Summary
Phospholipase Cε (PLCε) is a multifunctional enzyme implicated in cardiovascular, pancreatic and inflammatory functions. Here we show that conditional deletion of PLCε in mouse cardiac myocytes protects from stress-induced pathological hypertrophy. PLCε siRNA in ventricular myocytes decreases endothelin-1 (ET-1)-dependent elevation of nuclear calcium and activation of nuclear protein kinase D (PKD). PLCε scaffolded to muscle-specific A kinase anchoring protein (mAKAP), along with PKCε and PKD, localizes these components at or near the nuclear envelope and this complex is required for nuclear PKD activation. Phosphatidylinositol 4-phosphate (PI4P) is identified as a perinuclear substrate in the Golgi apparatus for mAKAP-scaffolded PLCε. We conclude that perinuclear PLCε, scaffolded to mAKAP in cardiac myocytes, responds to hypertrophic stimuli to generate DAG from PI4P in the Golgi apparatus, in close proximity to the nuclear envelope, to regulate activation of nuclear PKD, and hypertrophic signaling pathways.
Heart failure is a major global health problem and a leading cause of mortality. In response to high blood pressure or other cardiac stress the adult heart remodels enlarges in what is thought to be an initial compensatory mechanism, but which is maladaptive in the long term and leads to cardiac decompensation and heart failure. One major feature of cardiac remodeling is hypertrophic growth of cardiac myocytes, driven in part by increased levels of neurohumoral agonists such as norepinephrine and endothelin-1 (ET-1), acting on G protein-coupled receptors (GPCRs) in cardiac myocytes (Rockman et al., 2002).
Many hypertrophic agonists couple to activation of phosphoinositide-specific phospholipase C (PLC), to stimulate phosphatidylinositol 4,5-bisphosphate (PIP2) hydrolysis and produce inositol 1,4,5 trisphosphate (IP3) and diacylglycerol (DAG), a key reaction involved in mediating cardiomyocyte hypertrophy. Gαq, a GTP binding protein that directly activates PLC is a major participant in the hypertrophic process in mice (D'Angelo et al., 1997; Dorn II and Brown, 1999), and a splice variant of PLCβ, PLCβ1b, is required for cardiomyocyte hypertrophy in vitro driven by α-adrenergic receptor activation (Filtz et al., 2009; Grubb et al., 2011).
A novel isoform of PLC, PLCε, is downstream of both GPCRs and receptor tyrosine kinases by virtue of its ability to be regulated by small GTPases including Ras, Rho and Rap, and by heterotrimeric G protein βγ subunits (Kelley et al., 2001; Smrcka et al., 2012). We recently demonstrated that hypertrophy of neonatal rat ventricular myocytes (NRVMs) driven by endothelin-1 (ET-1), norepinephrine, or isoproterenol (Iso) was inhibited by siRNA-mediated knockdown of PLCε (Zhang et al., 2011). Furthermore, it was shown that PLCε scaffolds to muscle-specific A kinase anchoring protein (mAKAP) at the nuclear envelope (NE), and disruption of this scaffolding interaction prevents development of agonist-induced hypertrophy (Zhang et al., 2011). This suggests that PLCε generates second messengers at the nuclear envelope that are required for hypertrophy.
Canonical GPCR-mediated PIP2 hydrolysis by PLC occurs at the plasma membrane where PIP2 is preferentially enriched. PIP2 is not readily detectable in intracellular organelle membranes and PI - dependent signaling processes in intracellular membranes have not been well characterized. A phosphoinositide cycle involving PIP2 hydrolysis is present in the nuclear matrix (Divecha et al., 1993; Keune et al., 2011; Ramazzotti et al., 2011). Turnover of nuclear PIP2 is not subject to regulation by GPCRs, despite the presence of PLCβ in the nucleus of some cells, but IGF-1 has been shown to stimulate nuclear PIP2 hydrolysis (Cocco et al., 1989; Divecha et al., 1991). Surprisingly, nuclear PIP2 is not associated with nuclear envelope membranes but rather is in a poorly defined detergent resistant structure in the matrix (Keune et al., 2011). With no apparent PIP2 substrate at the NE, a role for a PLC hydrolytic activity associated with the NE is not obvious, although it is theoretically possible that a NE-scaffolded PLC could gain access to nuclear PIP2. Phosphatidylinositol 4-P (PI4P) is an alternate substrate for purified PLC in vitro that is found in intracellular membranes, but PI4P has not been shown to be a native physiological substrate for PLC in cells. PI4P has been thought of primarily as a precursor for replenishment of PI4,5P2 during active receptor-stimulated PIP2 hydrolysis but recent studies indicate that PI4P itself plays other important roles in cell function (Hammond et al., 2012).
One of the key signaling events that leads to expression of hypertrophic genes is phosphorylation of histone deacetylase (HDAC-5) and subsequent binding of phosphorylated HDAC to 14-3-3 proteins in the cytoplasm (Frey and Olson, 2003). Two key kinases that phosphorylate HDAC at the nucleus are calcium calmodulin-dependent protein kinase II (CamKII) and protein kinase D (PKD) (Frey and Olson, 2003; McKinsey, 2007). CamKII is regulated by Calcium, and PKD is regulated by DAG and phosphorylation by protein kinase C (PKC) (Rozengurt, 2011; Steinberg, 2012), suggesting that Ca2+ and DAG levels need to be elevated locally at the nucleus to maintain their activities. How plasma membrane GPCR generated signals regulate nuclear proteins has not been carefully examined, but it has been generally assumed that IP3 generated as a result of activation plasma membrane PLC activity, presumably PLCβ, can diffuse through the cytoplasm to the nuclear envelope where IP3 receptors are enriched in cardiac myocytes to release a local pool of Ca2+ (Higazi et al., 2009; Wu et al., 2006). DAG, on the other hand, is diffusible within membranes but not between membranes, so how PKD is activated at the nucleus is unclear. There is evidence that PKD can be activated at the plasma membrane (PM) and subsequently diffuse to the nucleus (Bossuyt et al., 2011) but whether PM signals are sufficient for this process has not been examined.
To determine the mechanistic role of PLCε in cardiac function and failure we deleted PLCε specifically in cardiac myocytes in mice after development and examined the subcellular scaffolded signals generated in cardiac myocytes by PLCε. We show that mice with cardiac myocyte-specific deletion of PLCε are protected from pressure overload-induced hypertrophy, clearly identifying a new gene critical for the development of heart failure. Further, we define a role of nuclear-scaffolded PLCε in regulating signaling proteins that drive hypertrophy. Notably, we identify PI4P in the perinuclear Golgi apparatus as the key substrate for PLCε at the nuclear envelope for generation of local DAG, and subsequent nuclear PKD activation.
Results
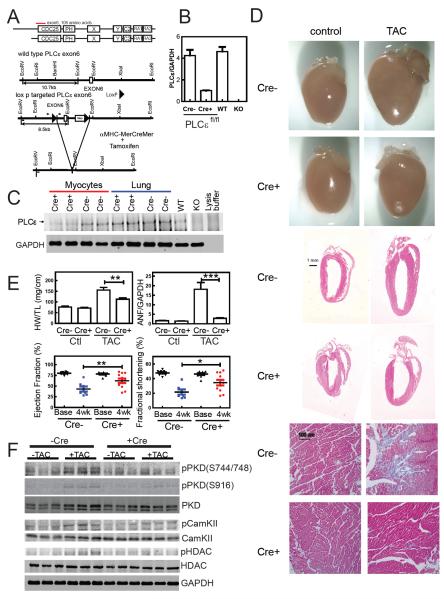
Cardiac-specific deletion of PLCε in PLCεflox/flox α-MHC-MerCreMer mice
Our recent results in NRVM's show that siRNA based depletion of PLCε prevents agonist-induced hypertrophy (Zhang et al., 2011). This contrasts with our previous work demonstrating that global knockout of PLCε exacerbates hypertrophy in response to isoproterenol infusion (Wang et al., 2005). To clarify the role of PLCε in cardiac function and failure, we deleted PLCε specifically in cardiac myocytes in mice after development (Figure 1A and methods). PLCεflox/flox mice (PLCεflox/flox Cre−) were bred with α-MHC-MerCreMer (α-MHC-MCM) mice to generate PLCεflox/flox Tg(α-MHC-MCM), (PLCεflox/flox Cre+) mice (Figure 1A) (Sohal et al., 2001). After one month of post-natal development, mice from both genotypes were given 3 daily injections of 40 mg/kg body weight tamoxifen i.p. PLCε mRNA was decreased by 80% in myocytes isolated from PLCεflox/flox Cre+ mice compared to either PLCεflox/flox Cre− or PLCε wt mice (Figure 1B). PLCε protein was significantly reduced (>70%) in myocytes isolated from PLCεflox/flox Cre+ mice relative to PLCεflox/flox Cre− mice, while in lung the level of PLCε protein was unchanged (Figure 1C). PLCε deletion in PLCεflox/flox Cre+ mice had no significant effect on ejection fraction, fractional shortening, ventricular wall dimensions or LV mass in the absence of stress (Figure 1E). Previous data from global PLCε−/− mice showed no change in levels of other PLCβ1, PLCβ3, PLCγ1 or PLCδ3 compared to PLCε+/+ mice (Wang et al., 2005).
Figure 1.
Conditional Deletion of PLCε in Cardiac Myocytes Prevents Development of Cardiac Hypertrophy. (A) Domain Structure of PLCε and Strategy for Conditional Deletion of PLCε. Exon 6 encodes the first common exon of two PLCε splice variants at the amino terminus of the CDC25 domain. Exon 6 was flanked by two LoxP sites as shown. Small arrows indicate location of primers for genotyping, small bars indicate the location of Southern blot probes. (B) 1 month old PLCεfl/flCre+ and PLCεfl/flCre− mice were injected with 40 mg/kg of tamoxifen once/day for three consecutive days. PLCε mRNA was measured by real time quantitative PCR and normalized to GAPDH levels. (C) PLCε protein was immunoprecipitated and analyzed by western blotting. GAPDH from the lysates was immunoblotted as a loading control. KO controls are from globally deleted PLCε−/− mice shown for comparison. (D) Anatomical view, histological HE stained sections, and trichome stained (for fibrosis, blue) sections from hearts from tamoxifen treated PLCεfl/fl mice with or without 4 weeks of transaortic constriction. (E) Quantitation of heart weight to tibia length (HW/TL), atrial natriuretic factor (ANF)/GAPDH mRNA levels by RT-PCR, Ejection Fraction (by echocardiography), and Fractional Shortening (by echocardiography) (+/− SEM) . Analyzed by One way ANOVA, 7 mice Cre−, 11 mice Cre+, *p<0.05, **p<0.01, ***p<0.005. (F) Western blots of PKD, phosphoPKD, CamKII, phosphoCamKII, HDAC, PhosphoHDAC and GAPDH from tamoxifen injected mice with and without TAC. See also Figure S1.
Cardiac-specific deletion of PLCε inhibits development of hypertrophy after transverse aortic constriction(TAC)
After four weeks of TAC, PLCεflox/flox Cre− mice had increased heart size, LV and RV wall thickness and interstitial fibrosis (Figure 1D, cre−). These morphological changes were significantly inhibited in the PLCεflox/flox Cre+ animals (Figure 1D, cre+). PLCεflox/flox Cre− mice developed a severe decrement in heart function with a drastic decrease in ejection fraction and fractional shortening, had a significant increase in HW to tibia length and showed large increases in hypertrophic gene expression (Figure 1E and Figure S1A and S1B). In contrast, PLCεflox/flox Cre+ mice were significantly protected from these hypertrophy related changes (Figure 1E and Figure S1A and S1B). This data indicates that cardiomyocyte PLCε plays a role in mediating cardiac hypertrophy, supporting the mechanistic data obtained in NRVMs.
Characterization of signaling pathways associated with PLCε deletion in cardiomyocytes
In the global PLCε knockout animal CamKII phosphorylation was basally increased (see Figure S5 in (Zhang et al., 2011)), but here in the cardiac specific knockout there was no detectable increase in CamKII phosphorylation nor PKD phosphorylation under basal conditions (Figure 1F and Figure S1C). TAC increased phosphorylation of PKD at S916 and S744/748 in PLCεflox/flox Cre− relative to control animals. This phosphorylation is significantly blunted in the PLCεflox/flox Cre+ animals (Figure 1F, Figure S1C). Similarly CamKII phosphorylation is increased by TAC in the PLCεflox/flox Cre− mice which is significantly blunted in the PLCεflox/flox Cre+ mice (Figure 1F, Figure S1C). These two kinases phosphorylate HDAC to cause translocation of HDAC from the nucleus to the cytoplasm leading to MEF dependent transcription of hypertrophic genes (Frey and Olson, 2003). HDAC phosphorylation was also blunted in the PLCεflox/flox Cre+ compared to PLCεflox/flox Cre− mice (Figure 1F, Figure S1C). Based on these data, we hypothesize that PLCε may be involved in regulation of PKD and CAMKII- dependent phosphorylation of HDAC in cardiac myocytes.
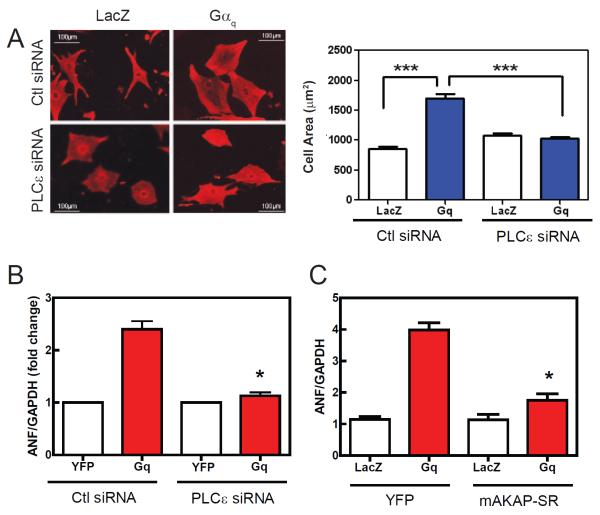
Gαq-dependent hypertrophy requires mAKAP bound PLCε
Gαq signaling has been shown to be a primary driver of hypertrophy both in NRVMs and mice (D'Angelo et al., 1997; Knowlton et al., 1993). To directly test the involvement of PLCε in Gαq-dependent hypertrophy, adenovirus expressing Gαq was used to infect NRVMs along with an adenovirus expressing either PLCε siRNA or a random siRNA control sequence. As previously reported, expression of Gαq caused an increase in NRVM cell area (Figure 2A) and ANF mRNA (Figure 2B) expression, relative to infection with control lacZ or YFP expressing viruses. Depletion of PLCε by siRNA completely inhibited the development of hypertrophy by Gαq assessed by both of these measures (Figure 2A, B).
Figure 2.
Gαq-stimulated hypertrophy in NRVMs is blocked by PLCε siRNA or disruption of nuclear envelope scaffolding via mAKAP. (A) NRVMs were transduced with 50 MOI of adenovirus expressing wild type Gαq or LacZ. Cells were co-transduced with either PLCε siRNA or control scrambled siRNA (Ctl) adenovirus. After 48 hours cells were fixed, permeabilized and stained for α-actinin. Cell area was calculated from 200 cells each treatment with NIH Image J and pooled from 3 separate experiments in the right panel (+/− SEM). (B) Same as A except ANF/GAPDH ratio was determined by quantitative real time PCR (+/− SEM). (C) NRVMs were cotransduced with adenoviruses expressing Gαq and mAKAP-SR domain. ANF/GAPDH ratio was measured after 48 h (+/− SEM). All experiments were repeated 3 times and were analyzed by One way ANOVA, *p<0.05; ***p<0.005.
To test whether mAKAP-PLCε scaffolding was important for Gαq-dependent hypertrophy we co-expressed Gαq with the mAKAP-SR1 domain, documented previously to disrupt mAKAP-PLCε interactions at the nuclear envelope (Zhang et al., 2011). Expression of the SR-1 domain almost completely inhibited hypertrophy driven by Gαq (Figure 2C), indicating that nuclear envelope scaffolding of PLCε via mAKAP is important for Gαq-dependent NRVM hypertrophy.
Scaffolding of PLCε, PKCε and PKD to mAKAP
mAKAP is a large scaffolding protein localized to the nuclear envelope in cardiac myocytes, that scaffolds to many partners including PLCε (Dodge-Kafka et al., 2005; Kapiloff et al., 2001; Zhang et al., 2011). We have shown that PLCε is localized to the nuclear fraction of cardiac myocyte lysates along with mAKAP by biochemical fractionation (Zhang et al., 2011). Expression of PLCε fused to mCherry was enriched at the periphery of the nucleus in NRVMs (Figure 3A) and some PLCε is localized to ER/SR like structures.
Figure 3.
PLCε is in a multicomponent signaling complex with mAKAP, Epac, PKCε, PKD and Ryr2 in the heart. (A) perinuclear localization of PLCε in NRVMs. mCherry tagged PLCε was expressed in NRVMs (left panel) and costained with DAPI (right panel). The boundaries of the cell are outlined with dashed lines. (B) Epac co-immunoprecipitates with PLCε and mAKAP from heart lysates. (C) Ryr2 immunoprecipitates with PLCε from heart lysates. (D) PKCε immunoprecipitates with PLCε and mAKAP from heart lysates. (E) PKD immunoprecipitates with PLCε and mAKAP from heart lysates. PLCε−/− mice were tested as a specificity control in B,C and D. See also Figure S2.
To identify other components in this complex potentially involved in PLCε signaling, we immunoprecipitated PLCε from mouse heart lysates and immunoblotted for key potential upstream regulators and downstream targets. First we looked for Epac1 association since Epac is upstream of PLCε-dependent regulation of cardiac contractile function and has been previously identified in mAKAP immunoprecipitates (Dodge-Kafka et al., 2005; Oestreich et al., 2009; Oestreich et al., 2007). Both PLCε and mAKAP immunoprecipitates contained Epac1 (Figure 3B). When PLCε was cotransfected into HEK293 cells with Epac1, Epac1 was detected in PLCε immunoprecipitates (Figure S2A). Co-expression of mAKAP did not alter the efficiency of the immunoprecipitation suggesting that PLCε directly binds to Epac1. To further characterize the interaction, purified GST-Epac1 was tested for binding to purified PLCε. GST-Epac1 specifically bound to PLCε confirming a direct interaction (Figure S2B). To determine whether interaction of Epac1 with PLCε was required for association with AKAP in the heart, we compared AKAP immunoprecipitates from PLCε−/− and PLCε+/+ mouse heart lysates (Figure S2C). Epac1 was detectable in the lysates from both genotypes but the level of Epac1 was significantly lower in immunoprecipitates from PLCε−/− mouse hearts suggesting that binding of Epac1 to PLCε contributes to scaffolding of Epac1 to mAKAP.
We had also previously shown the PLCε regulates phosphorylation of Ryr2 through a PKCe and CamKII dependent mechanism during acute β-adrenergic receptor (βAR) activation (Oestreich et al., 2009). We examined PLCε immunoprecipitates for the presence of Ryr2 and found Ryr2 immunoreactivity only in lysates from hearts isolated from PLCε+/+ mice (Figure 3C). Ryr2 has been shown to bind to mAKAP so it is possible this interaction is indirect. When co-expressed in HEK293 cells, Ryr2 weakly interacted with PLCε in a manner that was not affected by co-expression of mAKAP (data not shown). This suggests that mAKAP does not mediate Ryr2 scaffolding to PLCε, but since the PLCε-Ryr2 interaction is weaker in cotransfected cells than in heart lysates, another protein is likely to participate in PLCε-Ryr2 scaffolding. Immunodepletion of heart lysates with mAKAP antiserum reduces, but does not eliminate, Ryr2 immunoprecipitation with PLCε (Figure S2D). This indicates that a fraction of PLCε is scaffolded to Ryr2 but is not associated with mAKAP, supporting the existence of multiple pools of PLCε in cardiac myocytes.
Since PKCε and PKD are critical hypertrophic kinases that we propose are downstream of PLCε, we examined their scaffolding to mAKAP and PLCε. We had previously shown that PLCε mediates PKCε activation in adult cardiac myocytes during acute βAR stimulation (Oestreich et al., 2009). PLCε and mAKAP immunoprecipitates from heart lysates contained both PKCε and PKD immunoreactivity (Figure 3D, E). PLCε immunoprecipitates from hearts isolated from PLCε−/− mice did not contain PKCε immunoreactivity confirming the specificity of the immunoprecipitation. Together, these data indicate that many of the signaling components required for PLCε action in cardiac myocytes involved in either regulation of contraction or hypertrophy are assembled into a signaling complex with mAKAP. More importantly these data show that the key hypertrophic regulators, PKCε and PKD, are scaffolded with PLCε and mAKAP at the nuclear envelope where they play key functional roles analyzed below.
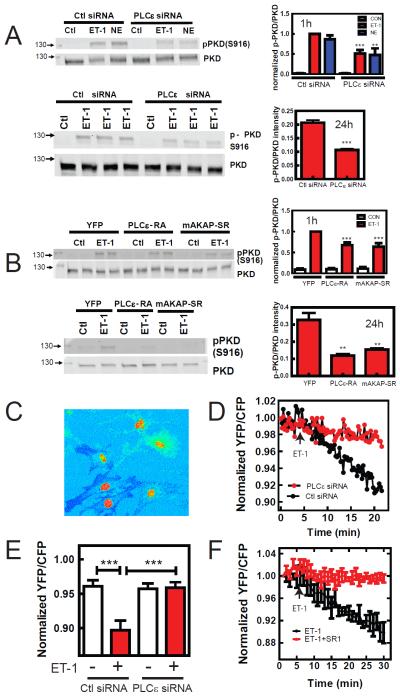
mAKAP bound PLCε is involved in nuclear PKD activation in NRVMs
To test the involvement of PLCε in ET-1-dependent PKD activation, PLCε was depleted in NRVMs using siRNA and PKD S916 phosphorylation was measured. PKD phosphorylation increases after 10 min of ET-1 stimulation and declines thereafter but remains active for at least 4h (Figure S3). ET-1 stimulation of PKD phosphorylation in PLCε siRNA treated NRVMs was blunted compared to Ctl siRNA treated cells at all times measured. We more carefully examined inhibition by PLCε depletion after stimulation with ET-1 and norepinephrine for 1 h and ET-1 for 24 h (Figure 4A). At both times ET-1-dependent PKD phosphorylation was inhibited in PLCε siRNA treated NRVMs by approximately 50%. This indicates that PLCε is important for activation of PKD, but since inhibition of total cellular PKD activation by PLCε depletion is partial, PLCε may be regulating a specific subcellular pool of PKD.
Figure 4.
PLCε regulates nuclear PKD activation. (A) NRVMs were infected with PLCε or control (Ctl) siRNA, followed by treatment with 100 nM ET-1 or 10 μM Norepinephrine (NE) for 1h and ET-1 for 24h, and assessment of PKD phosphorylation by Western blotting each repeated 3–4 times. Statistically different from Ctl siRNA **p<0.01, ***p<0.005 (B) NRVMs were infected with either control YFP, mAKAP-SR or PLCε-RA expressing adenoviruses to disrupt PLCε-mAKAP scaffolding, followed by treatment with 100 nM ET-1 for 1h or 24h and assessment of total PKD activation. Statistically different from Ctl siRNA ***p<0.005 (C) NRVMs were transduced with adenovirus expressing nuclear targeted D kinase activation reporter (nDKAR). The YFP to CFP ratio is shown as a pseudocolor image to emphasize the high level of FRET in the nucleus. D) a region in the nucleus from NRVMs expressing showing nDKAR FRET was selected and the YFP/CFP ratio was followed for the indicated times after 100 nM ET-1 addition in PLCε siRNA or ctl siRNA adenovirus treated NRVMs. E) Pooled data for the YFP/CFP ratio of nDKAR analyzed 20 min after addition of ET-1 (+) or vehicle (−) from 5 independent experiments in PLCε siRNA or Ctl siRNA expressing NRVMs (50–100 cells each condition) ***p<0.005. F) NRVMs were cotransfected with plasmids expressing nDKAR and mAKAP-SR1 or control LacZ. ET1 (100nM) was added at the indicated time. Data are pooled from 4 independent cells for each treatment from 2 separate NRVM preparations. All quantitative data is (+/− SEM). See also Figure S3.
To determine if mAKAP associated PLCε was involved in PKD activation, PLCε-mAKAP interactions were disrupted by expression of mAKAP-SR1 or PLCε-RA1 domains (Zhang et al., 2011) and ET-1-stimulated phosphorylation of PKD at 1h and 24h was measured. Expression of either PLCε-RA1 or mAKAP-SR1 domains inhibited ET-1 dependent global PKD phosphorylation at 1h (30%) and 24 h (70%) (Figure 4B). To determine if PLCε is required for activation of the nuclear pool of PKD, NRVMs were transduced with an adenovirus expressing a nuclear-targeted FRET based reporter of PKD activity, nuclear D kinase activity reporter (nDKAR) (Bossuyt et al., 2011; Kunkel et al., 2007). Figure 4C is a YFP/CFP ratiometric image from NRVMs expressing nDKAR, with CFP excitation showing basal FRET in the nucleus as previously reported (Bossuyt et al., 2011). NRVMs were treated with either vehicle or ET-1 and nuclear FRET was monitored over time. ET-1 treatment causes a significant decrease in nuclear FRET over the course of 20 min compared to vehicle control (Figure 4D and E). In NRVMs treated with PLCε siRNA, the ET-1 induced decrease in FRET was completely eliminated (Figure 4D and E). Expression of mAKAP-SR1 in NRVMs also completely inhibited the ET-1-dependent decrease in nDKAR FRET (Figure 4F). This indicates that ET-1 activates nuclear PKD, and that this nuclear PKD activation requires mAKAP-scaffolded perinuclear PLCε.
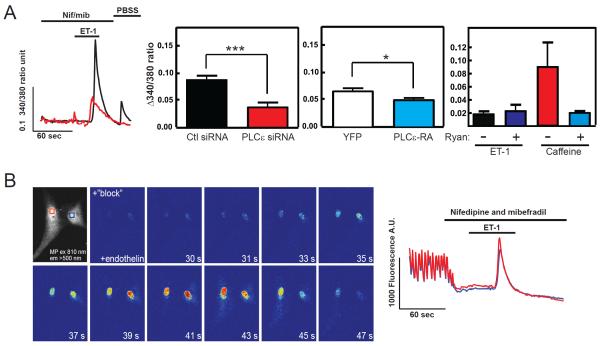
PLCε is important for localized nuclear calcium signals
ET-1 induces calcium increases in the nucleus of cardiac ventricular myocytes that is dependent on IP3 presumably binding to type II IP3 receptors localized at the nuclear envelope (Higazi et al., 2009; Wu et al., 2006). Several reports indicate the myocyte nuclear Ca2+ release via IP3 receptors can mediate cardiomyocyte hypertrophy (Arantes et al., 2012; Higazi et al., 2009; Nakayama et al., 2010). To monitor changes in nuclear calcium levels we utilized Fura-2-loaded NRVMs and specifically monitored the nuclear region of the cells by ratio imaging epifluorescence microscopy (Figure 5A). NRVMs were pretreated with nifedipine (L-type Ca2+ channel blocker) and mibefradil (L- and T-type Ca2+ channel blocker) to block Ca2+ entry and Ca2+ release associated with myocyte excitation-contraction (Higazi et al., 2009). ET-1 stimulated an increase in nuclear [Ca2+] that was inhibited by PLCε siRNA treatment (Figure 5A). Expression of the RA domain of PLCε to disrupt AKAP-dependent nuclear scaffolding also inhibited nuclear Ca2+ release but only by 10–20% (Figure 5A middle right panel). This nuclear Ca2+ release is likely IP3-dependent as previously reported (Higazi et al., 2009) because it not affected by blocking the other major Ca2+ release channel, Ryr2, with ryanodine (Figure 5A right panel). To confirm that the ET-1- dependent Ca2+ increases are localized to the nucleus we examined ET-1-dependent increases in nuclear Ca2+ signals in NRVMs using 2-photon microscopy. Treatment with ET-1 results in a robust Ca2+ release that is primarily restricted to the nucleus (Figure 5B, Supplemental Movie 1) as previously reported
Figure 5.
PLCε is involved in regulation of nuclear Ca2+ elevation. (A) NRVMs were loaded with Fura2 and the Fura2 ratio in the nucleus was measured with time after 200 nM ET-1 addition in the presence of a 10 μM nifedepine/ 2 μM mibefradil. Left panel is a representative trace from two individual cells. Middle left panel is the combined data from multiple cells comparing Ctl siRNA and PLCε siRNA treated NRVMs, ***p<0.005. Middle right panel is the combined data from cells expressing either YFP or the YFP-RA domain to disrupt mAKAP scaffolding. Both sets of data are from 4 independent experiments from cells isolated from 4 different NRVM preparations with >10 cells for each coverslip and 3 coverslip averages for each for each NRVM preparation, *p<0.05 calculated based on an average of 12 coverslip averages for each condition. Right panel shows that the ET-1-dependent nuclear Ca2+ response is not blocked by Ryanodine but the Caffeine response is (This experiment was repeated with 2 separate sets of NRVMs, data is representative of one experiment with 30 cells each condition). All data are +/− SEM. (B) 2-photon microscopy was used to measure nuclear Ca2+ levels in fluo-4 loaded NRVMs. Cells were perfused with imaging buffer containing 10 μM nifedepine and 1.8 μM mibefradil to block Ca2+ transients associated with voltage-dependent Ca2+ release followed by addition of 100 nM ET-1. Individual panels show the level of Fluo4 fluorescence at the indicated times (See Supplemental Movie 1). The boxes in the first panel correspond to the areas measured shown in the traces shown on the right.
These data indicate that IP3 generated by PLCε-dependent PIP2 hydrolysis contributes to nuclear Ca2+ signals that may be important for regulation of hypertrophy, but that PLCε scaffolded at the nuclear envelope is not a major participant in this release. This suggests that PLCε localized elsewhere, possibly at the plasma membrane, contributes to diffusible IP3 generation necessary for release Ca2+ at the nuclear envelope.
Perinuclear PI4P localization
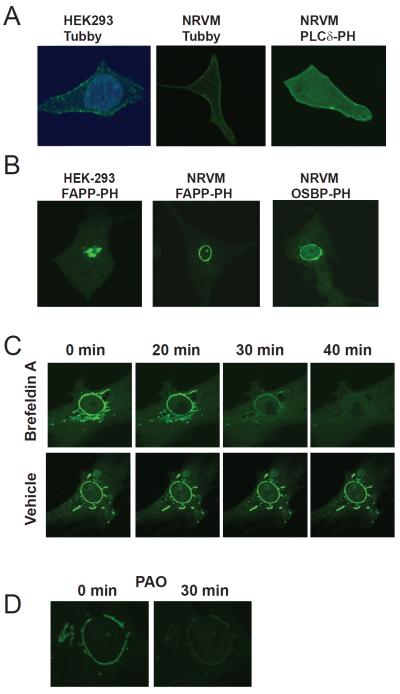
To identify potential perinuclear substrates for PLCε we utilized specific fluorescent probes containing GFP fused to domains that recognize specific phosphoinositides (Balla et al., 2009). To localize PIP2 we transfected cells with reporters containing GFP fused to the pleckstrin homology (PH) domain of PLCδ, or the PIP2 binding domain of Tubby, and examined the cells by confocal fluorescence microscopy. Both of these constructs bind to PIP2 specifically, but PLCδ-PH also binds IP3. Transfection of GFP-Tubby (Figure 6A) or GFP-PLCδ-PH (not shown) into HEK293 cells revealed a prominent PM distribution of PIP2 with some intracellular fluorescence, but no obvious localization near the nuclear envelope (Figure 6A). Transfection of NRVMs with the same constructs revealed a similar PM distribution of PIP2 (Figure 6A). Thus PIP2 is not detectable at the nuclear envelope in NRVMs and is likely not available as a substrate for NE-scaffolded PLCε activity.
Figure 6.
PI4P localizes to perinuclear Golgi surrounding the nuclear envelope in cardiac myocytes. (A) Detection of PI4,5P2 localization in NRVMs. Left panel: HEK-293 cells transfected with Tubby GFP and stained with DAPI, middle: NRVMs transfected with Tubby-GFP and right: NRVMs transfected with PLCδ-PH-GFP and analyzed by confocal microscopy. (B) Detection of PI4P at the nuclear envelope. The indicated cell types were transfected with either OSBP-PH-GFP or FAPP-PH-GFP and analyzed by confocal microscopy. (C) Inhibition of ARF eliminates perinuclear staining with FAPP-PH-GFP. NRVMs transfected with FAPP-PH-GFP were treated with 100 ng/mL Brefeldin A and GFP fluorescence monitored with time. Cells treated with vehicle are shown in the bottom panels indicate a lack of photobleaching in these experiments. (D) Inhibition of PI4kinase with PAO depletes perinuclear fluorescence associated with FAPP-PH-GFP. NRVMs transfected with FAPP-PH-GFP were treated with 10 μM PAO and fluorescence analyzed by confocal microscopy. All experiments were repeated a minimum of 3 times. See also Figure S4.
PI4P is also a substrate for PLCε and other mammalian PLC isoforms in vitro, but its utilization as a substrate in cells has not been convincingly demonstrated (Rhee et al., 1989; Seifert et al., 2004; Smrcka et al., 1991). To determine the localization of PI4P in NRVMs, cells were transfected with reporters containing GFP fused to the PH domains of either four-phosphate adapter protein (FAPP) or oxysterol binding protein (OSBP). These reporters selectively monitor PI4P subcellular localization in living cells, but this binding is co-dependent on interaction with the small GTPase ARF (Balla et al., 2005). Thus they readily detect PI4P at intracellular membranes containing ARF. As previously reported in other cultured cells, transfection of GFP-FAPP-PH into HEK293 cells leads to prominent fluorescence at asymmetrically localized structures that correspond to the Golgi apparatus (Figure 6B left panel). Surprisingly, and in striking contrast to other cultured cells, both FAPP-PH-GFP and OSBP-PH-GFP show strong a ring of perinuclear fluorescence surrounding the nucleus in NRVMs (Figure 6B middle and right panels, Figure S4A). In some cells other intracellular structures resembling either Golgi or SR were labeled (Figure 6C for example) but this was generally less prominent. Treatment with Brefeldin A to inhibit ARF, resulted in rapidly depletion of perinuclear FAPP-PH-GFP fluorescence (Figure 6C). Perinuclear FAPP-PH-GFP fluorescence was also completely disrupted by treatment a PI4-kinase inhibitor phenylarsine oxide (PAO) (Figure 6D).
Brefeldin A is an agent used to disrupt the Golgi apparatus through its ability to block Golgi localized ARF. ARF and PI4P are enriched in the Golgi in most cells and play important roles in Golgi membrane vesicle and lipid trafficking. This suggested that the perinuclear PI4P may be in Golgi closely associated with the outer surface of the nucleus. Treating cells with a BODIPY-TR ceramide to fluorescently label the Golgi revealed a perinuclear ring similar to what is observed with the PI4P reporters (Figure S4B). Indeed, previous studies have shown that the Golgi surrounds the nucleus in myocytes and is intimately associated with the nuclear envelope at a constant distance of 100–200 nm (Kronebusch and Singer, 1987; Tassin et al., 1985). Together these data indicate that PI4P is localized to the Golgi apparatus, in close proximity to the nuclear envelope, where it could be accessed by mAKAP-scaffolded PLCε.
PLCε activation at the nuclear envelope leads to PI4P depletion and DAG generation
To assess whether perinuclear PI4P is a substrate for PLCε we treated cells with a specific activator of Epac, 8-(4-chlorophenylthio)-2′-O-methyl-cAMP (cpTOME). Epac-dependent Rap activation stimulates PLCε but no other PLC isoforms and Epac is scaffolded at the nuclear envelope with PLCε and mAKAP. Thus, PLCε in this nuclear envelope signaling complex can be activated selectively with cpTOME. Treatment of NRVMs with cpTOME for 1 h caused a significant depletion of perinuclear Golgi FAPP-PH-GFP fluorescence (Figure 7A). Selected regions of perinuclear FAPP fluorescence were monitored in single live cells over time after addition of vehicle control, cpTOME or ET-1 (Figure 7B; Supplemental Movie 2). Both cpTOME and ET-1 treatment enhanced the rate of depletion of FAPP-PH-GFP fluorescence compared to vehicle control. ET-1-dependent decreases in PI4P were blocked by preincubation with the ET-1a receptor antagonist BQ-123 (Figure 7B). Total PI4P levels in NRVMs were measured after vehicle or cpTOME treatment using a PI4P strip mass assay (Dowler et al., 2002). Treatment with cpTOME lead to a significant decrease in cellular PI4P mass compared to vehicle treated NRVMs (Figure 7C). Similar studies examining PIP2 at the PM indicate that cpTOME does not stimulate PM dependent PIP2 depletion (Figure S5A). To show that ET-1 dependent perinuclear PI4P depletion was dependent on mAKAP-scaffolded PLCε, cells were cotransfected with GFP-FAPP-PH and either mAKAP-SR1 or control vector (Figure 7D). In cells transfected with mAKAP-SR1, ET-1-dependent depletion of PI4P was blocked. Additionally, as shown in Figure 7E, depletion of PLCε with PLCε-siRNA completely eliminated cpTOME-dependent depletion of perinuclear PI4P. These data strongly indicate that mAKAP-scaffolded PLCε is directly involved in perinuclear PI4P depletion consistent with its activity as a perinuclear enzyme that can hydrolyze PI4P (Figure S5B) to produce DAG.
Figure 7.
Perinuclear PI4P is a substrate for mAKAP-scaffolded PLCε, stimulated by either Epac or ET-1 receptors. (A) NRVMs transfected with FAPP-PH-GFP were analyzed by live cell confocal microscopy before and after treatment with 10 μM cpTOME for 1 h. (See also Supplemental Movie 2) (B) Individual regions GFP fluorescence in NRVMs transfected with FAPP-PH-GFP were monitored with confocal microscopy and followed with time after treatment with vehicle, 10 μM cpTOME, 50 nM ET-1 or 50 nM ET-1+ 100 nM BQ-123 (top panels are representative traces). Data was pooled from 4 experiments at 0 and 50 min for quantitation and statistics (Bottom panels). (C) NRVMs were treated with Vehicle or 10 μM cpTOME for 50 min followed by extraction of PI4P and assay using a PI4P protein-lipid overlay assay according to the manufacturer's instructions. Data from three separate experiments are quantitated in the bottom panel and analyzed by a student's t-test. (D) NRVMs were cotransfected with FAPP-PH-GFP and either mAKAP-SR1 or control plasmid. Perinuclear GFP fluorescence was monitored as in B. ET-1 experiments in B and D were done in parallel so the ET-1 alone representative traces are the same in both panels. Top panel is a representative trace and bottom panel is pooled data from 3 experiments analyzed by one way ANOVA. (E) NRVMs were transfect with FAPP-PH-GFP and transduced with viruses expressing PLCε siRNA or random control siRNA and perinuclear GFP fluorescence was monitored as in B and D. Data are pooled from 5 independent experiments each. (F) NRVMs were transfected with YFP-C1b-Y123W to detect DAG localization. Left panel shows localization of YFP-C1b-Y123W by confocal microscopy. Right panel: NRVMs transfected with YFP-C1b-Y123W were stimulated with 10 μM cpTOME and perinuclear regions and cytoplasm were imaged over time. The ratio of perinuclear fluorescence (Fpn) to the cytoplasmic fluorescence (Fc) was calculated and normalized to the starting Fpn/Fc before cpTOME addition. Data are pooled from 4 independent experiments, Fpn/Fc at 9–12 min were individually compared with Fpn/Fc at 0 min with a one way ANOVA. G) Golgi specific depletion of PI4P blocks ET-1-dependent nuclear PKD activation. Left panel; cells were transduced with GFP-tagged, Golgi targeted Sac-1 and imaged by confocal microscopy. Right panel: Cells were cotransfected with plasmids expressing nDKAR and Golgi targeted Flag-tagged Sac-1 plasmids and nDKAR FRET was monitored as in Fig. 4 D and F. 50 nM ET-1 was added at the indicated time. Data pooled from 4 independent cells for each treatment from 2 separate NRVM preparations. All data are +/− SEM. See also Figures S6 and S7.
One consideration is that although PIP2 is undetectable in the perinuclear Golgi in cardiac myocytes (Figure 6A), there could be a very low level of PIP2 that is the direct substrate for perinuclear PLCε. In this scenario PI4P might be indirectly depleted to replace the hydrolyzed PIP2 pool. To test this, a PI5 phosphatase was targeted to the Golgi or the plasma membrane using a rapamycin-recruitable Type IV PI5P ptase domain fused to FKBP12 (Varnai et al., 2006). This PI5 ptase cleaves the 5 phosphate from PI4,5P2 effectively depleting PIP2. Figure S6A shows that targeting of the PI5 ptase to the plasma membrane eliminates GFP-Tubby labeling of the PM indicating that the targeted PI5ptase depletes PM PIP2. Targeting of the PI5 ptase to the Golgi with rapamycin does not alter PI4P levels (Figure S6B, Figure S6C black squares). Treatment with cpTOME treatment depleted PI4P at the perinuclear Golgi at the same rate in the presence or absence of rapamycin-dependent targeting of the PI5P ptase to the Golgi (Figure S6C). This indicates that perinuclear Golgi PI4P depletion was not dependent on PIP2 hydrolysis in the Golgi, further supporting the idea that the direct substrate for perinuclear PLCε is PI4P.
To test for production of perinuclear DAG, cells were transfected with a DAG-binding domain (C1b domain of PKCβII with a Y123W mutation) fused to YFP (YFP-C1b-Y123W), that binds with high affinity to DAG and translocates to membranes where DAG is produced (Kunkel and Newton, 2010). In resting cells there was basal association of the reporter with perinuclear and other cellular membranes (Fig 7F). This could be due to basal turnover of lipids in the Golgi in the presence of serum, or the very high affinity of this reporter for DAG could stabilize a turning over DAG pool. Nevertheless, treatment with cpTOME increased the association of a DAG reporter with the perinuclear region of the cell. The increase was small (~10% increase), but reproducible, and consistent with what has been previously observed for this DAG reporter in the Golgi in other cell types (Kunkel and Newton, 2010). Thus DAG is generated in close proximity to the nuclear envelope by stimulation of PLCε.
To test whether PI4P is required for nuclear PKD activation, PI4P was depleted in NRVMs using PAO (as shown in Figure 6D) and ET-1-dependent nuclear PKD activity was monitored using nDKAR (Figure S7). PAO depletes cellular PI4P in cells without depleting the PIP2 pools necessary for conventional PLC mediated PIP2 hydrolysis (Hammond et al., 2012). We also found that PAO does not deplete NRVM PIP2 by examining the effect of PAO treatment on GFP-Tubby fluorescence (data not shown). PAO treatment completely eliminated ET-1-dependent nuclear PKD activation indicating that PI4P is a required substrate for nuclear PKD activation supporting the idea that Golgi PI4P is required for nuclear PKD activation.
To more directly show that Golgi localized PI4P is required for nuclear PKD activation we transfected cells with the PI 4-phosphatase, Sac 1 containing a mutation that leads to specific localization in the Golgi, Sac1-K2A (Blagoveshchenskaya et al., 2008; Rohde et al., 2003). In the left panel of Figure 7G is an NRVM transfected with GFP-Sac1-K2a showing perinuclear Golgi localization of Sac1-K2A. Sac1-K2A transfection completely eliminated the ET-1-dependent decrease in nDKAR FRET demonstrating that PI4P in the Golgi is required for nuclear PKD activation (Figure 7G, right panel). We conclude that mAKAP-scaffolded PLCε generates DAG from PI4P in close proximity to the nuclear envelope required for nuclear PKD activation.
Discussion
We previously demonstrated that PLCε integrates multiple hypertrophic stimuli in neonatal rat ventricular myocytes (Zhang et al., 2011). Here we demonstrate that cardiac specific deletion of PLCε after one month of development significantly inhibits cardiac hypertrophy development. This strongly suggests that PLCε signaling in the cardiac myocyte is important for hypertrophy development and that the neonatal myocyte analysis of PLCε function is largely relevant to whole heart function in an animal.
Our previously published data established that both phosphoinositide hydrolysis by PLCε, and nuclear envelope-scaffolding of PLCε via mAKAP, are required for PLCε-dependent hypertrophy in NRVMs (Zhang et al., 2011). This suggested that PLCε must be generating IP3, DAG, or both at the nuclear envelope to drive hypertrophic signaling cascades at the nucleus. IP3 generated at the PM is diffusible, so agonist-dependent nuclear Ca2+ release could be controlled by IP3 generated from PM PIP2 pools. DAG, on the other hand is a membrane bound lipid, not freely diffusible between membranes and therefore must be produced in the vicinity of the nucleus to regulate nuclear signaling proteins. Here we show that PLCε is important for agonist-dependent regulation of both nuclear Ca2+ release and PKD activation. PLCε scaffolded at the nuclear envelope is required to generate perinuclear DAG important for nuclear PKD activation, but surprisingly, the substrate for this reaction is in the uniquely localized perinuclear Golgi apparatus. PKD is recruited to the Golgi apparatus in a DAG-dependent manner in other cell types where it is involved in lipid and vesicle trafficking (Campelo and Malhotra, 2012). PLC, PKD and ARF are critical regulators of Golgi fission involved in the transport of cargo to the PM. Here, with the unique architecture of the Golgi apparatus in cardiac myocytes and the perinuclear scaffolding of PLCε by mAKAP, the process of PI hydrolysis and DAG generation appears to have been co-opted to recruit and activate PKD in the vicinity of the nucleus where it can regulate nuclear gene expression.
While we and others have shown that PI4P is a substrate for mammalian PLC isoforms, including PLCε, in vitro (Figure S5B), PI4P has not been shown to be a native physiological substrate for any mammalian PLC in cells. Thus, this is the first demonstration that PI4P hydrolysis is a physiologically relevant PLC reaction that likely performs a widespread function in cell biology. PI4P has generally been thought to function primarily as a precursor to replace PIP2 as it is depleted by receptor-stimulated PIP2 hydrolysis. Recently however, it has been shown that depletion of PI4P does not significantly alter the level of PIP2 in cells, nor does it affect acute receptor-stimulated PIP2 hydrolysis indicating that PI4P has other functional roles than simply serving as a PIP2 precursor (Hammond et al., 2012).
We also show that PLCε is involved in regulation of ET-1-dependent nuclear Ca2+ elevation. Previous studies have indicated that local Ca2+ signaling at the nucleus is IP3 receptor-dependent and regulates HDAC nuclear export via activation of CamKII (Wu et al., 2006). The source of IP3 was undefined in these experiments but has been suggested to diffuse from the PM. Hydrolysis of PI4P by PLC activity would generate DAG and inositol 1,4 bisphosphate (IP2) and thus would not be a relevant reaction for local IP3-dependent Ca2+ release since IP2 does not regulate Ca2+ release through IP3 receptors. Thus the role of PI4P hydrolysis by mAKAP-scaffolded PLCε at the Golgi appears to be DAG generation for PKD activation. We propose that a different pool of PLCε, perhaps at the PM in cooperation with PLCβ, generates IP3 from PIP2, which can diffuse to the nucleus and release Ca2+ via IP3 receptors in the nucleus. Overall, our model is that PLCε is located in different subcellular compartments allowing for regulation of hypertrophy and CICR via different mechanisms.
In conclusion, we have discovered a novel integrator of hypertrophic signals in the heart, PLCε, who's scaffolding at the nuclear envelope to generate DAG from the novel PLC substrate PI4P in the perinuclear Golgi apparatus, is critical for this function. This suggests that PLCε catalytic activity could be a novel target for heart failure. On the other hand PLCε is found in many cell types and has multiple functions (Smrcka et al., 2012). Global deletion of PLCε increases the propensity for heart failure (Wang et al., 2005). mAKAP has a more restricted distribution and thus so does the PLCε-mAKAP complex (Kapiloff et al., 1999). A more targeted strategy for treatment of heart failure could involve developing reagents that interfere with PLCε-scaffolding to mAKAP.
Experimental Procedures
See Supplemental Extended Experimental Procedures for detailed methodology.
Cardiomyocyte specific deletion of PLCε and induction and analysis of hypertrophy
PLCεflox/flox Cre+ and PLCεflox/flox Cre− mice were generated as described in extended experimental methods. At 30 days after birth mice were injected 3 times on consecutive days with 40 mg/kg tamoxifen to excise exon 6 of the PLCe1 gene. 30 days later mice were subjected to 4 weeks of TAC followed by echocardiographic, morphometric and biochemical parameters.
Isolation, culture and adenoviral infection, PLCε siRNA and hypertrophy measurements in NRVMs
Isolation of NRVMs, adenoviral mediated si-RNA of PLCε, hypertrophy induction and analysis was as previously described (Zhang et al., 2011).
Imaging of phosphoinositide and DAG reporters
Cells were maintained in DMEM containing 10% (v/v) fetal bovine serum (FBS), 100 IU/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine, and 2 μg/mL vitamin B-12 and 10 mM cytosine arabinoside. Cells were imaged 24–48 h after transfection with the appropriate plasmid at 1–2 mg. Transfection efficiency was 5%. Fluorescent cells were identified and imaged by confocal microscopy. During imaging and cpTOME, ET-1, BFA and PAO treatments, cells were in culture medium containing serum.
Live cell Imaging
Ca2+ was imaged by either two photon microscopy of fluo4 loaded NRVMs or by ratio imaging of Fura-2 loaded cells. All other imaging was by confocal microscopy.
Supplementary Material
Highlights
Conditional deletion of PLCε in mice protects against stress-induced hypertrophy.
PLCε is in a complex at the nuclear envelope with regulators and downstream targets.
PLCε is critical for regulation of local nuclear PKD activity and Ca2+ release.
PI4P in the perinuclear Golgi is the substrate for PLCε.
Acknowledgements
We would like to thank Tamas Balla for fluorescent phosphoinositide reporter fusion and PI5 ptase constructs and for valuable critical reading of this manuscript. We would also like to thank Peter Mayinger for providing the Sac1-K2A construct and Alexandra Newton for the nDKAR and DAG reporter constructs. This work was supported by National Institutes of Health grants 5R01GM053536 (AVS), R01DE041756 (DIY) and R01 HL089885, R01 HL091475 (BCB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arantes LAM, Aguiar CJ, Amaya MJ, Figueiró NCG, Andrade LM, Rocha-Resende C, Resende RR, Franchini KG, Guatimosim S, Leite MF. Nuclear inositol 1,4,5-trisphosphate is a necessary and conserved signal for the induction of both pathological and physiological cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2012;53:475–486. doi: 10.1016/j.yjmcc.2012.06.017. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Várnai P, Balla T. A Plasma Membrane Pool of Phosphatidylinositol 4-Phosphate Is Generated by Phosphatidylinositol 4-Kinase Type-III Alpha: Studies with the PH Domains of the Oxysterol Binding Protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Szentpetery Z, Kim YJ. Phosphoinositide Signaling: New Tools and Insights. Physiology. 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knödler A, Nicolson T, Boehmelt G, Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. The Journal of Cell Biology. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt J, Chang C-W, Helmstadter K, Kunkel MT, Newton AC, Campbell KS, Martin JL, Bossuyt S, Robia SL, Bers DM. Spatiotemporally Distinct Protein Kinase D Activation in Adult Cardiomyocytes in Response to Phenylephrine and Endothelin. J Biol Chem. 2011;286:33390–33400. doi: 10.1074/jbc.M111.246447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco L, Martelli AM, Gilmour RS, Ognibene A, Manzoli FA, Irvine RF. Changes in nuclear inositol phospholipids induced in intact cells by insulin-like growth factor I. Biochem Biophys Res Commun. 1989;159:720–725. doi: 10.1016/0006-291x(89)90054-5. [DOI] [PubMed] [Google Scholar]
- D'Angelo DD, Sakata Y, Lorenz JN, Boivin GP, Walsh RA, Liggett SB, Dorn GW., II Transgenic Gαq Overexpression Induces Cardiac Contractile Failure in Mice. Proc Natl Acad Sci U S A. 1997;94:8121–8126. doi: 10.1073/pnas.94.15.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 1991;10:3207–3214. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfić H, Irvine RF. Inositides and the nucleus and inositides in the nucleus. Cell. 1993;74:405–407. doi: 10.1016/0092-8674(93)80041-c. [DOI] [PubMed] [Google Scholar]
- Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW, II, Brown JH. Gq Signaling in Cardiac Adaptation and Maladaptation. Trends Cardiovasc Med. 1999;9:26–34. doi: 10.1016/s1050-1738(99)00004-3. [DOI] [PubMed] [Google Scholar]
- Dowler S, Kular G, Alessi DR. Protein Lipid Overlay Assay. Sci STKE. 2002;129:l6. doi: 10.1126/stke.2002.129.pl6. [DOI] [PubMed] [Google Scholar]
- Filtz TM, Grubb DR, McLeod-Dryden TJ, Luo J, Woodcock EA. Gq-initiated cardiomyocyte hypertrophy is mediated by phospholipase Cβ1b. FASEB J. 2009;23:3564–3570. doi: 10.1096/fj.09-133983. [DOI] [PubMed] [Google Scholar]
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Grubb DR, Iliades P, Cooley N, Yu YL, Luo J, Filtz TM, Woodcock EA. Phospholipase Cβ1b associates with a Shank3 complex at the cardiac sarcolemma. FASEB J. 2011;25:1040–1047. doi: 10.1096/fj.10-171470. [DOI] [PubMed] [Google Scholar]
- Hammond GRV, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 Are Essential But Independent Lipid Determinants of Membrane Identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higazi DR, Fearnley CJ, Drawnel FM, Talasila A, Corps EM, Ritter O, McDonald F, Mikoshiba K, Bootman MD, Roderick HL. Endothelin-1-Stimulated InsP3-Induced Ca2+ Release Is a Nexus for Hypertrophic Signaling in Cardiac Myocytes. Mol Cell. 2009;33:472–482. doi: 10.1016/j.molcel.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Kapiloff MS, Jackson N, Airhart N. mAKAP and the ryanodine receptor are part of a multicomponent signaling complex on the cardiomyocyte nuclear envelope. J Cell Sci. 2001;114:3167–3176. doi: 10.1242/jcs.114.17.3167. [DOI] [PubMed] [Google Scholar]
- Kapiloff MS, Schillace RV, Westphal AM, Scott JD. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. Journal of Cell Science. 1999;112:2725–2736. doi: 10.1242/jcs.112.16.2725. [DOI] [PubMed] [Google Scholar]
- Kelley GG, Reks SE, Ondrako JM, Smrcka AV. Phospholipase Ce: a novel Ras effector. EMBO J. 2001;20:743–754. doi: 10.1093/emboj/20.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keune W.j., Bultsma Y, Sommer L, Jones D, Divecha N. Phosphoinositide signalling in the nucleus. Advances in Enzyme Regulation. 2011;51:91–99. doi: 10.1016/j.advenzreg.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Knowlton KU, Michel MC, Itani M, Shubeita HE, Ishihara K, Brown JH, Chien KR. The α 1A-adrenergic receptor subtype mediates biochemical, molecular, and morphologic features of cultured myocardial cell hypertrophy. J Biol Chem. 1993;268:15374–15380. [PubMed] [Google Scholar]
- Kronebusch PJ, Singer SJ. The microtubule-organizing complex and the Golgi apparatus are co-localized around the entire nuclear envelope of interphase cardiac myocytes. J Cell Sci. 1987;88:25–34. doi: 10.1242/jcs.88.1.25. [DOI] [PubMed] [Google Scholar]
- Kunkel MT, Newton AC. Calcium Transduces Plasma Membrane Receptor Signals to Produce Diacylglycerol at Golgi Membranes. J Biol Chem. 2010;285:22748–22752. doi: 10.1074/jbc.C110.123133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel MT, Toker A, Tsien RY, Newton AC. Calcium-dependent Regulation of Protein Kinase D Revealed by a Genetically Encoded Kinase Activity Reporter. J Biol Chem. 2007;282:6733–6742. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA. Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovascular research. 2007;73:667–677. doi: 10.1016/j.cardiores.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Bodi I, Maillet M, DeSantiago J, Domeier TL, Mikoshiba K, Lorenz JN, Blatter LA, Bers DM, Molkentin JD. The IP3 Receptor Regulates Cardiac Hypertrophy in Response to Select Stimuli / Novelty and Significance. Circ Res. 2010;107:659–666. doi: 10.1161/CIRCRESAHA.110.220038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich EA, Malik S, Goonasekera SA, Blaxall BC, Kelley GG, Dirksen RT, Smrcka AV. Epac and Phospholipase Cε Regulate Ca2+ Release in the Heart by Activation of Protein Kinase Cε and Calcium-Calmodulin Kinase II. J Biol Chem. 2009;284:1514–1522. doi: 10.1074/jbc.M806994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich EA, Wang H, Malik S, Kaproth-Joslin KA, Blaxall BC, Kelley GG, Dirksen RT, Smrcka AV. Epac-mediated Activation of Phospholipase Cε Plays a Critical Role in β-Adrenergic Receptor-dependent Enhancement of Ca2+ Mobilization in Cardiac Myocytes. J Biol Chem. 2007;282:5488–5495. doi: 10.1074/jbc.M608495200. [DOI] [PubMed] [Google Scholar]
- Ramazzotti G, Faenza I, Fiume R, Matteucci A, Piazzi M, Follo MY, Cocco L. The physiology and pathology of inositide signaling in the nucleus. J Cell Physiol. 2011;226:14–20. doi: 10.1002/jcp.22334. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Suh PG, Ryu SH, Lee SY. Studies of inositol phospholipid-specific phospholipase C. Science. 1989;244:546–550. doi: 10.1126/science.2541501. [DOI] [PubMed] [Google Scholar]
- Rockman HA, Koch WJ, Lefkowitz RJ. Seven-transmembrane-spanning receptors and heart function. Nature. 2002;415:206–212. doi: 10.1038/415206a. [DOI] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. The Human Phosphatidylinositol Phosphatase SAC1 Interacts with the Coatomer I Complex. Journal of Biological Chemistry. 2003;278:52689–52699. doi: 10.1074/jbc.M307983200. [DOI] [PubMed] [Google Scholar]
- Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology (Bethesda) 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert JP, Wing MR, Snyder JT, Gershburg S, Sondek J, Harden TK. RhoA Activates Purified Phospholipase Cε by a Guanine Nucleotide-dependent Mechanism. J Biol Chem. 2004;279:47992–47997. doi: 10.1074/jbc.M407111200. [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Brown JH, Holz GG. Role of phospholipase Cε in physiological phosphoinositide signaling networks. Cell Signal. 2012;29:1333–1343. doi: 10.1016/j.cellsig.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally Regulated and Tissue-Specific Gene Manipulations in the Adult and Embryonic Heart Using a Tamoxifen-Inducible Cre Protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Steinberg SF. Regulation of protein kinase D1 activity. Mol Pharmacol. 2012;81:284–291. doi: 10.1124/mol.111.075986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Paintrand M, Berger EG, Bornens M. The Golgi apparatus remains associated with microtubule organizing centers during myogenesis. J Cell Biol. 1985;101:630–638. doi: 10.1083/jcb.101.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. The Journal of Cell Biology. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Oestreich EA, Maekawa N, Bullard TA, Vikstrom KL, Dirksen RT, Kelley GG, Blaxall BC, Smrcka AV. Phospholipase C ε Modulates β–Adrenergic Receptor- Dependent Cardiac Contraction and Inhibits Cardiac Hypertrophy. Circ Res. 2005;97:1305–1313. doi: 10.1161/01.RES.0000196578.15385.bb. [DOI] [PubMed] [Google Scholar]
- Wu X, Zhang T, Bossuyt J, Li X, McKinsey TA, Dedman JR, Olson EN, Chen J, Brown JH, Bers DM. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J Clin Invest. 2006;116:675–682. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Malik S, Kelley GG, Kapiloff MS, Smrcka AV. Phospholipase Cε Scaffolds to Muscle-specific A Kinase Anchoring Protein (mAKAPβ) and Integrates Multiple Hypertrophic Stimuli in Cardiac Myocytes. J Biol Chem. 2011;286:23012–23021. doi: 10.1074/jbc.M111.231993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.