7B). In the latter case, we envision that the installation of ubiquitin on nucleosomal H2B could have several consequences for H3K4 methylation by ySet1C (Figure 7C; see legend for additional details). One possibility is that, by modulating DNA-histone and/or histone-histone interactions, ubiquitin itself induces a conformational change(s) of the nucleosome that facilitates H3K4 methylation by the catalytic core. Earlier demonstrations of effects of H2B ubiquitylation on nucleosomal interactions and stability (Fierz et al., 2010; Chandrasekharan et al., 2010) and on H3 methylation by two different enzyme systems -- H3K79 methylation by hDot1L (McGinty et al., 2008) and H3K4 methylation by ySet1C (this study) -- lend support to this possibility.
Figure 7. Mechanistic Model for H2B Ubiquitylation-Dependent H3K4 Methylation by the Yeast Set1 Complex.
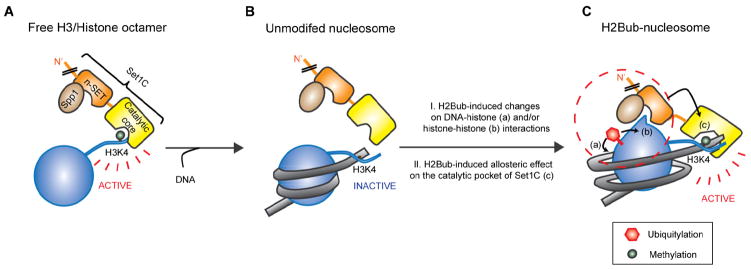
(A) Free histone H3 substrate. In this case, we envision that the ‘catalytic core’, consisting of the SET and post-SET domains of Set1 in association with Swd1, Swd3, Bre2, and Sdc1 subunits, is freely accessible to H3K4.
(B) Unmodified nucleosome substrate. In this more physiological case of a nucleosome with non-ubiquitylated histone H2B, and in contrast to the situation in (A), the active site of ySet1C (or the catalytic core) is not readily (or stably) accessed by H3K4. This parallels the in vivo situation involving H2Bub-independent recruitment of Set1C to genes (by other factors) without H3K4 methylation.
(C) H2B ubiquitylated nucleosome. Here we envision two possible models: (I) H2Bub-induced conformational changes (red-dashed circle) in the nucleosome by altering DNA-histone (a) and/or histone-histone (b) interactions and/or (II) H2Bub-induced allosteric changes in the active site of nucleosome-associated ySet1C that involve the Set1C catalytic core, the Spp1-associated n-SET domain, the RXXXRR motif within the n-SET domain and an internal interaction (black arrow) between the n-SET and SET domains (c). These conformational changes and interactions, which are not mutually exclusive, lead to the positioning (or stabilization) of H3K4 at a ‘favorable’ site for methylation by ySet1C.
H2Bub may also bring about a conformational change of the ySet1C active site that involves the catalytic core, the n-SET domain and Spp1 and, in turn, renders the active site accessible to H3K4 (Figure 7C). The selective n-SET domain and Spp1 requirements for H2Bub-dependent H3K4 methylation of nucleosomes relative to the H2Bub-independent H3K4 methylation of free and octameric H3, the H2Bub-independent recruitment of Set1C to chromatin, and the weak H2Bub-independent H3K4 mono-methylation of nucleosomal H3 provide hints at potential changes at the enzyme level. H2Bub-driven changes in either substrate or enzyme conformation are not mutually exclusive. For example, a ubiquitin-mediated nucleosomal conformational change might be a prerequisite for a conformational change in the ySet1C active site. Additional investigations, ideally involving structural analyses of the Set1 complex in association with H2Bub nucleosomes, should provide important information relevant to key aspects of these and other models. The present report, revealing key mechanistic aspects of H2Bub-dependent H3K4 methylation, sets the stage for such analyses with sharpened focus on the n-SET domain of Set1 and interacting subunits.
EXPERIMENTAL PROCEDURES
In Vitro Histone Methyltransferase Assays
Standard assays, detailed in Supplemental Experimental Procedures, involved either free histones, octamers, HeLa cell-derived oligonucleosomes, or recombinant chromatins as substrates; purified Set1 complexes; and detection of H3 methylation either by fluorography or by immunoblot.
Yeast Strains
All yeast strains used in this study are summarized in Table S1.
Chromatin Immunoprecipitation
Details of ChIP procedures are described in Supplemental Experimental Procedures. Primers for ChIP analyses are summarized in Table S2.
Supplementary Material
H2B ubiquitylation directly stimulates H3K4 methylation by the yeast Set1 complex
Swd2 is not directly required for H3K4 methylation activity of the yeast Set1 complex
The Set1 n-SET domain plays a critical role in H2Bub-dependent H3K4 methylation
Spp1 is conditionally involved in n-SET- and H2Bub-dependent H3K4 methylation
Acknowledgments
We thank Dr. P.L. Nagy for Set1, Bre2, Sdc1 antibodies and the swd2 deletion strain. This work was supported by NIH grants CA129325 (R.G.R.), DK071900 (R.G.R.) and CA148345 (R.G.R., C.D.A. and T.M.); GM40922 (C.D.A.); a Leukemia and Lymphoma Society SCOR grant (C.D.A. and R.G.R.); a Starr Foundation Cancer Consortium grant (R.G.R., C.D.A. and T.M.); and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A1014697, 2012M3A9B4027956 and 2012M3A9C6049938) (J.K.). J.K. is a recipient of the TJ Park Junior Faculty Fellowship and J.-A.K. was a fellow of The Rockefeller University ‘Women and Science’ program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Chen YC, Sun ZW. Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol Cell Biol. 2010;30:3216–3232. doi: 10.1128/MCB.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol Cell Biol. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Patel A. Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS J. 2010;277:1832–1842. doi: 10.1111/j.1742-4658.2010.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodríguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- Dehé PM, Géli V. The multiple faces of Set1. Biochem Cell Biol. 2006;84:536–548. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- Dehé PM, Pamblanco M, Luciano P, Lebrun R, Moinier D, Sendra R, Verreault A, Tordera V, Géli V. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J Mol Biol. 2005;353:477–484. doi: 10.1016/j.jmb.2005.08.059. [DOI] [PubMed] [Google Scholar]
- Dichtl B, Aasland R, Keller W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA. 2004;10:965–977. doi: 10.1261/rna.7090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman IM, Wu CL, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A, Zhang H, Wengi A, Jablonska Z, Gruber IM, Halbeisen RE, Dehé PM, Kemmeren P, Holstege F, Géli V, et al. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 2009;28:2959–2970. doi: 10.1038/emboj.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDOT1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Köhn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol. 2009;4:958–968. doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt JF, Nagy PL. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- Racine A, Page V, Nagy S, Grabowski D, Tanny JC. Histone H2B ubiquitylation promotes activity of the intact Set1 histone methyltransferase complex in fission yeast. J Biol Chem. 2012;287:19040–19047. doi: 10.1074/jbc.M112.356253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Aasland R, Shevchenko A, Stewart AF. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J Biol Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schlichter A, Cairns BR. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 2005;24:1222–1231. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell. 2005;19:271–27. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Soares LM, Buratowski S. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. J Biol Chem. 2012;287:15219–15231. doi: 10.1074/jbc.M112.341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Fingerman IM, Mersman DP, Du HN, Briggs SD. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J Biol Chem. 2010;285:595–607. doi: 10.1074/jbc.M109.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- Wang GG, Allis CD. “Misinterpretation” of a histone mark is linked to aberrant stem cells and cancer development. Cell Cycle. 2009;8:1982–1983. [PubMed] [Google Scholar]
- Wang Z, Cui B, Gorovsky MA. Histone H2B ubiquitylation is not required for histone H3 methylation at lysine 4 in tetrahymena. J Biol Chem. 2009;284:34870–34879. doi: 10.1074/jbc.M109.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.