Abstract
Staphylococcus lugdunensis is an opportunistic pathogen related to Staphylococcus aureus and Staphylococcus epidermidis. The genome sequence of S. lugdunensis strain N920143 has been compared with other staphylococci, and genes were identified that could promote survival of S. lugdunensis on human skin and pathogenesis of infections. Staphylococcus lugdunensis lacks virulence factors that characterize S. aureus and harbours a smaller number of genes encoding surface proteins. It is the only staphylococcal species other than S. aureus that possesses a locus encoding iron-regulated surface determinant (Isd) proteins involved in iron acquisition from haemoglobin.
Keywords: Staphylococcus lugdunensis, genome sequence, surface proteins, virulence factors
Introduction
Staphylococcus lugdunensis is a coagulase-negative Staphylococcus (CoNS). It is a commensal of the human skin that is found more frequently on the lower part of the body and the extremities, particularly in moist areas such as the perineum, the inguinal fold and under the large toenail (Bieber & Kahlmeter, 2010). Although it is regarded as an integral part of the normal human biota, S. lugdunensis is an opportunistic pathogen, causing serious skin and soft tissue infections, and is particularly associated with serious cases of infective endocarditis (IE), more akin to Staphylococcus aureus. Between 1% and 5% of IE cases are caused by S. lugdunensis. Thus, it is more virulent than expected for CoNS (Frank et al., 2008).
It is conceivable that the incidence of infections caused by S. lugdunensis is under-reported. In the clinical microbiology diagnostic laboratory, it could easily be mistaken for S. aureus because of its colony morphology, haemolytic activity and ability to agglutinate latex particles coated with fibrinogen (Zbinden et al., 1997).
The complete genome sequence of the S. lugdunensis strain HKU09-01 has been published (Tse et al., 2010). However, the annotation is incomplete and an in-depth analysis of potential colonization and virulence factors has not been carried out. This paper describes conclusions that were drawn from analysing the genome sequence of S. lugdunensis N920143 and comparing it with that of HKU09-01.
Materials and methods
Bacterial strain
Staphylococcus lugdunensis N920143 was isolated from a breast abscess in 1992. It was kindly supplied by Dr F. Vandenesch, Université Lyon, Lyon, France.
Genome sequencing
Staphylococcus lugdunensis N920143 genomic DNA was isolated using the PurElute Bacterial Genomic Kit (Edge Biosystems) with an additional incubation step with lysostaphin (25 μg mL−1) at 37 °C for 10 min. Genomic DNA was ethanol precipitated and dissolved in TE buffer for sequencing.
The genome of the S. lugdunensis strain N920143 was sequenced using both reversible terminator sequencing [on Illumina Genome Analysers (GAII)] and pyrosequencing (on 454 instruments; subsidiary of Roche Diagnostics Corporation, Branford, CT). A total of 40.1 Mb of Illumina sequence was produced from a 200-bp standard paired-end library run in one lane of a flow cell with 54-bp reads representing approximately 850-fold coverage. The 454 sequencing produced a 0.23-Mb sequence with an average length of 250 bp. The assembly of the Illumina reads using velvet 0.7.62 gave 142 contigs of >1 kb with a contig N50 of 22 kb.
A combined assembly of the 454 reads using newbler 2.1 and the Illumina consensus sequences from the velvet assembly produced 69 contigs >500 bp with an N50 of 72 kb. The length of the combined assembly was 2 588 004 bp in nine scaffolds. image (Tsai et al., 2010) and icorn (Otto et al., 2010), along with a further 1070 high-quality reads, were used to close gaps and to improve the quality of the sequence to the standard of improved high-quality draft (Chain et al., 2009).
The sequence and annotation of the S. lugdunensis strain N920143 genome has been deposited in the EMBL database with the accession number FR870271. The sequence was finished and annotated as described previously using artemis software (Holden et al., 2009). Comparison of the genome sequences was facilitated using the artemis comparison tool (act) (Carver et al., 2005). Orthologous proteins were identified as reciprocal best matches using fasta (Pearson & Lipman, 1988) with subsequent manual curation.
Results
Comparative genomics
The genome of S. lugdunensis N920143 comprises an approximately 2.6-Mbp chromosome. The sequence consist of six contigs and accordingly six gaps of approximately 1.5 –4 kb, occurring in repetitive DNA stretches like rRNA and the repeat region of a surface-anchored protein. The contigs were aligned according to their order in HKU09-01. Gap closing was not completed because no important features are encoded within the regions in HKU09-01. The genome contains a single prophage named φSL1 and 14 insertion sequences. It does not carry any integrated or replicating plasmids. Our analysis has identified the genomic differences that distinguish S. lugdunensis from S. aureus and other CoNS, and as a corollary of this, has addressed how the genome may influence its biology and ability to cause disease.
Phylogenetic analysis of 16S rRNA gene (Takahashi et al., 1999) and dnaJ (Shah et al., 2007) gene sequences places S. lugdunensis in a clade that includes Staphylococcus epidermidis, S. aureus and Staphylococcus haemolyticus. Comparative genomic analysis revealed that a large proportion of the S. lugdunensis N920143 genome is shared with these related pathogenic staphylococci. Of the 2447 coding sequences (CDSs) in the N920143 genome, 77.8% have reciprocal fasta matches to S. aureus MRSA252, 74.7% to S. epidermidis RP62a and 78.3% to S. haemolyticus. In comparison with the more distantly related staphylococci, 71.4% of all CDSs have matches to Staphylococcus saprophyticus, 71.3% to Staphylococcus carnosus and 54.4% to Macrococcus caseolyticus (Fig. 1).
Fig. 1.
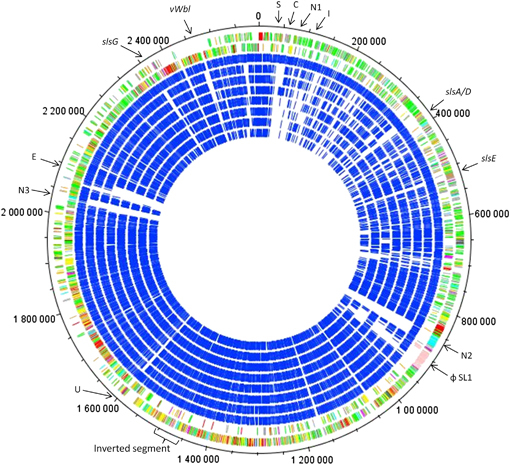
Schematic circular diagram of the Staphylococcus lugdunensis N920143 chromosome. Key for the circular diagram (outer to inner): annotated CDSs coloured according to predicted function are shown on a pair of concentric circles, representing both coding strands; blue circles show S. lugdunensis N920143 reciprocal fasta best matches shared with S. lugdunensis HKU09-01; MRSA252; MSSA476; Staphylococcus epidermidis; Staphylococcus haemolyticus; Staphylococcus saprophyticus; Staphylococcus carnosus; Macrococcus caseolyticus. Colour coding for S. lugdunensis CDS functions: neon green, pathogenicity/adaptation; dark gray, energy metabolism; red, information transfer; dark green, surface associated; sky blue, degradation of large molecules; dark pink, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, phage and IS elements; gray, miscellaneous. Arrows indicate regions of interest: C, CRISPR region; E, ESAT 6 toxin and secretion; I, Isd operon; N1, nonribosomal peptide synthetase 1; N2, nonribosomal peptide synthetase 2; N3, nonribosomal peptide synthetase 3; φSL1, prophage; S, streptolysin S-like toxin; U, gene cluster for sugar uptake and degradation.
Compared with all other staphylococci, there is an inversion of 60 375 bp located between 1 457 500 and 1 518 000 in a conserved region of the genome. Analysis of the boundaries of the inversion in N920143 has failed to identify flanking repeat sequences that could account for the inversion via recombination.
A pairwise comparison between N920143 and HKU09-01 identified that 95.4% of the chromosome is conserved (including the inversion), and that there are 125 unique CDSs in N920143. The major differences between S. lugdunensis N920143 and HKU09-01 are (1) the presence of two identical putative transposons in HKU09-01 encoding a β-lactamase (bla) and β-lactamase regulatory proteins, (2) a putative genomic island in HKU09-01 encoding resistance to cadmium, but lacking identifiable virulence factors, (3) the gene encoding one of the three SLUSH peptides (C) is missing in HKU09-01 and (4) the duplication in HKU09-01 of a 32-kb region comprising the locus encoding the iron-regulated surface-determinant locus (Isd). The duplication occurred by unequal recombination between SLGD_00058 (annotated as KdpA, potassium-transporting ATPase A chain) and SLGD_00116 (annotated as Na+ driven multidrug efflux pump). These ORFs flank the ∼16-kp isd locus and are separated by 32 kb in N920143. Recombination occurred between 19-bp sequences within the ORFs that are identical apart from one mismatch to create a hybrid gene SLGD_00087 in HKU09-01 (annotated asKdpA potassium-transporting ATPase A chain).
Putative virulence and colonization factors
We have identified several loci that might be of relevance to skin survival and virulence (Table 1). All staphylococci carry genes that encode a single iron-regulated ferric siderophore uptake system (sst) (Morrissey et al., 2000), which is duplicated in both S. lugdunensis strains, an accessory gene regulator (Agr) system (Otto, 2001) and systems for neutralizing the negatively charged cell envelope by adding d-alanine to teichoic acid (Dlt) (Peschel et al., 1999) and l-lysine to phosphatidyl glycerol (MrpF) (Peschel et al., 2001). Macrococcus caseolyticus and all sequenced staphylococci except S. haemolyticus and S. carnosus have the capacity to express poly-N-acetyl glutamine encoded by the ica operon (Goetz, 2002). Only the bovine strain of S. aureus RF122 along with S. lugdunensis has a locus encoding a putative streptolysin S-like toxin (Lee et al., 2008). Staphylococcus lugdunensis also carries a locus of four genes encoding lantibiotic resistance proteins similar to GdmG/E/F/H of Staphylococcus gallinarum (Siezen et al., 1996). Interestingly, no lantibiotic biosynthesis proteins are encoded within the locus. Furthermore, S. lugdunensis encodes a polysaccharide (O'Riordan & Lee, 2004) and a polyglutamic acid capsule (Kocianova et al., 2005), three nonribosomal peptide synthesis systems [one of the three synthetases is conserved in S. aureus and S. epidermidis (Wyatt et al., 2010)] and an ESAT-6 toxin secretion system (ess) with homology to ESAT-6 proteins of Mycobacterium tuberculosis (Burts et al., 2005). The ess loci of S. aureus MRSA252 and S. lugdunensis lack the genes encoding the cytoplasmic protein EsaC and the effector EsxB, but only S. lugdunensis N920143 contains a frameshift in the gene encoding the membrane-associated protein EssC. Among the sequenced staphylococci, only S. epidermidis and S. lugdunensis share a 12.5-kb CRISPR region, which is known to limit horizontal gene transfer (Marraffini & Sontheimer, 2008) and might give an explanation for the low number of mobile genetic elements within the S. lugdunensis genome.
Table 1.
Summary of notable Staphylococcus lugdunensis features and distribution of orthologues in other staphylococci
| S. lugdunensis N920143 | S. lugdunensis HKU09-01 | S. aureus MRSA252 | S. epidermidis RP62a | S. haemolyticus | S. saprophyticus | S. carnosus | M. caseolyticus | |
|---|---|---|---|---|---|---|---|---|
| NRPS 1 | + | + | + | + | − | − | − | − |
| NRPS 2 | + | + | − | − | − | − | − | − |
| NRPS 3 | + | + | − | − | − | − | − | − |
| isd locus | + | Duplicated | + | − | − | − | − | − |
| sst locus | Duplicated | Duplicated | + | + | + | + | + | + |
| cap locus (PS capsule) | + | + | + | − | + | + | + | + |
| cap locus (PGA capsule) | + | + | − | + | + | + | − | − |
| esx locus | + one gene FS | + one gene FS | + | − | − | − | − | − |
| Streptolysin S-like toxin | + one gene FS | + one gene FS | − (RF122) | − | − | − | − | − |
| Lantibiotic resistance locus | + | + | − | − | − | − | − | − |
| agr locus | + | + | + | + | + | + | + | − |
| ica locus | + | + | + | + | − | + | − | + |
| mprF/dlt | + | + | + | + | + | + | + | + |
| CRISPR region | + | + | − | + | − | − | − | − |
| β-Haemolysin | + | + | + | + | − | − | − | − |
| Putative haemolysin III | + | + | + | + | + | + | + | + |
FS, frameshift; NRPS, nonribosomal peptide synthetase; PGA, polyglutamic acid; PS, polysaccharide; RF122, sequenced Staphylococcus aureus bovine isolate.
Because S. lugdunensis is more virulent than other CoNS in its ability to cause SSSTIs and IE it is worthwhile to examine the differences in the repertoires of virulence factors. The S. aureus sphingomyelinase β-toxin (hlb) is conserved in S. lugdunensis and a putative haemolysin III is encoded as well. However, S. lugdunensis does not have genes encoding coagulase, protein A, superantigens, exfoliatins, β-barrel pore-forming toxins (hly, luk, hlg) or small secreted proteins involved in immune evasion viz map, efb, chp, scn, sak, ssl.
The isd locus
Staphylococcus lugdunensis is unique among CoNS by having a locus encoding iron-regulated surface determinant (Isd) proteins that have the potential to extract haem from haemoglobin and transport it across the cell wall using a series of wall-anchored proteins bearing near iron transporter (NEAT) motifs and into the cytoplasm using an ABC transporter. There, haem monooxygenases cleave the porphyrin ring to release the Fe2+. Staphylococcus aureus specifies three surface-exposed proteins that are anchored to peptidoglycan by processing at the C-terminal LPXTG-motif by sortase A. The IsdH, IsdB and IsdA proteins have three, two and one NEAT motifs, respectively. Staphylococcus lugdunensis has two putative LPXTG-anchored proteins, both with two NEAT motifs (Fig. 2 shows the isd loci of S. aureus and S. lugdunensis in comparison; Fig. 3 shows schematic diagrams of the surface-anchored proteins). One is an orthologue of IsdB with the sequence similarity to IsdB NEAT motifs being particularly high (50% and 55% identities). The second LPXTG-anchored protein named IsdJ has two NEAT motifs with 50% and 54% identity to the single NEAT motif of IsdA. The S. aureus Isd locus includes a novel sortase (SrtB) that recognizes the C-terminal NPQTN motif in IsdC when anchoring the protein to peptidoglycan. Staphylococcus lugdunensis has SrtB and IsdC orthologues and encodes a second putative SrtB substrate (IsdK) carrying a NKQPN motif in a gene located between isdC and isdE that replaces isdD in the S. aureus locus. In contrast to IsdD, the S. lugdunensis IsdK protein has a NEAT motif. Another major difference between S. aureus and S. lugdunensis Isd is the absence of the IsdI haem oxygenase and the cell wall-anchored protein IsdH. Finally, a putative autolysin is encoded downstream of isdG in the S. lugdunensis operon. No orthologue gene is present in S. aureus. Otherwise the overall gene organization is very similar apart from the presence of an insertion of genes encoding a membrane transporter located between the isdA-like gene and the isdB orthologue. Also a gene encoding an ABC transporter subunit is located between isdF and srtB. Whether these genes are Fur-regulated and whether they transport haem or another substrate is an open question.
Fig. 2.
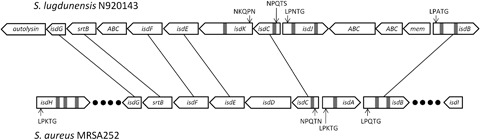
Comparison of the isd loci of Staphylococcus aureus and Staphylococcus lugdunensis. A schematic diagram of the isd loci is shown. The open boxes denote individual genes and the arrows the direction of their transcription. Encoded NEAT motifs are shown as small black boxes. Orthologous genes are linked by thin black lines. The % identities between the encoded proteins are as follows: IsdB 36.8%, IsdC 57.6%, IsdE, 74.7%, IsdF 57.7%, SrtB 58.2%, IsdG 68.2%. Cell wall sorting signals are indicated. The isdH and isdI genes of S. aureus are located outside the locus as indicated by black circles.
Fig. 3.
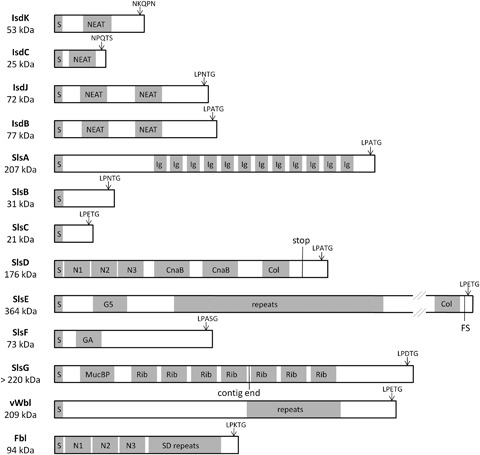
Schematic diagrams of the Staphylococcus lugdunensis MSCRAMMs. Predicted domains are indicated as grey boxes. NEAT, NEAT domain; GA, GA module; Ig, immunoglobulin-like fold; Col, collagen triple helix repeat; G5, G5 domain; MucBP, mucin-binding domain; Rib, Rib-like repeat; SD, serine–aspartate; FS, frame shift; see text for predicted functions of the various domains. S-labeled boxes indicate the putative signal sequences.
Of special interest is the location of the Isd locus with respect to the replication origin of the chromosome. The S. aureus MRSA252 locus is located in the middle of the chromosome (nucleotide 1146876–1155059). In contrast, the S. lugdunensis locus (nucleotide 96442–111616) is located in a different genomic context, close to the origin of replication.
Surface-anchored proteins
Proteins that are covalently anchored to the cell wall surface by sortase A-mediated processing of LPXTG are of particular interest because of their possible roles in adhesion to skin and host tissue, immune evasion and biofilm formation. Apart from the two Isd proteins described above, the previously described fibrinogen-binding protein Fbl that is related to ClfA of S. aureus (Geoghegan et al., 2010) and the von Willebrand factor-binding protein vWbl (Nilsson et al., 2004), S. lugdunensis has seven genes with the potential to encode wall-anchored proteins ranging in size from 20.8 to 380 kDa. These proteins have been called S. lugdunensis surface proteins (Sls). Figure 3 shows schematic diagrams of these proteins. Analysis using the Pfam database (Sammut et al., 2008) has revealed several interesting features. (1) The 1930 residue SlsA protein has 12 nonidentical repeats of an IgG-like fold located between the LPXTG sequence and the 734 residue N-terminal domain. (2) SlsE (3459 residues) has an 1240-residue N-terminal domain containing a 78-residue G5 motif (Ruggiero et al., 2009) followed by a collagen-like sequence (24 residues), 46 repeats of a 31 residue motif and a C-terminal collagen-like sequence (50 residues). The slsE gene of N920143 has a frameshift between the region encoding the collagen-like domain and the LPXTG sequence while that of HKU09-01 is intact. (3) SlsG has a 874-residue N-terminal domain with a 125-residue MucBP domain (Du et al., 2011) followed by repeats of a Rib-like domain (Wastfelt et al., 1996) seen in group B streptococci surface-anchored proteins. There are 18 Rib repeats in the HKU09-01 protein, whereas in the N920143 protein the number of repeats is uncertain because the gene is located on two contigs in our sequence. (4) The SlsD protein has a 575-residue N-terminal domain that has sequence and putative structural similarity to the fibrinogen-binding domain of SdrG of S. epidermidis (Ponnuraj et al., 2003). Located C-terminally to this are two repeats with similarity to the B repeats of the collagen-binding protein CNA of S. aureus and a 45-residue collagen-like domain. The slsD gene contains a nonsense codon located just 5′ to the region encoding LPXTG so it is unlikely that the protein would remain anchored to the cell wall. (5) The 659-residue SlsF protein has a 41-residue GA albumin-binding domain similar to that of Peptostreptococcus magnus (Johansson et al., 1997).
All MSCRAMMs are highly conserved in the strain HKU09-01 with only minor differences in the number of repeats within the stalk regions. The only major difference is that slsE does not contain a frameshift in HKU09-01. Although both strains represent isolates form distant geographical origins, the nonsense mutation in slsD is present in both strains.
Discussion
The first in-depth analysis of an S. lugdunensis genome sequence and the comparison with several other staphylococci revealed a multitude of interesting characteristics, making S. lugdunensis an outstanding member of the staphylococci. The core genome of all species included in the evaluation is highly conserved and encodes housekeeping functions like DNA replication, RNA synthesis, sugar and amino acid degradation/biosynthesis and metabolite transport. However, several features are apparent in S. lugdunensis that make it unique and place it in between S. aureus and the other CoNS. The presence of three nonribosomal peptide synthetases is remarkable, considering that most staphylococci encode not a single one. One can only speculate about the products, but they might support S. lugdunensis while colonizing the human skin by inhibiting other skin commensals or by facilitating the uptake of rare ions or other substrates. Furthermore S. lugdunensis encodes a plethora of surface-anchored proteins with sizes and domain organizations unusual for staphylococci.
Staphylococcus lugdunensis is known to be an important pathogen although it causes invasive infections only infrequently. Paradoxically, once S. lugdunensis gets established in a thrombus on a heart valve or is encased in an abscess it appears to be as virulent as S. aureus (Frank et al., 2008). The genome sequence revealed only very few putative virulence factors, verifying earlier reports about the absence of many typical S. aureus toxin genes in S. lugdunensis (Fleurette et al., 1989). This might explain the low infectivity of the organism. However, S. lugdunensis has the potential to encode a streptolysin S-like toxin and secretion system, which is well described for different bacterial species, but among the staphylococci it is found only in the bovine S. aureus isolate RF122 (Lee et al., 2008). Further, S. lugdunensis is the only CNS carrying an ESAT-6 system similar to the one encoded by S. aureus. A striking difference from S. aureus is the absence of any obvious immune evasion molecules. In S. aureus, immune evasion molecules are located on mobile genetic elements, pathogenicity islands or prophages. The lack of these elements seems to be a characteristic for S. lugdunensis and might in part explain its low infectivity.
The most interesting linkage between S. aureus and S. lugdunensis is the presence of an isd locus. In S. aureus, the locus has been shown to be important for pathogenicity. Isd proteins of S. aureus are not just involved in the acquisition of iron. In particular, IsdA is an important virulence factor because its C-terminal part confers resistance to antimicrobial fatty acids and lantibiotics (Clarke et al., 2007). Furthermore IsdA binds to fibronectin, fibrinogen, lactoferrin, transferrin, feutin, involucrin, loricrin and cytokeratin 10 and is important for nasal colonization (Clarke et al., 2004, 2009; Clarke & Foster, 2008). IsdB promotes binding to and activation of platelets (Miajlovic et al., 2010). Interestingly, IsdB and the N-terminal domain of IsdA are conserved in S. lugdunensis, perhaps suggesting similar functions of the proteins. In addition, the duplication of the locus in HKU09-01 strengthens the hypothesis of an important function of the locus. The ability of the bacterium to adhere to certain ligands or to resist against antimicrobial agents might be enhanced by a gene dosage effect.
In S. lugdunensis and S. aureus, the Isd loci are located in different chromosomal contexts, suggesting that they were acquired independently from different sources. This explains the apparent differences between the loci including the absence of isdI and isdG in S. lugdunensis. In S. aureus these genes are located outside the isd operon and must have been acquired independently of the other genes to support the function of the operon. The functions of these proteins are probably dispensable in S. lugdunensis. However, the presence of an independently acquired isd operon in S. lugdunensis and S. aureus suggests convergent evolution towards invasive behaviour that has not been described for any other CoNS.
The conclusions of the genome analysis give a picture showing S. lugdunensis to be a skin commensal that is well equipped for the survival and competition on human skin. Nevertheless, provided with capsules, different toxins, putative haemolysins and a plethora of surface-anchored proteins, the aggressive, S. aureus-like behaviour of S. lugdunensis that distinguishes it from the other CoNS might be explained. The properly annotated sequence provides the starting point for projects to investigate the mechanistic basis of skin colonization and pathogenesis by facilitating cloning and expression of genes for biochemical studies, and for generating site-specific mutants by allelic exchange to allow testing of Koch's Postulates at the molecular level.
Acknowledgments
The Sanger Institute is core funded by the Wellcome Trust. We would like to thank the Sanger Institute's Pathogen Production Group for shotgun and finishing sequencing, and the core Informatics Group for support. Research in Dublin is supported by the IRCSET Embark Scholarship and by Science Foundation Ireland (Programme Investigator grant 08/IN.1/B1854).
Statement
Data deposition: The sequence and annotation of the N920143 genome has been deposited in the EMBL database under the accession number FR870271.
References
- Bieber L, Kahlmeter G. Staphylococcus lugdunensis in several niches of the normal skin flora. 2010;16:385–388. doi: 10.1111/j.1469-0691.2009.02813.x. [DOI] [PubMed] [Google Scholar]
- Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. P Natl Acad Sci USA. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Chain PS, Grafham DV, Fulton RS, et al. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SR, Foster SJ. IsdA protects Staphylococcus aureus against the bactericidal protease activity of apolactoferrin. Infect Immun. 2008;76:1518–1526. doi: 10.1128/IAI.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SR, Wiltshire MD, Foster SJ. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Mol Microbiol. 2004;51:1509–1519. doi: 10.1111/j.1365-2958.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- Clarke SR, Mohamed R, Bian L, et al. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe. 2007;1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Clarke SR, Andre G, Walsh EJ, Dufrene YF, Foster TJ, Foster SJ. Iron-regulated surface determinant protein A mediates adhesion of Staphylococcus aureus to human corneocyte envelope proteins. Infect Immun. 2009;77:2408–2416. doi: 10.1128/IAI.01304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, He YX, Zhang ZY, et al. Crystal structure of the mucin-binding domain of Spr1345 from Streptococcus pneumoniae. J Struct Biol. 2011;174:252–257. doi: 10.1016/j.jsb.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Fleurette J, Bes M, Brun Y, et al. Clinical isolates of Staphylococcus lugdunensis and S. schleiferi: bacteriological characteristics and susceptibility to antimicrobial agents. Res Microbiol. 1989;140:107–118. doi: 10.1016/0923-2508(89)90044-2. [DOI] [PubMed] [Google Scholar]
- Frank KL, Del Pozo JL, Patel R. From clinical microbiology to infection pathogenesis: how daring to be different works for Staphylococcus lugdunensis. Clin Microbiol Rev. 2008;21:111–133. doi: 10.1128/CMR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Ganesh VK, Smeds E, Liang X, Hook M, Foster TJ. Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. J Biol Chem. 2010;285:6208–6216. doi: 10.1074/jbc.M109.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Holden MT, Hauser H, Sanders M, et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 2009;4:e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MU, de Chateau M, Wikstrom M, Forsen S, Drakenberg T, Bjorck L. Solution structure of the albumin-binding GA module: a versatile bacterial protein domain. J Mol Biol. 1997;266:859–865. doi: 10.1006/jmbi.1996.0856. [DOI] [PubMed] [Google Scholar]
- Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-dl-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Mitchell DA, Markley AL, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. P Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miajlovic H, Zapotoczna M, Geoghegan JA, Kerrigan SW, Speziale P, Foster TJ. Direct interaction of iron-regulated surface determinant IsdB of Staphylococcus aureus with the GPIIb/IIIa receptor on platelets. Microbiology. 2010;156:920–928. doi: 10.1099/mic.0.036673-0. [DOI] [PubMed] [Google Scholar]
- Morrissey JA, Cockayne A, Hill PJ, Williams P. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect Immun. 2000;68:6281–6288. doi: 10.1128/iai.68.11.6281-6288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Bjerketorp J, Wiebensjo A, Ljungh A, Frykberg L, Guss B. A von Willebrand factor-binding protein from Staphylococcus lugdunensis. FEMS Microbiol Lett. 2004;234:155–161. doi: 10.1016/j.femsle.2004.03.024. [DOI] [PubMed] [Google Scholar]
- O'Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides. 2001;22:1603–1608. doi: 10.1016/s0196-9781(01)00495-8. [DOI] [PubMed] [Google Scholar]
- Otto TD, Sanders M, Berriman M, Newbold C. Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics. 2010;26:1704–1707. doi: 10.1093/bioinformatics/btq269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. P Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Goetz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- Peschel A, Jack RW, Otto M, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuraj K, Bowden MG, Davis S, et al. A ‘dock, lock, and latch’ structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Tizzano B, Pedone E, Pedone C, Wilmanns M, Berisio R. Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis. J Mol Biol. 2009;385:153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10 000 families and still growing. Brief Bioinform. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- Shah MM, Iihara H, Noda M, et al. dnaJ gene sequence-based assay for species identification and phylogenetic grouping in the genus Staphylococcus. Int J Syst Evol Micr. 2007;57:25–30. doi: 10.1099/ijs.0.64205-0. [DOI] [PubMed] [Google Scholar]
- Siezen RJ, Kuipers OP, de Vos WM. Comparison of lantibiotic gene clusters and encoded proteins. Antonie van Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Satoh I, Kikuchi N. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol. 1999;49:725–728. doi: 10.1099/00207713-49-2-725. [DOI] [PubMed] [Google Scholar]
- Tsai IJ, Otto TD, Berriman M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H, Tsoi HW, Leung SP, Lau SK, Woo PC, Yuen KY. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J Bacteriol. 2010;192:1471–1472. doi: 10.1128/JB.01627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastfelt M, Stalhammar-Carlemalm M, Delisse AM, Cabezon T, Lindahl G. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem. 1996;271:18892–18897. doi: 10.1074/jbc.271.31.18892. [DOI] [PubMed] [Google Scholar]
- Wyatt MA, Wang W, Roux CM, Beasley FC, Heinrichs DE, Dunman PM, Magarvey NA. Staphylococcus aureus nonribosomal peptide secondary metabolites regulate virulence. Science. 2010;329:294–296. doi: 10.1126/science.1188888. [DOI] [PubMed] [Google Scholar]
- Zbinden R, Müller F, Brun F, von Graevenitz A. Detection of clumping factor-positive Staphylococcus lugdunensis by Staphaurex Plus®. J Microbiol Meth. 1997;31:95–98. [Google Scholar]


