Abstract
Monoclonal antibodies with specificity for HLA class I determinants of HLA were originally characterized using serological assays in which the targets were cells expressing 3-6 HLA class I variants. Because of this complexity, the specificities of the antibodies were defined indirectly by correlation. Here we use a direct binding assay, in which the targets are synthetic beads coated with one of 111 HLA class I variants, representing the full range of HLA-A, -B and -C variation. We studied one monoclonal antibody with monomorphic specificity (W6/32) and four with polymorphic specificity (MA2.1, PA2.1, BB7.2 and BB7.1) and compared the results with those obtained previously. W6/32 reacted with all HLA class I variants. MA2.1 exhibits high specificity for HLA-A*02, -B*57 and -B*58, but also exhibited cross-reactivity with HLA-A*11 and -B*15:16. At low concentration (1μg/ml) PA2.1 and BB7.2 were both specific for HLA-A*02 and -A*69, and at high concentration (50μg/ml) exhibited significant cross-reactions with HLA-A*68, -A*23, and -A*24. BB7.1 exhibits specificity for HLA-B*07 and -B*42, as previously described, but reacts equally well with HLA-B*81, a rare allotype defined some 16 years after the description of BB7.1. The results obtained with cell-based and bead-based assays are consistent and, in combination with amino acid sequence comparison, increase understanding of the polymorphic epitopes recognized by the MA2.1, PA2.1, BB7.2 and BB7.1 antibodies. Comparison of two overlapping but distinctive bead sets from two sources gave similar results, but the overall levels of binding were significantly different. Several weaker reactions were observed with only one of the bead sets.
Keywords: HLA class I, monoclonal antibodies, epitope, polymorphism
Introduction
Since first being reported in 19781, monoclonal antibodies with specificity for HLA class I molecules have been invaluable tools for both basic and clinical research in human immunology. These antibodies can be divided into two groups according to the types of epitope they recognize2. Monomorphic antibodies, such as W6/32, the antibody described by Barnstable et al (1), recognize monomorphic determinants that are common to all HLA class I variants, whereas polymorphic antibodies recognize determinants carried by a subset of such variants. Well-studied examples of polymorphic antibodies are PA2.1, BB7.1, BB7.2 and MA2.1. Originally, PA2.1 and BB7.2 were seen to be specific for HLA-A22–4, but with more extensive characterization they were also shown to recognize and define HLA-A*69, a variant that is a recombinant of HLA-A*02 and HLA-A*685. In a similar fashion, BB7.1 was originally seen to be specific for HLA-B*072 but was subsequently shown to recognize HLA-B*426, a recombinant of HLA-B*07 and HLA-B*087 that is characteristic of African populations8. MA2.1, which was originally described as recognizing HLA-A2 and HLA-B17 antigens9, has been shown to react with both the B*57 and B*58 components of the HLA-B1710, but no additional reactivities have been reported.
In large part, the HLA class I specificity of monoclonal antibodies has been determined using panels of cells each of which minimally expresses one HLA-A, one HLA-B and one HLA-C allotype and more commonly express two allotypes for HLA-A, -B and -C. This complexity means that binding reactions cannot be directly attributed to particular HLA class I variants but must be inferred through various types of correlation. As a consequence, there are limitations in the extent to which data can be interpreted and thus in the resolution and accuracy of the data. An initial approach to address these limitations was the use of mutant cell lines that lacked endogenous HLA class I expression and could be transfected to express a single HLA class I allotype of choice11. A more recent approach has been to replace cells as the target antigen with synthetic beads each of which is coated with a single HLA class I allotype12,13. Elimination of cells from the assay facilitated the commercial development of panels of >90 different beads that provide a representation of the range of HLA-A, B and C diversity. In using such beads to determine the HLA class I specificities of Fc-fusion proteins made from killer-cell immunoglobulin-like receptors (KIR), we have achieved results of higher resolution that are more reproducible and insightful than was possible with cell-based assays14,15. Here we have reexamined the HLA class I specificities of the MA2.1, PA2.1, BB7.2 and BB7.1 monoclonal antibodies using two panels of beads coated with HLA class I molecules.
Materials and Methods
Binding of MA2.1, PA2.1, BB7.2 and BB7.1 antibodies to beads, each coated with a representative range of HLA-A, HLA-B and HLA-C allotypes was assessed in a multiplex assay on the Luminex platform (Austin, TX). The bead panels tested were (a) LabScreen single-antigen beads (One Lambda, Canoga Park, CA) and (b) LifeCodes single-antigen beads (Gen-Probe, San Diego, CA). Antibodies (1μg/ml and 50μg/ml) were incubated with each set of beads for 60 mins at 4°C, washed four times and then labeled with anti-mouse Fc antibody conjugated with phycoerythrin and incubated for a further 60 mins at 4°C. The fluorescent intensity and identification of individual labels of the beads were visualized on a Luminex 100 reader. In each experiment data was collected from a minimum of 100 antigen-coated beads for each combination of bead and monoclonal antibody. The results presented are the mean fluorescence relative to the fluorescence intensity obtained with the W6/32 antibody obtained from three replicate experiments performed for each monoclonal antibody.
Results
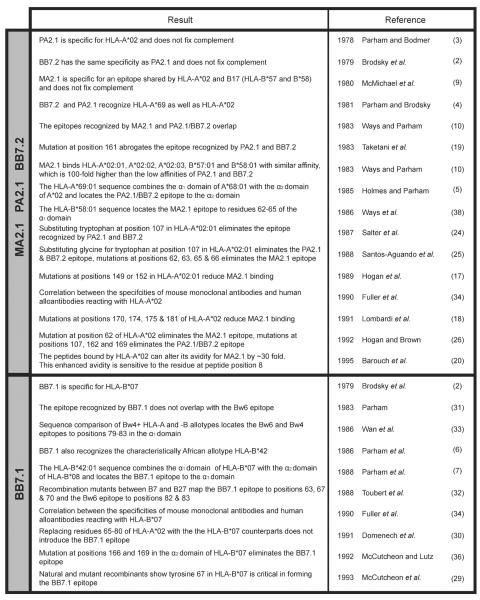
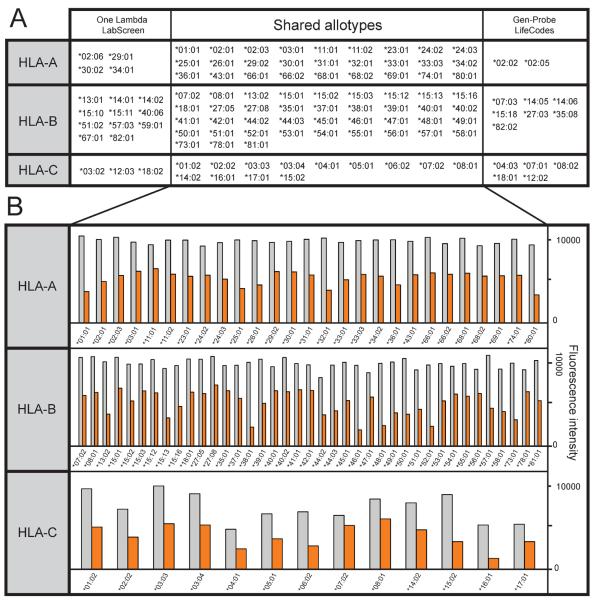
Figure 1 provides a distillation of the results obtained from previous work using cell-based assays to study the HLA class I specificities of the MA2.1, PA2.1, BB7.2 and BB7.1 mouse monoclonal antibodies and the epitopes of HLA class I molecules that these antibodies recognize. In this study, these four antibodies were tested for binding to beads coated with single HLA-A, -B or -C allotypes. Two sets of commercially available beads (from One Lambda and Gen-Probe) were tested and compared. Together the two bead sets allowed us to test a total of 111 HLA class I allotypes (33 HLA-A, 66 HLA-B and 21 HLA-C allotypes). The HLA class I allotypes represented in each bead set are shown in Figure 2A.
Figure 1.
Summary of studies investigating the monomorphic antibodies MA2.1, PA2.1, BB7.2 (top panel) and BB7.1 (bottom panel). Listed is the main finding from each study, the study's author and year of publication. The specific reference as listed in the current publication is noted to the right.
Figure 2.
(A) HLA class I allotypes represented by the One Lambda Labscreen and Gen-Probe LifeCodes beadsets. (B) Binding of the monomorphic HLA class I antibody W6/32 to beads from One Lambda LabScreen (grey bars) and Gen-Probe LifeCodes (orange bars). The allotypes shown are those common to both beadsets.
The assay used to measure the binding of the monoclonal antibodies is essentially the same as the one that we designed and have used for measuring the binding of KIR-Fc fusion products to beads coated with HLA class I14. In this assay the binding of the antibodies recognizing polymorphic epitopes was normalized to that of the W6/32 monoclonal antibody that recognizes an epitope common to all HLA class I epitopes1,16. This normalization corrects for differences in the absolute amount of HLA class I on the various beads. As measured by the binding of W6/32, the amount of HLA class I on the beads varied with allotypes, but a more general property was that the One Lambda Labscreen beads consistently bound more W6/32 than the Gen-Probe LifeCodes beads (Figure 2B). Gen-Probe beads showed a mean reduction in W6/32 binding of 54% (range 35–69%), 52% (range 20–71%) and 50% (range 23–75%) for HLA-A, -B and -C allotypes respectively as compared to One Lambda beads.
Specificity of the MA2.1 monoclonal antibody
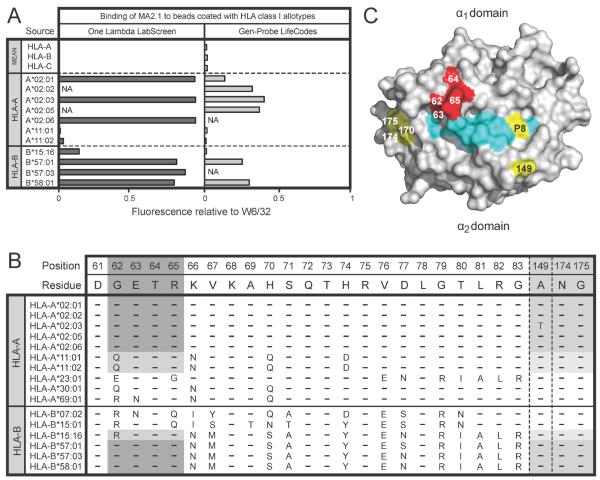
The MA2.1 antibody bound strongly to the six HLA-A*02 subtypes tested (A*02:01, :02, :03, :05 and :06) and also to HLA-B*57:01, B*57:03 and B*58:01 (corresponding to the serological HLA-B17 antigen) (Figure 3A). These results are consistent with the previously defined specificity of MA2.1 for the HLA-A2 and -B17 antigens9. In addition, we observed a weak reactivity of MA2.1 with HLA-B*15:16 and an even weaker one with HLA-A*11. These weak cross-reactions were detected only with the One Lambda beads.
Figure 3.
(A) Binding of MA2.1 (1μg/ml) to beads coated with HLA class I allotypes from the One Lambda LabScreen (left panel) and Gen-Probe LifeCodes (right panel) beadsets. (B) Alignment of HLA class I allotypes showing selected residues in the α1 and α2 domains. Residues from allotypes that form the epitope recognized by MA2.1 are shaded in grey. (C) Space-filling model of the binding surface of HLA-A*02 (grey) with associated peptide (cyan). Residues highlighted in yellow fall within the footprint recognized by MA2.1. Residues 62–65 are critical for formation of the epitope recognized by MA2.1 and are highlighted in red.
Results from several groups of investigators are consistent with the GETR motif at residues 62–65 in the helix of the α1 domains of HLA-A*02, B*57 and B*58 being critical for forming the epitope recognized by MA2.1 (Figure 1). Within this motif, glycine 62 is the only residue that is not found in any of the other 89 HLA-A, -B and -C allotypes tested. HLA-B*15:16 and HLA-A*11 differ only at position 62, having the RETR and QETR motifs respectively, which correlates with them having some avidity for MA2.1. The replacement of glycine 62 with arginine in B*15:16 is seen to be more favorable for binding MA2.1 than the glutamine residue at this position in A*11 (Figure 3B). Other HLA class I allotypes having the QETR motif (e.g HLA-A*03, A*30 and A*36) are not recognized by MA2.1, suggesting that further residues, away from this motif in the α1 domain also influence recognition of HLA class I by MA2.1.
In addition to the GETR motif at residues 62–65 in the α1 domain, mutation at several positions in the α2 domain (149, 152, 170, 174, 175 and 181) has been shown to influence MA2.1 binding17–19, as has the residue at position 8 in the peptide bound to HLA-A220 (Figure 3C). These influences could involve direct contact with the antibody or be a consequence of indirect effects that alter the conformation of residues 62–65.
Specificity of the PA2.1 and BB7.2 monoclonal antibodies
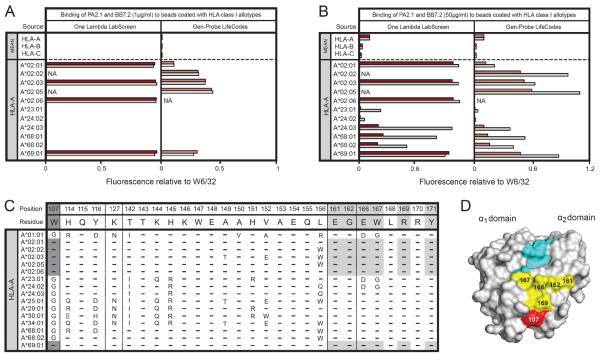
When used at a concentration of 1μg/ml the PA2.1 and BB7.2 antibodies gave similarly strong reactions with five subtypes of HLA-A*02 and with HLA-A*69:01 (Figure 4A). For each of the these reactions the Gen-Probe beads gave weaker reactions than the One Lambda beads, the difference being greatest for HLA-A*02:01, as was also the case for MA2.1 (Figure 3A). These results are consistent with those obtained with the W6/32 antibody (Figure 2B).
Figure 4.
(A) Binding of PA2.1 (1μg/ml) and BB7.2 (1μg/ml) to beads coated with HLA class I allotypes from the One Lambda LabScreen (left panel) and Gen-Probe LifeCodes (right panel) beadsets. (B) Binding of PA2.1 (50μg/ml) and BB7.2 (50μg/ml) to beads coated with HLA class I allotypes from the One Lambda LabScreen (left panel) and Gen-Probe LifeCodes (right panel) beadsets. (C) Alignment of HLA class I allotypes showing selected residues in the α2 domain. Residues from allotypes that form the epitope recognized by PA2.1 and BB7.2 are shaded in grey. (D) Space-filling model of HLA-A*02 (grey) with associated peptide (cyan). Residues highlighted in yellow fall within the footprint recognized by PA2.1 and BB7.2. Tryptophan at position 107 is considered critical for formation of the epitope recognized by PA2.1 and BB7.2 and is highlighted in red.
When a much higher concentration of antibody was used (50μg/ml), cross-reactions of the PA2.1 and BB7.2 antibodies were observed with HLA-A*23:01, A*24:02, A*24:03, A*68:01, A*68:02 and A*69:01 (Figure 4B). These cross-reactions correspond well with the cross-reactivity first described in the 1960s21 between the serological A2, A28 (A*68 and A*69) and A9 (A*23 and A*24) antigens and more recently described for cell-binding assays with the I-145 monoclonal antibody22. These cross-reactions were stronger for BB7.2 than PA2.1, a feature observed for both sets of HLA class I coated beads. The cross-reactivity was much stronger for A*24:03 than either A*24:02 or A*23:01. This must be due to the two substitutions that distinguish A*24:03 from both A*24:02 and A*23:0123 and which are at positions 166 and 167 in the α2 domain (Figure 3C). Consistent with the relative binding to PA2.1 and BB7.2, HLA-A*24:03 shares the glutamate 166, tryptophan 167 motif with HLA-A*02, whereas HLA-A*24:02 and A*23:01 share the aspartate 166, glycine 167 motif with HLA-A*01:01, which binds neither PA2.1 nor BB7.2.
The combination of sequence comparisons2,4,5,10 and site-directed mutagenesis24–26 has shown that tryptophan 107 in the α2 domain is a critical factor in forming the epitope recognized by PA2.1 and BB7.2 (Figure 4B, Figure 1). In addition, mutations at positions 161, 163, 166, 167 and 169 can lead to loss of binding by PA2.1 and BB7.2, showing that these residues also contribute to forming the epitope19,26 (Figure 4C).
Specificity of the BB7.1 monoclonal antibody
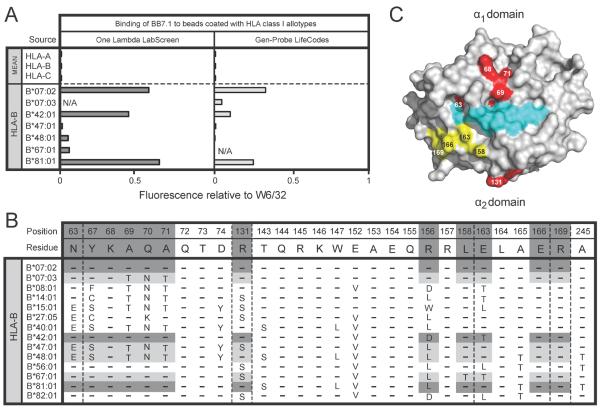
As previously reported, we find that BB7.1 reacts with HLA-B*07:02 and HLA-B*42:01 (Figure 5A). In addition we found that HLA-B*81:01, an African allotype27,28, also reacts strongly with BB7.1, which fits with both HLA-B*81:01, B*42:01 and B*0701 having α1 domains with identical sequence (Figure 5B) and residues 63–70 of the α1 domain being critical for the BB7.1 epitope2,6,29–34 (Figure 1). Comparably strong reactions for HLA-B*07:02, B*42:01 and B*81:01 were observed with the One Lambda beads, but the binding of BB7.1 to the Gen-Probe B*42:01 bead was relatively weak. Even weaker was the binding to the B*07:03 bead, which may reflect an inherent difference between B*07:02 and B*07:03 in their avidities for BB7.1. That B*07:03 differs from B*07:02 by non-conservative substitutions at positions 69, 70 and 71 is also evidence that favors this interpretation35.
Figure 5.
(A) Binding of BB7.1 (1μg/ml) to beads coated with HLA class I allotypes from the One Lambda LabScreen (left panel) and Gen-Probe LifeCodes (right panel) beadsets. (B) Alignment of HLA class I allotypes showing selected residues in the α1 and α2 domains. Residues from allotypes that form the epitope recognized by BB7.1 are shaded in grey. (C) Space-filling model of the binding surface of HLA-B*07 (grey) with associated peptide (cyan). Residues highlighted in yellow fall within the footprint recognized by BB7.1. Residues 63–71 in the a1 domain and position 131 in the α2 domain are critical for formation of the epitope recognized by BB7.1 and are highlighted in red.
Both HLA-B*56:01 and B*82:01 have α1 domains with identical sequence to that of HLA-B*07:02, B*42:01 and B*81:01 but these allotypes are not bound by BB7.1 suggesting that residues in the α2 domain abrogate their recognition. HLA-B*07:02, B*42:01 and B*81:01 encode arginine at position 131 whereas B*56:01 and B*82:01 encode serine at this position. Similarly, both B*56:01 and B*82:01 encode leucine at position 163 whereas HLA-B*07:02 and B*81:01 both encode glutamic acid. Interestingly, HLA-B*42:01 encodes a threonine at position 131 which may explain its weaker recognition by BB7.1 as detected on both the One Lambda and Gen-Probe beads. That BB7.1 interacts with residues in the α2 domain is further supported by the observation that mutation at positions 166 and 169 of HLA-B*07 eliminates the BB7.1 epitope36.
Therefore, whilst the α1 domain residues between positions 63–71 likely form the dominant epitope for recognition by BB7.1, binding is influenced by polymorphism in the α2 domain, suggesting that this antibody binds both the α1 and α2 regions of HLA class I. Given that this footprint would span the peptide binding cleft, peptide variability is likely to influence BB7.1 recognition of HLA class I.
In summary, we find that the patterns of antibody reactivity observed here are entirely consistent with those obtained previously, and extend those results by being able to detect and distinguish reactions and cross-reactions in a quantitative manner. All the observed reactions are with HLA-A and -B variants, with no cross-reactivity for HLA-C further emphasizing the distinctive properties of HLA-C37.
Discussion
Consistent with previous studies we showed that MA2.1 bound to HLA-A*02 subtypes and to HLA-B*57 and HLA-B*58 allotypes9,26. Experimental data and sequence analysis suggest that the α1 residues GETR at positions 62–65 are critical in forming the epitope recognized by MA2.126,38. In this study we showed weak interactions with MA2.1 are also formed when glycine at position 62 is substituted for arginine, as in HLA-B*15:16 and glutamine as in HLA-A*11:02. This finding contrasts with results of a cellular assay in which substitution of glycine for arginine at position 62 abrogated recognition of HLA-A*02 by MA2.126. HLA-B*15:16 has a high degree of sequence homology with HLA-B*57 which appears sufficient to confer reactivity with MA2.1 despite the introduction of a residue apparently less favorable for recognition by MA2.1 at position 62. That glutamine at position 62 results in weak binding to HLA-A*11 and no detectable binding to other HLA-A allotypes with glutamine at this position (e.g HLA-A*03, A*30 and A*36) would suggest that this residue is less favorable for binding than both glycine and arginine. An alternative explanation is that the range of peptides presented by either HLA-B*15:16 or HLA-A*11 collectively form an epitope with a high affinity for MA2.1, sufficient to overcome the reduction in affinity incurred by the presence of either arginine or glutamine at position 62.
Several lines of evidence suggest that peptide variability influences recognition of HLA class I by MA2.1. Although the residues critical for formation of the epitope recognized by MA2.1 are located in the α1 domain, mutational experiments indicate that the binding footprint extends to residues in the α2 domain of the HLA class I molecule17,18. This is supported by the finding that MA2.1 recognition of HLA-A2 is prevented in a competitive binding assay with PA2.110, an antibody whose epitope is formed exclusively by residues in the α2 domain. Together these studies suggest that MA2.1 binds to an area spanning the peptide binding groove on the face of HLA class I molecule. Direct evidence of the effect of peptide variability is provided from the observation that MA2.1 recognizes HLA-A2 complexed with the HIV-1 p17 epitope (SLYNTVATL) at least 30 times more strongly than all other complexes studied and this enhanced reactivity was sensitive to point mutation of threonine to alanine at position 8 in the peptide20. Therefore there exist two possible explanations for the ability of the bound peptide to influence MA2.1 reactivity. Either MA2.1 is sensitive to peptide induced conformational changes of the helices, or it directly contacts certain peptides in the groove of HLA class I. The identity of the bound peptides presented by HLA class I on beads manufactured by One Lambda and Gen-Probe is unknown but presumed to be highly heterogeneous.
The weaker interactions between MA2.1 and HLA-B*15:16 and A*11:02 were evident only in the One Lambda LabScreen beadset. Similarly, weak interactions between BB7.1 and HLA-B*48:01 and B*67:01 were identified on the One Lambda beads but these interactions were not observed in the Gen-Probe beadset. Our data and that of others suggests that the residues critical for formation of the epitope recognized by BB7.1 are found in the α1 and α2 domains suggesting that, like MA2.1, the identity of the peptide might play a role in the recognition of HLA class I by this antibody. Therefore, the weak interactions observed with BB7.1 might reflect the different peptide pools found in the One Lambda and Gen-Probe beadsets with those from One Lambda collectively having higher binding affinity than those found in the Gen-Probe beadset. Such weak interactions were not evident for either PA2.1 or BB7.2, antibodies thought to recognize an epitope formed exclusively by residues found in the α2 domain and as such they are unlikely to be subject to the same changes in peptide pool that we have hypothesized to influence recognition of MA2.1 and BB7.1.
An alternative explanation for the lower affinity of MA2.1 and BB7.1 for HLA class I allotypes presented on the Gen-Probe beadset is the lower antigen density present on these beads. We have shown that the antigen density present on the Gen-Probe beadset is approximately 50% of that present on the One Lambda beadset as evidenced by binding of the monomorphic HLA class I antibody W6/32. As a result the weak interactions noted on the One Lambda beadset might not reach the lower level of detection of the assay with the Gen-Probe beadset.
Acknowledgements
The authors thank Professor Rene Duquesnoy, University of Pittsburgh Medical Center for suggesting this study. The work was supported by National Institutes of Health Grant A122039 (to P.P.). H.G.H. was supported by the March of Dimes Prematurity Center at Stanford University School of Medicine, Clinical and Translational Science Awards Grant UL1 RR025744 and a Stanford University School of Medicine Dean's Postdoctoral Fellowship.
Footnotes
Conflict of Interest Statement The authors have no conflicting interests.
References
- 1.Barnstable CJ, Bodmer WF, Brown G, et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky FM, Parham P, Barnstable CJ, et al. Monoclonal antibodies for analysis of the HLA system. Immunol Rev. 1979;47:3–61. doi: 10.1111/j.1600-065x.1979.tb00288.x. [DOI] [PubMed] [Google Scholar]
- 3.Parham P, Bodmer WF. Monoclonal antibody to a human histocompatibility alloantigen, HLA-A2. Nature. 1978;276:397–399. doi: 10.1038/276397a0. [DOI] [PubMed] [Google Scholar]
- 4.Parham P, Brodsky FM. Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum Immunol. 1981;3:277–299. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 5.Holmes N, Parham P. Exon shuffling in vivo can generate novel HLA class I molecules. EMBO J. 1985;4:2849–2854. doi: 10.1002/j.1460-2075.1985.tb04013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parham P, Antonelli P, Herzenberg LA, et al. Further studies on the epitopes of HLA-B7 defined by murine monoclonal antibodies. Hum Immunol. 1986;15:44–67. doi: 10.1016/0198-8859(86)90316-2. [DOI] [PubMed] [Google Scholar]
- 7.Parham P, Lomen CE, Lawlor DA, et al. Nature of polymorphism in HLA-A, -B, and -C molecules. Proc Natl Acad Sci USA. 1988;85:4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao K, Hollenbach J, Shi X, et al. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001;62:1009–1030. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 9.McMichael AJ, Parham P, Rust N, et al. A monoclonal antibody that recognizes an antigenic determinant shared by HLA A2 and B17. Hum Immunol. 1980;1:121–129. doi: 10.1016/0198-8859(80)90099-3. [DOI] [PubMed] [Google Scholar]
- 10.Ways JP, Parham P. The binding of monoclonal antibodies to cell-surface molecules. A quantitative analysis with immunoglobulin G against two alloantigenic determinants of the human transplantation antigen HLA-A2. Biochem J. 1983;216:423–432. doi: 10.1042/bj2160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu Y, Koller B, Geraghty D, et al. Transfer of cloned human class I major histocompatibility complex genes into HLA mutant human lymphoblastoid cells. Mol Cell Biol. 1986;6:1074–1087. doi: 10.1128/mcb.6.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei R, Lee JH, Shih NJ, et al. Single human leukocyte antigen flow cytometry beads for accurate identification of human leukocyte antigen antibody specificities. Transplantation. 2003;75:43–49. doi: 10.1097/00007890-200301150-00008. [DOI] [PubMed] [Google Scholar]
- 13.El-Awar N, Lee J, Terasaki PI. HLA antibody identification with single antigen beads compared to conventional methods. Hum Immunol. 2005;66:989–997. doi: 10.1016/j.humimm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Moesta AK, Norman PJ, Yawata M, et al. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 15.Hilton HG, Vago L, Older Aguilar AM, et al. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol. 189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 17.Hogan KT, Clayberger C, Bernhard EJ, et al. A panel of unique HLA-A2 mutant molecules define epitopes recognized by HLA-A2-specific antibodies and cytotoxic T lymphocytes. J Immunol. 1989;142:2097–2104. [PubMed] [Google Scholar]
- 18.Lombardi G, Matsui M, Moots R, et al. Limited regions of the alpha 2-domain alpha-helix control anti-A2 allorecognition: an analysis using a panel of A2 mutants. Immunogenetics. 1991;34:149–156. doi: 10.1007/BF00205817. [DOI] [PubMed] [Google Scholar]
- 19.Taketani S, Krangel MS, Pious D, et al. Structural analysis of HLA-A2 antigen from immunoselected mutant 8.6.1: further definition of an HLA-A2-specific serological determinant. J Immunol. 1983;131:2935–2938. [PubMed] [Google Scholar]
- 20.Barouch D, Davenport M, McMichael A, et al. A mAb against HLA-A2 can be influenced both positively and negatively by the associated peptide. Int Immunol. 1995;7:1599–1605. doi: 10.1093/intimm/7.10.1599. [DOI] [PubMed] [Google Scholar]
- 21.Svejgaard A, Kissmeyer-Nielsen F. Cross-reactive human HL-A isoantibodies. Nature. 1968;219:868–869. doi: 10.1038/219868a0. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno S, Yako F, Ohta H, et al. A new murine lymphocytotoxic monoclonal antibody recognizing HLA-A2, -A28 and -A9. Tissue Antigens. 1996;48:224–227. doi: 10.1111/j.1399-0039.1996.tb02635.x. [DOI] [PubMed] [Google Scholar]
- 23.Little AM, Madrigal JA, Parham P. Molecular definition of an elusive third HLA-A9 molecule: HLA-A9.3. Immunogenetics. 1992;35:41–45. doi: 10.1007/BF00216625. [DOI] [PubMed] [Google Scholar]
- 24.Salter RD, Clayberger C, Lomen CE, et al. In vitro mutagenesis at a single residue introduces B and T cell epitopes into a class I HLA molecule. J Exp Med. 1987;166:283–288. doi: 10.1084/jem.166.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos-Aguado J, Barbosa JA, Biro PA, et al. Molecular characterization of serologic recognition sites in the human HLA-A2 molecule. J Immunol. 1988;141:2811–2818. [PubMed] [Google Scholar]
- 26.Hogan KT, Brown SL. Localization and characterization of serologic epitopes on HLA-A2. Hum Immunol. 1992;33:185–192. doi: 10.1016/0198-8859(92)90070-4. [DOI] [PubMed] [Google Scholar]
- 27.Ellexson ME, Zhang G, Stewart D, et al. Nucleotide sequence analysis of HLA-B*1523 and B*8101. Dominant alpha-helical motifs produce complex serologic recognition patterns for the HLA-B“DT” and HLA-B“NM5” antigens. Hum Immunol. 1995;44:103–110. doi: 10.1016/0198-8859(95)00082-f. [DOI] [PubMed] [Google Scholar]
- 28.Vilches C, Sanz L, de Pablo R, et al. Molecular characterization of the new alleles HLA-B*8101 and B*4407. Tissue Antigens. 1996;47:139–142. doi: 10.1111/j.1399-0039.1996.tb02527.x. [DOI] [PubMed] [Google Scholar]
- 29.McCutcheon JA, Smith KD, Valenzuela A, et al. HLA-B*0702 antibody epitopes are affected indirectly by distant antigen residues. Hum Immunol. 1993;36:69–75. doi: 10.1016/0198-8859(93)90108-d. [DOI] [PubMed] [Google Scholar]
- 30.Domenech N, Santos-Aguado J, Lopez de Castro JA. Antigenicity of HLA-A2 and HLA-B7. Loss and gain of serologic determinants induced by site-specific mutagenesis at residues 62 to 80. Hum Immunol. 1991;30:140–146. doi: 10.1016/0198-8859(91)90083-l. [DOI] [PubMed] [Google Scholar]
- 31.Parham P. Antigenic determinants of the HLA-B7 molecule; Bw6- and B7-specific determinants are spatially separate. Immunogenetics. 1983;18:1–16. doi: 10.1007/BF00401351. [DOI] [PubMed] [Google Scholar]
- 32.Toubert A, Raffoux C, Boretto J, et al. Epitope mapping of HLA-B27 and HLA-B7 antigens by using intradomain recombinants. J Immunol. 1988;141:2503–2509. [PubMed] [Google Scholar]
- 33.Wan AM, Ennis P, Parham P, et al. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. 1986;137:3671–3674. [PubMed] [Google Scholar]
- 34.Fuller AA, Trevithick JE, Rodey GE, et al. Topographic map of the HLA-A2 CREG epitopes using human alloantibody probes. Hum Immunol. 1990;28:284–305. doi: 10.1016/0198-8859(90)90058-w. [DOI] [PubMed] [Google Scholar]
- 35.Bergmans AM, Tijssen H, Lardy N, et al. Complete nucleotide sequence of HLA-B*0703, a B7 variant (BPOT) Hum Immunol. 1993;38:159–162. doi: 10.1016/0198-8859(93)90533-7. [DOI] [PubMed] [Google Scholar]
- 36.McCutcheon JA, Lutz CT. Mutagenesis around residue 176 on HLA-B*0702 characterizes multiple distinct epitopes for anti-HLA antibodies. Hum Immunol. 1992;35:125–131. doi: 10.1016/0198-8859(92)90020-n. [DOI] [PubMed] [Google Scholar]
- 37.Zemmour J, Parham P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J Exp Med. 1992;176:937–950. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ways JP, Rothbard JB, Parham P. Amino acid residues 56 to 69 of HLA-A2 specify an antigenic determinant shared by HLA-A2 and HLA-B17. J Immunol. 1986;137:217–222. [PubMed] [Google Scholar]