Summary
TXNIP is an α-arrestin family protein that is induced in response to glucose elevation. It has been shown to provide a negative feedback loop to regulate glucose uptake into cells, though the biochemical mechanism of action has been obscure. Here, we report that TXNIP suppresses glucose uptake directly by binding to the glucose transporter, Glut1, inducing Glut1 internalization through clathrin coated pits, as well as indirectly by reducing the level of Glut1 mRNA. In addition, we show that energy stress results in phosphorylation of TXNIP by AMP-dependent protein kinase (AMPK), leading to its rapid degradation. This suppression of TXNIP results in an acute increase in Glut1 function and an increase in Glut1 mRNA (hence total protein levels) for long-term adaptation. The glucose influx through GLUT1 restores ATP/ADP ratios in the short run and ultimately induces TXNIP protein production to suppress glucose uptake once energy homeostasis is reestablished.
Introduction
Thioredoxin interacting protein (TXNIP) has many reported functions. As its name implies, TXNIP can form intermolecular disulfide bonds with reduced thioredoxin (Nishiyama et al., 1999; Patwari et al., 2006). This interaction has been interpreted as inhibition of thioredoxin function, leading to cellular oxidative stress as well as perturbing activities of proteins that rely on the presence of thioredoxin for their function, such as PTEN, ASK-1 and NLRP3 inflammasome (Hui et al., 2008; Hui et al., 2004; Jeon et al., 2005; Junn et al., 2000; Schulze et al., 2004; Zhou et al., 2010). On the other hand, independent of its thioredoxin binding property, TXNIP overexpression represses cellular glucose uptake while knocking down TXNIP increases glucose uptake in peripheral tissues in both insulin-dependent and insulin-independent manners (Parikh et al., 2007). Metabolically, Txnip knock-out animals exhibit phenotypes of familial combined hyperlipidemia, consistent with enhanced glucose uptake (Bodnar et al., 2002; Chutkow et al., 2008; Sheth et al., 2005). Since glucose availability affects ROS production in mitochondria, the diverse effects of TXNIP on thioredoxin functions and on glucose uptake suggest a unifying mechanism for maintaining homeostasis.
Since the initial identification of TXNIP as a Vitamin D3 upregulated protein (VDUP1) in HL-60 cells (Chen and DeLuca, 1994), many factors were found to regulate TXNIP mRNA level (Baker et al., 2008; Billiet et al., 2008; Kim et al., 2004; Lerner et al., 2012; Oslowski et al., 2012; Parikh et al., 2007; Wang et al., 2006; Yu et al., 2009). Most notable are the transcription complexes of chREBP/Mlx and MondoA/Mlx that bind to the carbohydrate response element (ChoRE) on the TXNIP promoter (Cha-Molstad et al., 2009; Stoltzman et al., 2008), resulting in a very tight correlation between TXNIP expression level and elevation in glucose influx into cells. As discussed above, the elevation in TXNIP protein suppresses glucose uptake, thus it appears to play a central role in maintaining glucose homeostasis.
TXNIP belongs to the arrestin superfamily, which has 14 members in human: 6 alphas (ARRDC1-5 and TXNIP), 4 visual/betas, and 4 Vps26 genes (Alvarez, 2008). The β-arrestins are well studied in their regulation of GPCR signaling. Recently, one member of the a-arrestin family, ARRDC3, was reported to down-regulate β2-adrenergic receptor and β-4 integrin, much like the β-arrestins (Draheim et al., 2010; Nabhan et al., 2010). With the exception of ARRDC5, a-arrestins have PPxY motifs in their C-terminal tails, and these motifs have been implicated in binding to WW-domains, such as ones in the HECT family of ubiquitin E3 ligases (Alvarez, 2008). One HECT member, Itch, reportedly binds to a PPxY motif in TXNIP and mediates its degradation under basal conditions (Zhang et al., 2010).
AMP-activated protein kinase (AMPK) monitors cellular energy status in response to nutritional variation in the environment. A low energy state results in activation of AMPK which, in turn, phosphorylates a host of key cellular proteins to suppress ATP consumption and increase in ATP production to restore energy homeostasis (Mihaylova and Shaw, 2011; Viollet et al., 2009). A recent study indicated that AMPK regulates chREBP/Mlx activity through phosphorylation-dependent nuclear translocation, thus indirectly regulating TXNIP protein level (Kawaguchi et al., 2002). In the course of examining the effect of glucose deprivation on chREBP/Mlx function we found that activation of AMPK leads to phosphorylation and degradation of TXNIP. We also found that TXNIP binds to Glut1 and facilitates its endocytosis via clathrin-coated pits (CCP). In addition, TXNIP expression causes a reduction in Glut1 mRNA level, thus reducing long-term Glut1 protein level. We propose that TXNIP is a dynamic sensor modulating the cell’s demand for energy together with its need to avoid the negative effects of nutrient overload.
Results
TXNIP is phosphorylated in an AMPK-dependent manner
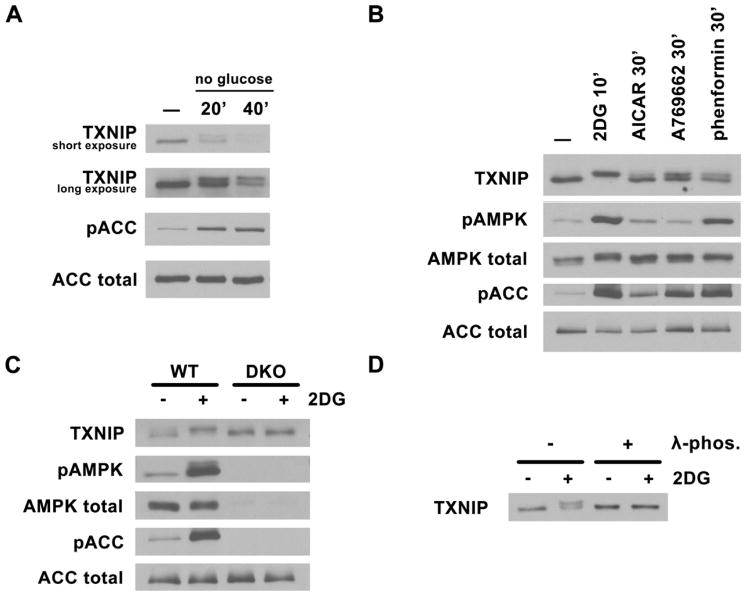
While investigating the mechanism of chREBP/Mlx function using TXNIP as a reporter, we discovered that under acute glucose withdraw, TXNIP protein is modified such that its mobility on SDS-PAGE is reduced (Fig. 1A). Since AMPK is acutely activated by glucose withdrawal, we explored the possibility that other mechanisms for activating AMPK would induce this altered migration of TXNIP. As shown in Fig. 1B (see also Fig. S1), treatment of various cell lines (HepG2, MCF7, ACHN and T47D) with 2-deoxyglucose (2DG), AICAR, A769662 or phenformin (agents known to either directly or indirectly activate AMPK) resulted in decreased migration of TXNIP on SDS-PAGE; this shift in migration correlated well with the extent of phosphorylation of ACC at a known AMPK site. More importantly, this shift in migration was not observed when AMPK α1/α2 double knockout MEFs were treated with 2DG (Fig. 1C), indicating that AMPK activation was required for the shift. Treatment of cell lysates with -phosphatase prior to SDS-PAGE abrogated the shift in migration (Fig. 1D), indicating that this difference was due to phosphorylation of TXNIP. Therefore, the activation of AMPK induces a phosphorylated species of TXNIP that migrates slower on SDS-PAGE.
Fig. 1. AMPK-dependent TXNIP phosphorylation.
Western blots of total cell lysates probed with TXNIP, phosphoAMPK (T172), total AMPK, phosphoACC (S79), and total ACC antibodies under various conditions. Activation of AMPK causes an upshift in TXNIP mobility. (A) HepG2 cells were glucose starved for 20′ and 40′ in DMEM with no glucose and 10% dialyzed FBS. (B) HepG2 cells were treated with AMPK activators: 25mM 2DG for 10′, 2mM AICAR for 60′, 1mM A769662 for 30′ and 2mM phenformin for 30′. (C) AMPK MEFs, WT and DKO (double knock-out for both α1 and α2) were treated with 25mM 2DG for 10′. (D) phosphatase treatment of endogneous TXNIP immunoprecipitated from HepG2 cells treated with 25mM 2DG abolishes the upshift.
AMPK-dependent phosphorylation of TXNIP on Ser308 leads to its accelerated degradation
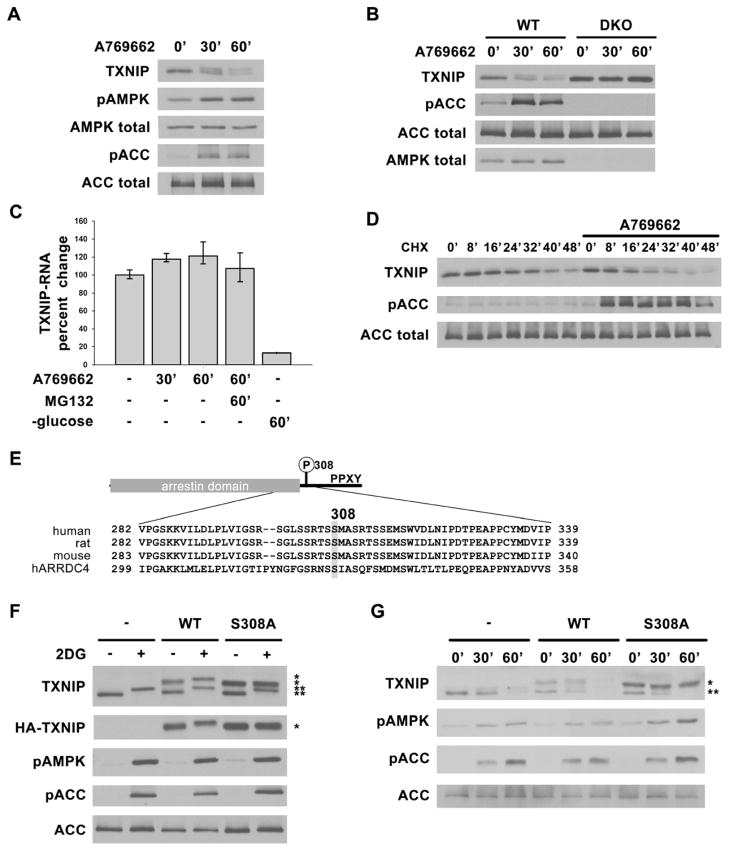
We observed that prolonged treatment of rat hepatocytes with AMPK activators resulted in a decrease in the total amount of TXNIP protein (Fig. 2A). These activators also caused a decrease in TXNIP protein in AMPK WT MEFs, but not in AMPK double knockout MEFs (Fig. 2B). The decrease in TXNIP protein did not appear to be due to an acute decrease in mRNA, since quantitative RT-PCR revealed no change in TXNIP mRNA within the time frame when protein levels dropped (Fig. 2C). After inhibiting protein synthesis with cycloheximide, TXNIP protein levels dropped more rapidly in the presence of A769662 than in its absence (Fig. 2D), further supporting the idea that TXNIP protein degradation was accelerated by AMPK activators. Finally, we found that the proteosome inhibitor MG132 blocked the A769662-dependent degradation of TXNIP and essentially all the TXNIP accumulated in the upper band (Fig. S2A). MG132 did not affect TXNIP mRNA level in A769662-treated cells. Altogether, these results consistently support the concept that a phosphorylation event leads to the shift of TXNIP on SDS-PAGE and that this phosphorylation event correlates with proteosome-dependent degradation.
Fig. 2. AMPK phosphorylation of TXNIP on S308 accelerates its degradation.
Western blots showing decreasing TXNIP levels after AMPK activation in (A) primary rat hepatocytes and (B) AMPK WT and DKO MEFs treated with 1mM A769662 for 0′, 30′ and 60′. (C) qRT-PCR analysis of TXNIP mRNA from RNA isolated from HepG2 cells after various treatments indicating short-term activation of AMPK (e.g. using A769662) does not decrease TXNIP mRNA level while glucose starvation does as reported before. The values are the average of triplicates ±STDV. (D) HepG2 cells were pretreated with cycloheximide (CHX) for 20′ to stop protein synthesis, then stimulated with A769662 to activate AMPK. Lysates harvested at indicated time points show increased rate of TXNIP protein degradation with AMPK activation. (E) Domain structure of TXNIP and multiple sequence alignment of human, rat, mouse TXNIP and human ARRDC4 around Ser308. (F) HepG2 cells stably expressing vector control, HA-WT, or HA-S308A TXNIP treated with 25mM 2DG for 10′ show that S308A mutation abolishes the phosphorylation induced protein mobility upshift after AMPK activation. (G) HepG2 cells stably expressing vector control, HA-WT or HA-S308A TXNIP were treated with 1mM A769662 for 0′, 30′ and 60′. HA-S308A TXNIP protein is degraded at a slower rate than both HA-WT and endogenous TXNIP. *: HA-tagged protein, **: endogenous protein.
We investigated the possibility that AMPK might directly phosphorylate TXNIP. Incubation of bacterially-expressed GST-TXNIP with AMPK and ATP resulted in an increase in phosphorylation at Ser308 as assessed by mass spectrometry (Fig. S2B). The sequence surrounding this site (Fig. 2E) is consistent with the general selection of AMPK for sites with Arg at the -3 position and surrounded by hydrophilic residues (Ser and Thr), though the site is not optimal as it has a hydrophobic residue at -6 rather than -5 (Gwinn et al., 2008; Scott et al., 2002). Importantly, mutation of Ser308 to Ala dramatically impaired the ability of AMPK to phosphorylate TXNIP in vitro, as judged by incorporation of 32P-phosphate from γ-32P-ATP. (Fig. S2C). We synthesized a peptide based on the sequence surrounding Ser308 of TXNIP and found that this peptide had a Vmax/Km ratio that was approximately 60% that of the optimized AMPK peptide substrate SAMS (Fig. S2D). Remarkably, despite the presence of 9 Ser/Thr residues in this synthetic peptide, mutation of the Ser308 residue to Ala almost completely eliminated phosphorylation, indicating that this position is strongly selected by AMPK. Furthermore, we found that an HA-tagged TXNIP S308A mutant does not undergo the shift in SDS-PAGE migration in response to 2DG treatment in HepG2 cells while both the HA-TXNIP WT and the endogenous TXNIP protein do exhibit the migration shift (Fig. 2F). Importantly, the HA-S308A mutant appears more stable compared to HA-WT and endogenous TXNIP in a time course of treatment with A769662 (Fig. 2G). These results indicate that phosphorylation at S308 is critical for AMPK-dependent degradation of TXNIP. In addition, phosphorylated TXNIP seems to be degraded through HECT domain containing E3 ligases interacting with the PPxY motifs. When these two PPxY motifs are mutated (P332A, Y334A, P376A and Y378A altogether), the AMPK facilitated degradation is impaired (Fig. S2E). It is likely that phosphorylation on S308 induces a conformational change that exposes the PPxY motifs for binding to E3 ligases.
Localization of TXNIP at clathrin coated pits
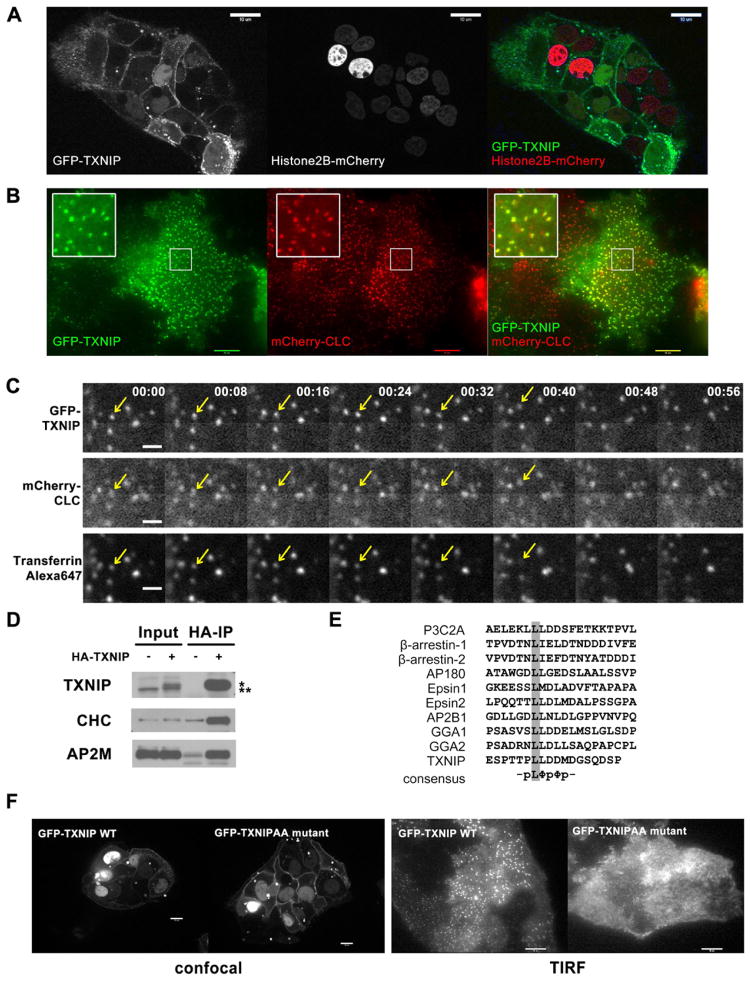
To investigate how TXNIP controls glucose uptake, we first examined the cellular location of GFP-TXNIP in HepG2 cells. Consistent with previous studies (Nishinaka et al., 2004), we found a fraction of TXNIP in the nucleus, but we also observed a significant fraction of GFP-TXNIP at the plasma membrane (Fig. 3A). Moreover, the localization on the plasma membrane was characterized by dynamic speckles (Video S1). With TIRF imaging, these dynamic speckles periodically disappear from the cell surface, suggesting that they may be sites of endocytosis (Video S2). Consistent with this idea, we found that the TXNIP speckles co-localize with clathrin coated pits (CCP) on the plasma membrane as visualized by TIRF (Fig. 3B). In addition, using Alexa647-labeled transferrin as a marker for clathrin-dependent endocytosis, we observed a subset of CCPs that contained both transferrin and TXNIP and observed that TXNIP, transferrin and clathrin simultaneously disappeared from the plasma membrane in time-lapse studies (Fig. 3C, Video S3).
Fig. 3. Dual localization of TXNIP.
(A) Confocal live-cell images of HepG2 cells stably expressing GFP-TXNIP and Histone2B-mCherry, show plasma membrane localization of TXNIP in addition to its nuclear localization as reported before. (B) TIRF live-cell images of HepG2 cells stably expressing GFP-TXNIP and mCherry-clathrin light chain (CLC) show some TXNIP protein localization in clathrin-coated pits (CCP). (C) HepG2 cells stably expressing GFP-TXNIP and mCherry-CLC were labeled with Alexa647-transferrin at 4°C. After rinsing off excess transferrin, time-lapse images were taken at room temperature to capture endocytosis events using a confocal microscope. For every time point, there was a 2–3 seconds delay between each fluorophore: GFP was followed by mCherry and then Alexa647. The arrow points to an endocytosed CCP that contained both GFP-TXNIP and Alexa647-transferrin. This sequence is also shown in Video S3. The scale bar is 1 μM. (D) Western blot of HA IP of lysates from mouse liver expressing adenoviral HA-TXNIP probed with clathrin heavy chain (CHC) and adaptor AP2 μ subunit (AP2M) antibodies. *: HA-tagged protein, **: endogenous protein (E) Multiple sequence alignment of di-leucine motif in various clathrin-interacting proteins. (F) Confocal and TIRF live-cell images of HepG2 cells stably expressing GFP-WT or GFP-L351AL352A TXNIP show that the LL to AA mutation abolished TXNIP localization to the CCP, but not to the plasma membrane.
To provide further evidence that TXNIP binds to clathrin complexes in vivo, we used an adenovirus delivery system to express HA-TXNIP in the mouse liver, harvested the liver, and immunoprecipitated TXNIP using an HA antibody. Both clathrin heavy chain and the clathrin adaptor, AP2, co-precipitated with TXNIP (Fig. 3D). Examining the TXNIP sequence carefully, we identified a di-leucine motif that likely serves as either a clathrin or an AP2 binding site (Fig. 3E). Mutating these two leucine residues (L351 and L352) to alanines abolished CCP localization of TXNIP, but did not prevent the plasma membrane localization (Fig. 3F). Using mCherry-Rab5A as a marker for endosomes, we have not observed convincing co-localization of TXNIP and Rab5, suggesting that TXNIP falls off the coated vesicles after internalization rather than trafficking to endosomes (data not shown). From these data, we conclude that TXNIP, like other arrestin family members, is involved in protein trafficking through CCPs.
TXNIP regulation of Glut1
Given the localization of TXNIP on plasma membrane and clathrin coated pits and prior evidence for its role in suppressing glucose uptake, we investigated whether TXNIP is directly involved in regulating glucose transporter activity. There are 14 family members of facilitative glucose transporters in humans, some of which transport metabolites other than glucose (Augustin, 2010). Glut1 drew our attention since it has a wide tissue distribution like TXNIP itself, and is upregulated in many cancer cells.
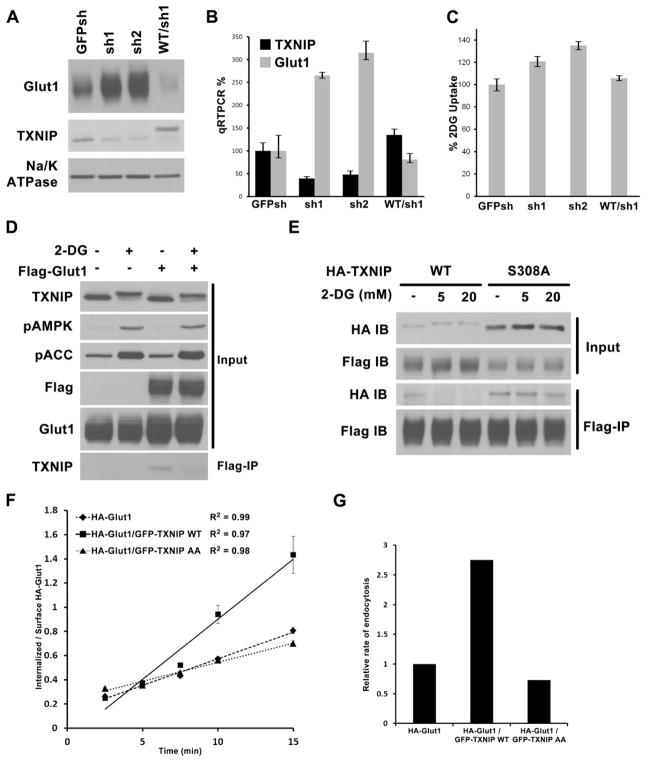
First, we discovered that stable knockdown of TXNIP in HepG2 cells with shRNA resulted in a dramatic increase in total Glut1 protein levels; this could be suppressed by adding back shRNA-resistance HA-TXNIP construct (Fig. 4A). This result could, in part, be explained by an increase in GLUT1 mRNA upon TXNIP knockdown (Fig. 4B), and this is likely to be mediated by the TXNIP fraction that localizes in the nucleus. Consistent with the increased total Glut1 protein and mRNA, 3H-2DG uptake increased upon knocking down TXNIP and this change could be reversed by the HA-TXNIP add-back (Fig. 4C). The enhancement in 3H-2DG uptake was less dramatic than the increase in Glut1 levels suggesting either that other glucose transporters dominate glucose uptake in these cells or that other homeostatic mechanisms (suppression of hexokinase activity or suppression of recycling of Glut1 to the plasma membrane) are induced in response to the large increase in Glut1 levels to bring the cells back to glucose homeostasis.
Fig. 4. TXNIP regulation of Glut1.
Stable HepG2 cells were generated expressing control shRNA construct (GFPsh), or TXNIP knock-down shRNAs (sh1 and sh2), or knock-down cells reconstituted with HA- WT TXNIP construct that is resistant to sh1 (WT/sh1). Cells were examined for (A) Glut1 protein levels by Western blot, (B) Glut1 mRNA levels by qRT-PCR normalized to GFPsh control cells, and (C) relative rate of glucose uptake in media with 2mM glucose using trace amount of 3H-2DG, normalized to GFPsh control cells. The average values of triplicate experiments (±STDV) are reported. (D) HepG2 control cells and cells stably expressing Flag-Glut1 were treated with either water or 25mM 2DG for 10′. Flag–tag IP was carried out with the cell lysates to test interaction with endogenous TXNIP. (E) HepG2 cells stably expressing both Flag-Glut1/HA-TXNIP WT or both Flag-Glut1/HA-TXNIP S308A mutant were treated for 10′ with water, 5mM 2DG, or 20mM 2DG. Flag-tag IP was carried out to check for the effect of S308 phosphoryaltion on Glut1/TXNIP interaction. (F). HA-Glut1 endocytosis assay in TRVb-1 cells transiently transfected with HA-Glut1, HA-Glut1/GFP-TXNIP WT or HA-Glut1/GFP-TXNIP AA constructs. Cells were incubated with antibody against HA-tag on the first exofacial loop of Glut1 at 37°C and fixed after each time point. The cell surface HA-Glut1 is labeled with Cy5 secondary antibody while the endocytosed HA-Glut1 is labeled with Cy3 secondary antibody. The ratio of Cy3-to-Cy5 fluorescence signal was reported as the ratio of internalized-to-surface HA-Glut1 and plotted versus time ± SEM. R2 of the linear regression lines are shown. The data are from a representative experiment. (G). Relative endocytosis rates calculated from the slopes of linear regression from data in (F) normalized to HA-Glut1 alone control.
Second, we found TXNIP directly interacts with Glut1. When co-expressing HA-TXNIP and Myc-Glut1 in 293T cells, immunoprecipitation of TXNIP results in co-precipitation of Glut1 and vice versa (Fig. S3A). We also generated HepG2 cells stably expressing double-Flag-tagged Glut1 at levels comparable to endogenous Glut1 and observed that endogenous TXNIP co-precipitated with Flag-tagged Glut1, indicating that the proteins interact when at endogenous levels (Fig. 4D). Importantly, the phosphorylated form of TXNIP that appears upon treatment with 2DG failed to co-precipitate with Glut1 (notice the absence of the slowly migrating form of TXNIP in the Flag IP in the far right lane of the lower panel of Fig. 4D, despite its presence in the TXNIP western blot of the total cell lysate in the top panel of Fig. 4D). When the same IP was performed with cells stably expressing both Flag-Glut1 and HA-TXNIP, the HA-S308A mutant retained its interaction with Glut1 longer than the HA-WT protein after AMPK stimulation (Fig. 4E), further supporting the idea that AMPK-dependent phosphorylation of TXNIP inhibits its interaction with Glut1.
Since TXNIP binds to Glut1 and to coated pits we examined the possibility that it facilitates Glut1 endocytosis. We adapted the endocytosis assay developed for Glut4 (Blot and McGraw, 2008) to examine Glut1 endocytosis in TRVb-1 cells(McGraw et al., 1987) (Fig. 4F and 4G). Briefly, TRVb-1 cells were transfected with HA-Glut1 construct alone, or together with either GFP-TXNIP WT or the GFP-TXNIP LL to AA construct that impairs localization to coated pits. The cells were incubated with anti-HA tag antibody for various periods of time at 37°C and then fixed. The cell surface HA-Glut1 was labeled with Cy5 secondary antibody first, then cells are permeabilized to label the internalized HA-Glut1 with Cy3 secondary antibody. The ratio of Cy3-to-Cy5 signal is calculated as a measure of internalization. For early time points, the linear rate of increase in internalization is taken as the rate constant of endocytosis. In this assay, HA-Glut1 is endocytosed at a much faster rate when GFP-TXNIP WT was present compared to the control or GFP-TXNIP LL to AA mutant. The incomplete suppression of Glut1 endocytosis by the GFP-TXNIP LL to AA mutant on Glut1 endocytosis could be due to the presence of endogenous TXNIP or to other endocytotic pathways for Glut1, such as non-concentrative bulk endocytosis.
To evaluate if the effect of TXNIP on Glut1 endocytosis is reflected in cellular glucose uptake independent of its effects on Glut1 mRNA, we used shRNA to knock down endogenous TXNIP and introduced either shRNA-resistant HA-WT or HA-S308A TXNIP in HepG2 cells. Cells were then treated with phenformin to activate AMPK and 3H-2DG uptake was monitored. The acute treatment with phenformin led to preferential degradation of the HA-WT compared to the HA-S308A mutant of TXNIP, and had no significant effect on total Glut1 protein over the one hour time period (Fig. S3B). However, it caused a smaller increase in glucose uptake in the HA-S308A cells than the HA-WT cells (Fig. S3C). These results support our model in which the plasma membrane located TXNIP binds to and suppresses Glut1 function by facilitating its endocytosis; TXNIP degradation resulting from AMPK-dependent phosphorylation relieves this inhibition.
Altogether, both populations of TXNIP affect Glut1: the plasma membrane localized TXNIP likely mediates the acute suppression of Glut1 function partly through endocytosis, while the nuclear localized TXNIP likely mediates the overall suppression of Glut1 mRNA and protein levels to provide a long-term suppression of glucose uptake.
Discussion
In this study, we characterize an auto-inhibitory cellular response loop to extracellular glucose levels. Our findings suggest glucose induces TXNIP expression to down-regulate Glut1 function in order to suppress further glucose uptake (Fig. S4). TXNIP accomplishes this by affecting Glut1 on multiple levels, ensuring a response both in speed and amplitude. Energy stress additionally modulates this feed-back loop through AMPK-induced degradation of TXNIP, resulting in upregulation of glucose transport through Glut1.
The results presented here, along with previous studies (Cha-Molstad et al., 2009; Stoltzman et al., 2008) indicate that glucose homeostasis is mediated by three major components that mediate feedback control: Glut1, TXNIP and chREBP/Mlx (or MondoA/Mlx). Metabolites of glucose that build up due to high rates of glucose influx activates the transcription factors ChREBP/Mlx (or MondoA/Mlx). These factors then drive the expression of TXNIP which in turn, induces internalization of Glut1 to acutely suppress glucose uptake. In addition, TXNIP suppresses Glut1 mRNA levels by an unknown mechanism to cause longer term decreases in Glut1 protein and glucose uptake. Under conditions of energy stress AMPK is activated and phosphorylates TXNIP to acutely block TXNIP-dependent Glut1 endocytosis. This phosphorylation also leads to degradation of TXNIP to provide a prolonged enhancement of glucose uptake, thereby allowing cells to utilize glucose as a fuel for restoring energy homeostasis.
While our data support a model wherein TXNIP serves as an adaptor that recruits Glut1 into clathrin coated pits, facilitating its endocytosis, they do not exclude the possibility that TXNIP binding to Glut1 directly inhibits its ability to transport glucose at the plasma membrane. Previous studies have indicated that activation of AMPK increases the Vmax of Glut1-dependent glucose uptake (Abbud et al., 2000; Barnes et al., 2002; Fryer et al., 2002) and this could potentially be explained if AMPK-dependent phosphorylation of TXNIP released TXNIP from Glut1, thereby acutely increasing Glut1 intrinsic activity. It is also possible that TXNIP contributes to ubiquitination and lysosomal-dependent degradation of Glut1. Glut1 is continuously endocytosed under basal conditions, employing both clathrin-mediated and clathrin-independent pathways (Eyster et al., 2009; Reed et al., 2005). AMPK regulation of cellular glucose uptake, both through Glut4 and Glut1, has long been reported (Hardie et al., 2012). However, the only previously known direct substrate of AMPK that has an immediate effect on glucose uptake is AS160/TBC1D4, a Rab GTPase-activating protein (GAP) that regulates exocytosis of intracellular Glut4-storage vesicles(Zeigerer et al., 2004). Our data provide a missing link between AMPK and Glut1 function.
In S. cerevisiae, ART (arrestin-related trafficking adaptor) family proteins regulate ubiquitin-mediated endocytosis of many nutrient transporters through Rsp5, a yeast HECT E3 ligase (Becuwe et al., 2012; Macgurn et al., 2012; MacGurn et al., 2011). Recently, Becuwe et al. (2012) reported that when yeast cells are grown in lactate media, Snf1, the yeast homologue of AMPK, phosphorylates Art4/Rod1. This phosphorylation allows binding of 14-3-3 to Art4 and prevents Art4 from facilitating the endocytosis of the lactate transporter Jen1. To our knowledge, TXNIP is the first mammalian arrestin protein shown to directly regulate a nutrient transporter. It appears that AMPK regulation of nutrient transporters through arrestin proteins is conserved through evolution.
Finally, many cancer cells express high levels of Glut1 to maintain their high levels of glycolysis and anabolic metabolism (Vander Heiden et al., 2009). Interestingly, TXNIP has been reported to have a negative role on tumor development: its expression is reported to be reduced in many tumors, while overexpression of TXNIP inhibits tumor growth and metastasis (Han et al., 2003; Ikarashi et al., 2002; Yan et al., 2011; Zhou et al., 2011). In one mouse model, animals lacking TXNIP had markedly increased incidence of hepatocellular carcinoma (Sheth et al., 2006). The possibility that TXNIP suppresses tumor formation by restricting glucose uptake is intriguing but not adequately explored.
Overall, TXNIP appears to be a key regulator of both acute and long-term glucose influx in response to energy stress. Its importance in cellular homeostasis is just beginning to be recognized.
Experimental procedures
Materials
2-deoxyglucose, cycloheximide and phenformin were purchased from Sigma, A769662 from Tocris Bioscience, AICAR from Toronto Research Chemicals, MG132 from EMD4Biosciences. Antibodies used are: TXNIP (B2) and Myc (9E10) from Santa Cruz Biotechnology; AMPK, ACC, phosphoAMPK(T172) and phosphoACC(S79) from Cell Signaling Technology; Glut1 from Millipore; Flag antibody from Sigma; HA.11 from Convance.
Cell Culture
Primary rat hepatocytes were purchased from Invitrogen. AMPK MEFs were kind gifts from Dr. Benoit Viollet, France. HepG2 cells were purchased from ATCC and maintained according to their instruction.
Cell lysis, IP, western blots
Cells were washed three times with cold PBS and lysed in 1% NP40 buffer (30mM Tris7.5, 120mM NaCl, 1mM vanadate, 20mM NaF, with protease inhibitors and Calyculin A). The supernatant was used for running western blots or IP. For co-IP studies involving Glut1, 0.5% β-OG was used instead of 1% NP40 as the detergent.
qRT-PCR
For quantitative RT-PCR, RNA was isolated with Qiagen RNeasy kit and analyzed with Real time PCR core facility at BIDMC using Taqman assay.
Live cell imaging and TIRF
All images were collected and analyzed at the Nikon Imaging Center at Harvard Medical School. See the supplementary material for details.
HA-Glut1 endocytosis assay
The assay was adapted from Blot and McGraw, 2008 (Blot and McGraw, 2008). TRVB-1 cells transiently transfected with HA-Glut1 and GFP-TXNIP constructs were plated on glass-bottom dishes. Cells were labeled with HA.11 antibody (Convance) for indicated time point at 37°C, then fixed after washing quickly with cold HBSS. Secondary Cy5 antibody incubation was carried out at 37°C for 30min without permeablization. After washing with PBS, cells were fixed again, permeablized with saponin and labeled with Cy3 secondary antibody at 37°C for 30min. The cells were washed and stored in PBS at 4°C until imaging the next day. All imaging and data analysis were carried out with Metamorph. Cells were selected for good morphology and positive for GFP signal when GFP-TXNIP construct is present. Average Cy3-to-Cy5 ratio of around 50 cells was reported as internalized-to-total HA-Glut1 ratio for each time point.
Supplementary Material
Highlights.
TXNIP is an AMPK substrate
TXNIP negatively regulates Glut1 protein and mRNA levels
Acknowledgments
The authors thank Ken Swanson and the Nikon Imaging Center at Harvard Medical School for their help with the imaging studies; Wade Harper for pLentiGFP construct; Benoit Viollet for AMPK MEFs. The authors acknowledge Susanne Breitkopf and Min Yuan for help with mass spectrometry experiments. This research was supported by NIH R01-GM56302 (LCC), NCI P01-CA120964 (LCC), NIH 5P01CA120964 (JMA), NIH DF/HCC Cancer Center Support Grant 5P30CA006516 (JMA), R00-CA133245 (BZ), NIH DK52852 (TEM), NIH R37 DK43051 (BBK), NIH R01 DK098002 (BBK and TEG), and an American Diabetes Association mentor-based fellowship grant (BBK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez CE. On the origins of arrestin and rhodopsin. BMC evolutionary biology. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin R. The protein family of glucose transport facilitators: It’s not only about glucose after all. IUBMB life. 2010;62:315–333. doi: 10.1002/iub.315. [DOI] [PubMed] [Google Scholar]
- Baker AF, Koh MY, Williams RR, James B, Wang H, Tate WR, Gallegos A, Von Hoff DD, Han H, Powis G. Identification of thioredoxin-interacting protein 1 as a hypoxia-inducible factor 1alpha-induced gene in pancreatic cancer. Pancreas. 2008;36:178–186. doi: 10.1097/MPA.0b013e31815929fe. [DOI] [PubMed] [Google Scholar]
- Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Leon S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. The Journal of cell biology. 2012;196:247–259. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiet L, Furman C, Cuaz-Perolin C, Paumelle R, Raymondjean M, Simmet T, Rouis M. Thioredoxin-1 and its natural inhibitor, vitamin D3 up-regulated protein 1, are differentially regulated by PPARalpha in human macrophages. Journal of molecular biology. 2008;384:564–576. doi: 10.1016/j.jmb.2008.09.061. [DOI] [PubMed] [Google Scholar]
- Blot V, McGraw TE. Use of quantitative immunofluorescence microscopy to study intracellular trafficking: studies of the GLUT4 glucose transporter. Methods Mol Biol. 2008;457:347–366. doi: 10.1007/978-1-59745-261-8_26. [DOI] [PubMed] [Google Scholar]
- Bodnar JS, Chatterjee A, Castellani LW, Ross DA, Ohmen J, Cavalcoli J, Wu C, Dains KM, Catanese J, Chu M, et al. Positional cloning of the combined hyperlipidemia gene Hyplip1. Nature genetics. 2002;30:110–116. doi: 10.1038/ng811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Saxena G, Chen J, Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. The Journal of biological chemistry. 2009;284:16898–16905. doi: 10.1074/jbc.M109.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KS, DeLuca HF. Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochimica et biophysica acta. 1994;1219:26–32. doi: 10.1016/0167-4781(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Chutkow WA, Patwari P, Yoshioka J, Lee RT. Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. The Journal of biological chemistry. 2008;283:2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- Draheim KM, Chen HB, Tao Q, Moore N, Roche M, Lyle S. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 2010;29:5032–5047. doi: 10.1038/onc.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyster CA, Higginson JD, Huebner R, Porat-Shliom N, Weigert R, Wu WW, Shen RF, Donaldson JG. Discovery of new cargo proteins that enter cells through clathrin-independent endocytosis. Traffic. 2009;10:590–599. doi: 10.1111/j.1600-0854.2009.00894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Jeon JH, Ju HR, Jung U, Kim KY, Yoo HS, Lee YH, Song KS, Hwang HM, Na YS, et al. VDUP1 upregulated by TGF-beta1 and 1,25-dihydorxyvitamin D3 inhibits tumor cell growth by blocking cell-cycle progression. Oncogene. 2003;22:4035–4046. doi: 10.1038/sj.onc.1206610. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature reviews. Molecular cell biology. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui ST, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui TY, Sheth SS, Diffley JM, Potter DW, Lusis AJ, Attie AD, Davis RA. Mice lacking thioredoxin-interacting protein provide evidence linking cellular redox state to appropriate response to nutritional signals. The Journal of biological chemistry. 2004;279:24387–24393. doi: 10.1074/jbc.M401280200. [DOI] [PubMed] [Google Scholar]
- Ikarashi M, Takahashi Y, Ishii Y, Nagata T, Asai S, Ishikawa K. Vitamin D3 up-regulated protein 1 (VDUP1) expression in gastrointestinal cancer and its relation to stage of disease. Anticancer research. 2002;22:4045–4048. [PubMed] [Google Scholar]
- Jeon JH, Lee KN, Hwang CY, Kwon KS, You KH, Choi I. Tumor suppressor VDUP1 increases p27(kip1) stability by inhibiting JAB1. Cancer research. 2005;65:4485–4489. doi: 10.1158/0008-5472.CAN-04-2271. [DOI] [PubMed] [Google Scholar]
- Junn E, Han SH, Im JY, Yang Y, Cho EW, Um HD, Kim DK, Lee KW, Han PL, Rhee SG, et al. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164:6287–6295. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. The Journal of biological chemistry. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- Kim KY, Shin SM, Kim JK, Paik SG, Yang Y, Choi I. Heat shock factor regulates VDUP1 gene expression. Biochemical and biophysical research communications. 2004;315:369–375. doi: 10.1016/j.bbrc.2004.01.047. [DOI] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell metabolism. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgurn JA, Hsu PC, Emr SD. Ubiquitin and Membrane Protein Turnover: From Cradle to Grave. Annual review of biochemistry. 2012 doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- McGraw TE, Greenfield L, Maxfield FR. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. The Journal of cell biology. 1987;105:207–214. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO reports. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaka Y, Masutani H, Oka S, Matsuo Y, Yamaguchi Y, Nishio K, Ishii Y, Yodoi J. Importin alpha1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein 1. The Journal of biological chemistry. 2004;279:37559–37565. doi: 10.1074/jbc.M405473200. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y, Yodoi J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. The Journal of biological chemistry. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- Oslowski CM, Hara T, O’Sullivan-Murphy B, Kanekura K, Lu S, Hara M, Ishigaki S, Zhu LJ, Hayashi E, Hui ST, et al. Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell metabolism. 2012;16:265–273. doi: 10.1016/j.cmet.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS medicine. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT. The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. The Journal of biological chemistry. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BC, Cefalu C, Bellaire BH, Cardelli JA, Louis T, Salamon J, Bloecher MA, Bunn RC. GLUT1CBP(TIP2/GIPC1) interactions with GLUT1 and myosin VI: evidence supporting an adapter function for GLUT1CBP. Molecular biology of the cell. 2005;16:4183–4201. doi: 10.1091/mbc.E04-11-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze PC, Yoshioka J, Takahashi T, He Z, King GL, Lee RT. Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. The Journal of biological chemistry. 2004;279:30369–30374. doi: 10.1074/jbc.M400549200. [DOI] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. Journal of molecular biology. 2002;317:309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- Sheth SS, Bodnar JS, Ghazalpour A, Thipphavong CK, Tsutsumi S, Tward AD, Demant P, Kodama T, Aburatani H, Lusis AJ. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene. 2006;25:3528–3536. doi: 10.1038/sj.onc.1209394. [DOI] [PubMed] [Google Scholar]
- Sheth SS, Castellani LW, Chari S, Wagg C, Thipphavong CK, Bodnar JS, Tontonoz P, Attie AD, Lopaschuk GD, Lusis AJ. Thioredoxin-interacting protein deficiency disrupts the fasting-feeding metabolic transition. Journal of lipid research. 2005;46:123–134. doi: 10.1194/jlr.M400341-JLR200. [DOI] [PubMed] [Google Scholar]
- Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet B, Athea Y, Mounier R, Guigas B, Zarrinpashneh E, Horman S, Lantier L, Hebrard S, Devin-Leclerc J, Beauloye C, et al. AMPK: Lessons from transgenic and knockout animals. Frontiers in bioscience: a journal and virtual library. 2009;14:19–44. doi: 10.2741/3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rong YP, Malone MH, Davis MC, Zhong F, Distelhorst CW. Thioredoxin-interacting protein (txnip) is a glucocorticoid-regulated primary response gene involved in mediating glucocorticoid-induced apoptosis. Oncogene. 2006;25:1903–1913. doi: 10.1038/sj.onc.1209218. [DOI] [PubMed] [Google Scholar]
- Yan GR, Xu SH, Tan ZL, Liu L, He QY. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics. 2011;11:912–920. doi: 10.1002/pmic.201000539. [DOI] [PubMed] [Google Scholar]
- Yu FX, Goh SR, Dai RP, Luo Y. Adenosine-containing molecules amplify glucose signaling and enhance txnip expression. Mol Endocrinol. 2009;23:932–942. doi: 10.1210/me.2008-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer A, McBrayer MK, McGraw TE. Insulin stimulation of GLUT4 exocytosis, but not its inhibition of endocytosis, is dependent on RabGAP AS160. Molecular biology of the cell. 2004;15:4406–4415. doi: 10.1091/mbc.E04-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wang C, Gao K, Wang D, Mao J, An J, Xu C, Wu D, Yu H, Liu JO, et al. The ubiquitin ligase itch regulates apoptosis by targeting thioredoxin-interacting protein for ubiquitin-dependent degradation. The Journal of biological chemistry. 2010;285:8869–8879. doi: 10.1074/jbc.M109.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu Q, Chng WJ. TXNIP (VDUP-1, TBP-2): a major redox regulator commonly suppressed in cancer by epigenetic mechanisms. The international journal of biochemistry cell biology. 2011;43:1668–1673. doi: 10.1016/j.biocel.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nature immunology. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.