Abstract
Perturbation of ribosomal biogenesis has recently emerged as a relevant p53-activating pathway. This pathway can be initiated by depletion of certain ribosomal proteins, which is followed by the binding and inhibition of MDM2 by a different subset of ribosomal proteins that includes L11. Here, we report that depletion of L37 leads to cell cycle arrest in a L11- and p53-dependent manner. DNA damage can initiate ribosomal stress, although little is known about the mechanisms involved. We have found that some genotoxic insults, namely UV light and cisplatin, lead to proteasomal degradation of L37 in the nucleoplasm and to the ensuing L11-dependent stabilization of p53. Moreover, ectopic L37 overexpression can attenuate the DNA damage response mediated by p53. These results support the concept that DNA damage-induced proteasomal degradation of L37 constitutes a mechanistic link between DNA damage and the ribosomal stress pathway, and is a relevant contributing signaling pathway for the activation of p53 in response to DNA damage.
Keywords: p53, DNA damage, ribosomal stress, nucleolus, ribosome
Introduction
Mammalian cells are endowed with multiple mechanisms to prevent tumor development. The p53 transcription factor is key in cancer protection due to its pivotal role in preventing damaged cells from becoming malignant.1,2 Hence, loss of function of p53 is a frequent event in human cancers. In normal cells, p53 activity must be tightly controlled since its deregulated activity is incompatible with cell survival and proliferation. This regulation is accomplished primarily by the proto-oncoprotein MDM2, a ubiquitin-ligase that binds and inhibits p53 under physiological conditions.3,4 Conversely, inhibition of MDM2 function is a universal requirement for p53 activation. A number of well-understood stress signaling pathways lead to MDM2 inhibition. In particular, DNA damage induces phosphorylation of both MDM2 and p53, which reduces their mutual affinity and activates p53. Additionally, oncogenic signals upregulate the ARF tumor suppressor, which binds and inhibits MDM2, consequently stabilizing p53.
During the last few years, a new p53-activating stress signaling pathway has emerged in response to disruption of ribosomal biogenesis.5,6 This new type of p53-activating stimulus is known as nucleolar or ribosomal stress, and it involves the association of a subset of ribosomal proteins (RPs), including L11,7,8 L23,9,10 L59 and S7,11,12 to MDM2. Upon binding to MDM2, these effector RPs inhibit MDM2-mediated p53 ubiquitylation and degradation. Knockdown of each of these effector RPs attenuates p53 stabilization in response to ribosomal stress suggesting that all of them may contribute additively to p53 activation.
Cellular insults that result in ribosomal stress include the inhibition of RNA polymerase I and, hence, of ribosomal RNA production by low doses of actinomycin D (ActD). Similarly, impairment of rRNA processing by expression of a dominant negative mutant of Bop113 or treatment with mycophenolic acid14 or with 5-flourouracil15 induce the ribosomal stress pathway. Depletion of certain nucleolar and ribosomal proteins also activates the ribosomal stress pathway, as it has been shown for nucleostemin,16 S6,17 S9,18 L7,17 L23,10 L2919 and L30.19 All these insults trigger a p53-mediated checkpoint, generally dependent on the effector RPs and, particularly, on L11, which is the most studied effector RP. The activation of p53 by the ribosomal stress pathway allows cells to halt proliferation under conditions of impaired ribosomal biogenesis, thus coupling cell growth and cell division.
It is well established that DNA damage activates p53 through a number of kinase signaling cascades, notably including ATM, ATR and CHK1,2 kinases.2 However, in addition to this, there is evidence that DNA damage may also disrupt ribosomal biogenesis,20,21 and also that L11 and S7 contribute to the full stabilization of p53 in response to DNA damage.12 In this study we show that depletion of L37 engages the ribosomal stress pathway that results in p53 activation. In addition, we report that L37 undergoes proteasomal degradation in response to certain p53-activating DNA damage insults. Lastly, we find that the p53 response to DNA damage is attenuated either by L37 overexpression or by L11 depletion. Taken together these results suggest that the regulation L37 stability by specific DNA damage agents could contribute to p53-mediated DNA damage response.
Results
Knockdown of L37 results in p53 stabilization and activation
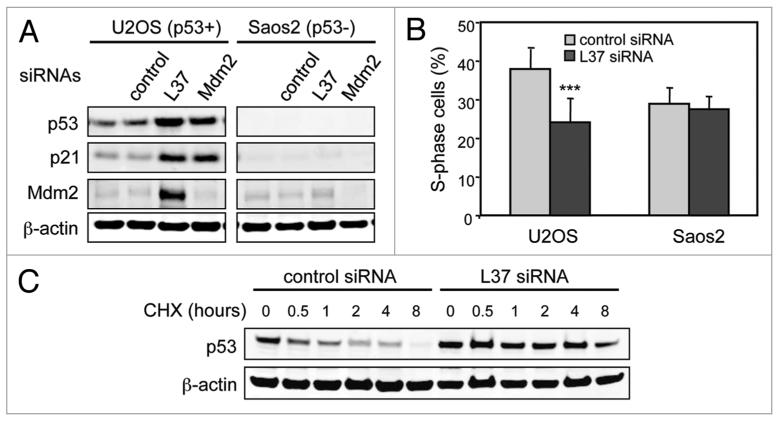
We have previously performed an unbiased large-scale screening of an shRNA library to identify negative regulators of p53.22 Ribosomal protein L37 was among the initially identified genes. Knockdown of L37 significantly induced p53 activity on a p53-responsive promoter (data not shown). Based on this, we sought to analyze the effect of L37 depletion on p53 levels. A GFP-L37-expressing U2OS cell line allowed us to visualize L37 in living cells. GFP-L37 fusion protein localized exclusively in the nucleolar compartment, and 48 hours after siRNA transfection most cells showed a drastic reduction of GFP-L37 levels and fluorescence (Sup. Fig. 1A and B). The efficiency of the L37 siRNA was further confirmed by co-transfection of a V5-tagged L37 plasmid (Sup. Fig. 1C). L37 siRNA reduced the endogenous levels of L37 mRNA as well (see below, Fig. 3C). Interestingly, silencing of endogenous L37 resulted in increased protein levels of p53 and its targets p21 and MDM2 in p53-positive U2OS cells, but not in p53-negative Saos2 cells (Fig. 1A). In addition, depletion of L37 led to cell cycle arrest 48 hours after siRNA transfection in U2OS cells but not in Saos2 cells (Fig. 1B and 2B). Ribosomes have a half-life of 5 days23 and, therefore, this p53-dependent response at 72 hours is probably independent of a decreased ribosomal content. Indeed, when analyzed 5 days post-transfection, silencing of L37 resulted in inhibition of cell proliferation in Saos2 cells, probably due to a reduction in ribosomes (data not shown). The upregulation of p53 following knockdown of L37 was due to p53 stabilization since, in the presence of cycloheximide, L37 siRNA transfection prolonged the half-life of p53 compared to control siRNA (Fig. 1C). These observations indicate that L37 depletion leads to p53 stabilization and activation.
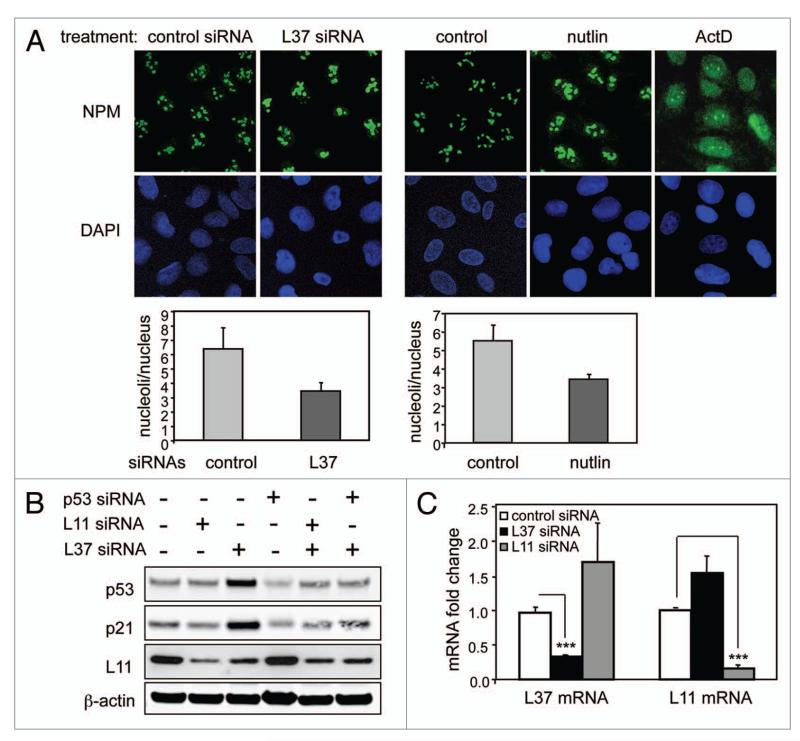
Figure 3.
L37 depletion does not result in nucleolar breakdown. (A) U2OS cells were transfected with control or L37 siRNAs for 72 h (left) or treated with nutlin for 72 h or with actinomycin D for 16 h (right). NpM expression was detected by immunofluorescence using a polyclonal antibody against B23 (top). Histograms showing the number of nucleoli per cellular nucleus are shown (bottom). (B) U2OS cells were interfered with the indicated siRNAs, and the levels of the indicated proteins were determined 72 h later by immunoblotting. (C) U2OS cells were interfered with the indicated siRNAs, and 72 h later mRNA abundance was quantified by qRt-pCR. Data are shown as means ± SD of three independent experiments assayed in triplicate. the Ct values were corrected by β-actin and fold expression values are relative to control siRNA. ***Student’s t-test, p < 0.001.
Figure 1.
Depletion of L37 stabilizes and induces p53. (A) U2OS and Saos2 cells were transfected with the indicated siRNAs, and the levels of the indicated proteins were determined 72 h later by immunoblotting. (B) percentage of S-phase cells in control (non-targeting) and L37 siRNA treated cells. Data were obtained 48 h after siRNA treatment. Data are shown as means ± SD of four independent experiments. ***Student’s t-test, p < 0.001. (C) U2OS cells were transfected with control (non-targeting) or L37 siRNAs and, 48 h later, cells were treated with cycloheximide (CHX) for the indicated periods of time. the indicated proteins were detected by immunoblotting.
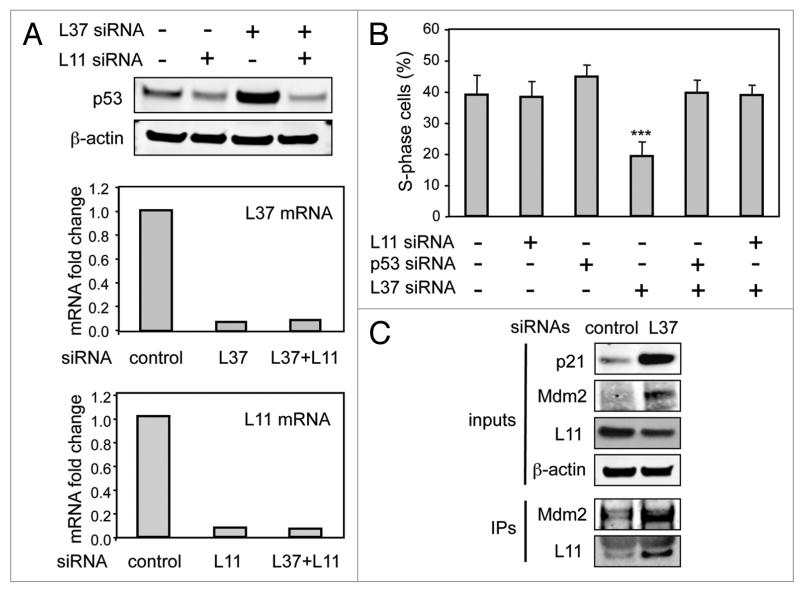
Figure 2.
L11 mediates the effects of L37 depletion. (A) U2OS cells were transfected with control, L37 or L11 siRNAs, and 72 h after, protein levels of p53 were determined by immunoblotting (upper part), and changes in L37 and L11 mRNA levels were quantified by qRt-pCR (lower parts). For qRt-pCR, the Ct values were corrected by β-actin and fold expression values are relative to control siRNA. (B) U2OS cells were transfected with the indicated siRNAs and, 48 h later, S-phase cells were analyzed by flow cytometry. Data corresponds to four independent experiments ***Student’s t-test compared to each of the other conditions, p < 0.001. (C) U2OS cells were transfected with control or L37 siRNAs and, 48 h later, identical amounts of cellular lysates were immunoprecipitated with anti-MDM2 antibody, and protein levels of the indicated proteins were determined by immunoblotting in the inputs and in the immunoprecipitates (Ips).
Silencing of L37 elicits ribosomal stress
To explore whether p53 activation by L37 silencing was a consequence of the engagement of the ribosomal stress pathway, we analyzed its dependence on L11. We observed that simultaneous knockdown of L37 and L11 impaired p53 upregulation by L37 siRNA (Fig. 2A and 3B). Similarly, L11 depletion prevented cell cycle arrest induced by L37 downregulation (Fig. 2B). Next we sought to analyze L11 and MDM2 interaction in control and L37 siRNA transfected cells. Knockdown of L37 led to an increase in the total levels of MDM2 as well as in the levels of MDM2/L11 complexes (Fig. 2C). This indicates that in L37-depleted cells, MDM2 is maintained inactive by association with L11. These results demonstrate that the ribosomal stress pathway mediates the activation of p53 by L37 knockdown.
Nucleolar effects of L37 knockdown
Perturbation of ribosomal biogenesis may impinge on nucleolar morphology in multiple manners. In the case of actinomycin D and certain DNA damage agents, there is a dramatic disorganization of the nucleolar structure.20,21 In other cases, such as depletion of ribosomal proteins L23, L29 or L30, there is nucleolar enlargement and release of nucleolar proteins to the nucleoplasm.10,19 These disparate effects on nucleolar morphology are often referred to as nucleolar disruption. Visualization of nucleoli by immunofluorescence detection of nucleophosmin (NPM) indicated that L37 depletion results in enlarged nucleoli concomitant with a reduction in the number of nucleoli per nucleus (Fig. 3A). Similar results were obtained using fibrilarin to detect nucleoli (not shown). As a positive control, nucleolar disruption was readily detected upon treatment with actinomycin D (Fig. 3A). Taking into account that L37 depleted cells have an impaired proliferation (see Figs. 1B and 2B), we wondered whether the morphological changes observed in nucleoli could be secondary to a proliferative arrest induced by L37 knockdown. With this in mind we treated cells with nutlin, a synthetic MDM2 inhibitor that induces a p53-dependent G1 arrest.24 Interestingly, the nucleolar alterations observed in nutlin-treated cells were identical to those caused by L37 depletion (Fig. 3A), suggesting that p53-mediated cell cycle arrest, in the absence of ribosomal stress, may result in nucleolar enlargement.
It has been recently reported that depletion of S6 has no effect on nucleolar integrity, but it results in increased levels of L11 and L11-mediated p53 activation.17 In the case of L37 knockdown, we observed a decrease in the protein levels of L11 (Fig. 3B). Reduction of L11 protein levels was not due to transcriptional regulation because its mRNA abundance remained unchanged upon L37 depletion (Fig. 3C). Similar to our observations, depletion of L7 or S9 also results both in decreased L11 and in L11-dependent stabilization of p53.17,18 Taken together these results suggest that L37-depletion triggers the ribosomal stress pathway in the absence of nucleolar breakdown without increasing total L11 levels.
L37 degradation induced by DNA damage
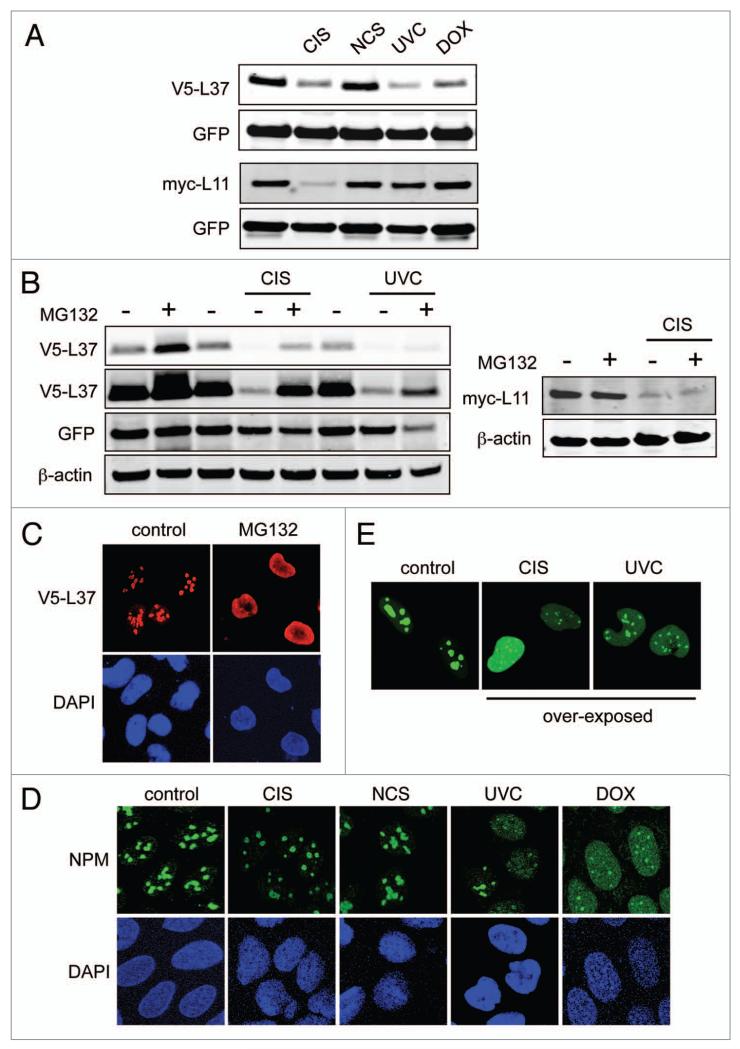
Recent evidence has shown that a variety of DNA damage agents impair ribosomal biogenesis21 and induce L11- and S7-dependent p53 stabilization.12 Ribosomal protein S27L is a transcriptional target of p53 and, consequently, it is induced by certain DNA damaging agents.25,26 However, beyond S27L, little is known about the impact of DNA damage on the levels of ribosomal proteins. Interestingly, we observed that cisplatin (CIS), UV light (UVC) and doxorubicin (DOX), but not neocarzinostatin (NCS), decreased ectopically expressed L37 (Fig. 4A). By contrast, transfected L11 was only affected by CIS, but not by NCS, UVC or DOX (Fig. 4A). This indicates that the stability of L37 and L11 is differentially regulated in response to DNA damage. In support of this, we observed that L37, but not L11, was significantly stabilized in the presence of the proteasomal inhibitor MG132, even after treatment with CIS or UVC (Fig. 4B). L37 degradation by DNA damage was not secondary to p53 activation because neither nutlin nor overexpression of p53 or MDM2 affected L37 levels (Sup. Fig. 2A and B). Moreover, L37 degradation was also observed in p53-null cells (Sup. Fig. 2C). Ribosomal proteins are degraded in the nucleoplasm27 and, in agreement with this, we observed a substantial nucleoplasmic accumulation of exogenous L37 when proteasomal degradation was inhibited (Fig. 4C). Interestingly, cells treated with CIS, UVC or DOX, but not NCS, showed nucleolar breakdown (Fig. 4D) and low, albeit detectable, nucleoplasmic L37 (Fig. 4E). These results suggest that DNA damage-induced nucleolar disruption could contribute to L37 degradation. However, nucleolar disruption by actinomycin D (ActD) did not promote L37 degradation (Sup. Fig. 2D and E). Therefore, L37 degradation by severe DNA damage involves additional events concurrent with nucleolar breakdown. Together, these observations indicate that L37, but not L11, undergoes proteasomal degradation in the nucleoplasm that is accelerated by certain DNA-damaging insults.
Figure 4.
L37 undergoes proteasomal degradation in the nucleoplasm. (A) U2OS cells were transfected with V5-L37 or myc-L11 together with peGF-C3 plasmid. twenty-four hours later cells were either irradiated (UVC) or treated with cisplatin (CIS), neocarzinostatin (NCS) or doxorubicin (DOX) for an additional period of 16 hours. protein abundance was determined by western blotting. (B) U2OS cells were transfected with V5-L37 (left) or myc-L11 (right), and 24 hours later they were either irradiated (UVC) or treated with cisplatin (CIS) in the presence or absence of MG132 for an additional period of 16 hours. protein levels were analyzed by immunobloting. (C) U2OS cells were transfected with V5-L37 expressing-plasmid, and 24 hours later they were treated with MG132 for an additional period of 16 hours. V5-L37 protein was detected with an antibody to V5 tag. (D) U2OS cells were treated with the indicated DNA-damaging agents for 16 hours. NpM detection was used as a marker for nucleolar integrity. (e) GFp-L37 expressing U2OS cells were irradiated (UVC) or treated with cisplatin (CIS) for 16 hours and then analyzed by immunofluorescence.
L37 modulates the DNA damage response
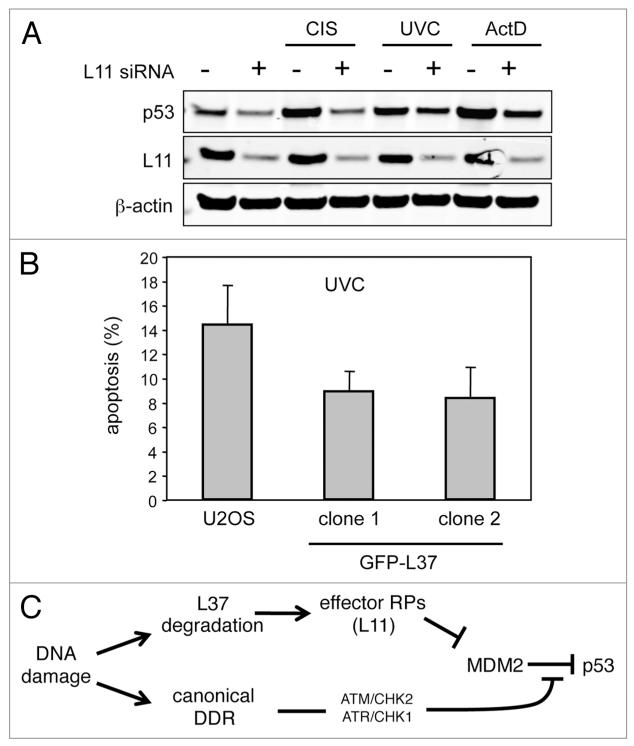
The above observations linking L37 with p53 activity and with the DNA damage response, led us to investigate whether L37 degradation by DNA damage could contribute to p53 activation in response to genotoxic stress. Previous investigators have shown that downregulation of L11 attenuates p53 activation upon treatment with certain DNA damage agents.12 We confirmed and extended these observations by using CIS and UVC, whose ability to stabilize p53 was significantly impaired in cells depleted of L11 (Fig. 5A). Once it wasestablished that CIS and UVC activate p53 in part through the ribosomal stress pathway mediated by L11, we wondered whether L37 overexpression would attenuate p53 response to these agents. Compared to L11, our data with L37 predict that it should have the opposite effect on p53. Interestingly, U2OS clones stably expressing GFP-L37 showed a reduced sensitivity to UVC-induced apoptosis (Fig. 5B). Taken together these results indicate that L37 protein levels can modulate p53 cellular response to DNA damage.
Figure 5.
L37 and L11 levels modulate DNA damage response. (A) U2OS cells were transfected with control or L11 siRNAs, and 48 hours later they were treated with the indicated DNA-damaging agents for an additional period of 16 hours. protein abundance was analyzed by western blotting. (B) U2OS cells and U2OS clones stably-expressing GFp-L37 were UVC irradiated. Apoptosis was analyzed by flow cytometry 24 hours after irradiation. Data corresponds to the means ± SD of three independent experiments. (C) Model summarizing our data. We propose that the ribosomal stress pathway contributes to the activation of p53 by DNA damage through the canonical DNA damage response (DDR) pathway. DNA damage may trigger the ribosomal stress pathway by degrading L37.
Discussion
We report here that depletion of ribosomal protein L37, similar to depletion of other ribosomal proteins (RPs), namely L7,17 L29,19,28 L30,19,28 S617 and S9,29 induces the ribosomal stress pathway. This pathway involves the inhibition of MDM2 through its association with a subset of effector RPs, including L11,30 L5,9 S712 and S3.31 According to their role in this pathway, ribosomal proteins can be classified into those whose depletion can initiate the ribosomal stress pathway, and those that act as effectors of the pathway by binding and inhibiting MDM2. Exceptions are known, such as protein L23, that belong to both categories.10 Our data here indicate that L37 belongs to the group of RPs able to initiate the ribosomal stress pathway when their levels decrease. Moreover, we present evidence linking L37 degradation to DNA damage and, consequently, to p53 activation by genotoxic stress (Fig. 5C).
Ribosomal stress is often associated with the physical breakdown of the nucleolus, which results in the dispersion of ribosomal proteins into the nucleoplasm where they can interact with MDM2. However, nucleolar breakdown does not seem to be an obligatory step for activation of the ribosomal stress pathway. In particular, S6 depletion does not produce nucleolar breakdown, but rather augments the translation of L11 whose increased levels lead to inhibition of MDM2.17 In the case of L37 knockdown, we have not found evidence either of nucleolar breakdown or of increased L11 levels. In particular, depletion of L37 results in enlarged nucleoli and reduced number of nucleoli per nucleus. These morphological changes can be mimicked by the synthetic MDM2-inhibitor nutlin, which induces a robust p53-dependent cell cycle arrest without interfering with ribosomal biogenesis. Therefore, the enlarged nucleoli induced by L37 depletion or by nutlin are likely secondary to the cellular proliferation arrest associated with both conditions. Regarding L11, interference of L37 produced a reduction of L11 levels, an effect that has also been observed upon depletion of L717 or S9.18 Nonetheless, despite a reduction in L11 levels, we also show that the total amount of L11/MDM2 complexes is increased upon L37 depletion. These observations suggest the existence of a complex regulation of the L11/MDM2 interaction. In this regard, for example, modification of L11 by NEDDylation has recently been involved in L11 signaling to p53.19,32
Most DNA damaging agents cause nucleolar breakdown20,21 and inhibit ribosomal biogenesis.21 In addition, we and others12 have observed that depletion of L11 or S7 drastically reduces p53 stabilization in response to genotoxic stress. Conversely, we also report that overexpression of L37 attenuates the response of p53 to DNA damage. Together, these observations strongly implicate the ribosomal stress pathway as an integral part of p53 activation by DNA damage.
The mechanisms by which DNA damage impair ribosomal biogenesis are beginning to be unraveled. In particular, DNA damage activates JNK2, which phosphorylates and inhibits the transcription factor TIF-IA involved in the transcription of rDNA.33 Adding another possible connection between DNA damage and ribosomal biogenesis, we have found that L37 is degraded in response to some DNA damaging agents (cisplatin and UVC). Other RPs, such as S712 and L26,34 are ubiquitylated by MDM2. However, we have found that L37 degradation is mediated by the proteasome independently of MDM2. In contrast to this, L11 was not subjected to proteasomal degradation either in the presence or absence of DNA damage, suggesting that the stability of each individual ribosomal protein is regulated in a specific manner. In conclusion, given that depletion of L37 leads to a robust activation of p53, DNA damage-mediated L37 degradation could constitute a relevant mechanism for the activation of p53 in response to genotoxic stress (Fig. 5C).
Materials and Methods
Plasmids
L37 cDNA was cloned into a pcDNA vector in which a duplicated V5 tag sequence (Glu-Lys-Pro-Ile-Pro-Asn-Leu-Leu-Gly-Leu-Asp-Ser-Thr) was inserted into the BamH1-EcoRI restriction sites. In addition, L37 was cloned into the pEGFP-C3 plasmid (Invitrogen) in order to generate a GFP-L37 fusion protein. The pcDNA3-myc3-L11-expressing plasmid was purchased from Addgene. Plasmids are available upon request.
RNA interference
The siRNA duplexes were synthesized by Dharmacon and transfected with Dharmafect (Dharmacon) following the manufarturer’s instructions. RNA, protein extraction or immunofluorescence analysis were carried out 24 to 72 hours after siRNA transfections. The following siRNA sequences were used to knockdown endogenous genes: L37, GCG CAA GAG AAA GUA UAA CUU; p53, GAC UCC AGU GGU AAU CUA CUU; MDM2, CCA CCU CAC AGA UUC CAG CUU. Non-targeting siRNAs (Dharmacon) were used as negative controls.
Apoptosis and cell cycle analysis
Cells were seeded at sub-confluency in 60 mm plates, and 24 h later they were UVC irradiated (10 J/m2). After 24 hours treatment, floating and attached cells were collected and fixed in 70% ethanol. Fixed cells were treated with RNAse 100 μg/ml (Qiagen) and stained with propidium iodide (Sigma). Subsequently the cell cycle distribution was analyzed by flow cytometry in FACSscalibur (BD Biosciences, Franklin Lakes, NJ) and quantified with the program MoFit. Interfered cells were seeded at sub-confluency 24 hours after siRNA transfection, and flow cytometry analysis was performed 24 h later.
Immunoblotting
Cells were interfered with siRNAs for 24 to 72 hours or treated with p53-inducing agents (UVC 10 J/m2, 40 μM cisplatin, 10 μM nutlin, 200 μg/ml neocarzinostatin or 0.9 μM doxorubicin) for the indicated times and then harvested in lysis buffer containing 1% SDS and 1% Triton X-100. Identical amounts of protein were resolved by SDS/PAGE and electro-transferred to nitrocellulose membranes. Blots were incubated first with the following primary antibodies: anti-p53 DO-1, Santa Cruz, anti-β-actin (AC-15, Sigma), anti-V5 (Invitrogen), anti-p21 (C-19, Santa Cruz), anti-MDM2 (2A10, Abcam and SMP14, Santa Cruz), anti-myc tag (9E10, Santa Cruz) and anti-GFP (Roche) and subsequently with the corresponding Alexa-fluor 680 secondary antibodies (Invitrogen). Proteins were detected with Li-Cor Oddyssey Infrared imager. The anti-L11 antibody was a generous gift from Dr. Y. Zhang.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated with Trizol reagent (Invitrogen). cDNA synthesis was performed using 5 μg total RNA, Superscript II Reverse transcriptase (Invitrogen) and random primers (Invitrogen). For either mRNA or ChIP experiments real-time PCR was performed using SybrGreenER Master Mix in a 7500 Fast Real-Time PCR System (Applied Biosystems). All the reactions were performed in triplicate and normalized by β-actin. The mRNA levels were analyzed using the following specific primers: L37-F, TCG CAA TAA GAC GCA CAC GTT; L37-R, ACC AGT TCC GGT GGT ATT TCG; L11-F, GCG CAG GAT CAA GGT GAA AAG; L11-R, GCT CGC GTC AGT CTG TCT C.
Immunofluorescence
The immunofluorescence procedure was previously described.35 Nucleophosmin (NPM) was detected using a rabbit polyclonal antibody (C-19, Santa Cruz), and a mouse monoclonal antibody to V5 (Invitrogen) was used to detect V5-L37. Alexa-488-conjugated goat anti-rabbit (Molecular Probes) and Cy3-conjugated goat anti-mouse (Jackson Immunoresearch) were used as secondary antibodies.
Immunoprecipitation
Immunoprecipitations were performed as previously described.36 Endogenous MDM2 was immunoprecipitated with 1 μg of antibody to MDM2 (SMP14, Santa Cruz) and TrueBlot anti-mouse IP beads (eBioscience). The immunoprecipitated proteins were detected with SMP14 antibody to MDM2, a polyclonal antibody to L11 (a generous gift from Dr. Zhang and Dr. Volarevic) and the secondary antibodies Alexa fluor 680 anti-mouse (Invitrogen) and TrueBlot anti-rabbit (eBioscience). Proteins were detected with both ECL and Li-Cor Oddyssey Infrared imager.
Acknowledgements
We thank Drs. Yanping Zhang and Siniša Volarevic for generously providing us with critical reagents. S.L. is recipient of a grant from the MICINN (SAF2009-09343). Work in the laboratory of M.S. is funded by the CNIO and by grants from the Spanish Ministry of Science (SAF and CONSOLIDER), the Regional Government of Madrid (GsSTEM), the European Union (PROTEOMAGE), the European Research Council (ERC Advanced Grant), and the “Marcelino Botin” Foundation.
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/LlanosCC9-19-sup.pdf
References
- 1.Junttila MR, Evan GI. p53—a jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–9. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 2.Meek DW. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer. 2009;9:714–23. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 3.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 5.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: Probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–60. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–77. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 10.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–80. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: Binding to MDM2, stabilization of p53 protein and activation of p53 function. Oncogene. 2007;26:5029–37. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316–26. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–55. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem. 2008;283:12387–92. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052–9. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 16.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28:4365–76. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–8. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindstrom MS, Nister M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PLoS One. 2010;5:e9578. doi: 10.1371/journal.pone.0009578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun XX, Wang YG, Xirodimas DP, Dai MS. Perturbation of 60S ribosomal biogenesis results in ribosomal protein L5 and L11-dependent p53 activation. J Biol Chem. 2010;285:25812–21. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–77. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burger K, Muhl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–25. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llanos S, Efeyan A, Monsech J, Dominguez O, Serrano M. A high-throughput loss-of-function screening identifies novel p53 regulators. Cell Cycle. 2006;5:1880–5. doi: 10.4161/cc.5.16.3140. [DOI] [PubMed] [Google Scholar]
- 23.Caldarola S, De Stefano MC, Amaldi F, Loreni F. Synthesis and function of ribosomal proteins—fading models and new perspectives. FEBS J. 2009;276:3199–210. doi: 10.1111/j.1742-4658.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- 24.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 25.He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–16. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Tan J, Zhuang L, Banerjee B, Yang X, Chau JF, et al. Ribosomal protein S27-like, a p53-inducible modulator of cell fate in response to genotoxic stress. Cancer Res. 2007;67:11317–26. doi: 10.1158/0008-5472.CAN-07-1088. [DOI] [PubMed] [Google Scholar]
- 27.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–60. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu JJ, Huang BH, Zhang J, Carson DD, Hooi SC. Repression of HIP/RPL29 expression induces differentiation in colon cancer cells. J Cell Physiol. 2006;207:287–92. doi: 10.1002/jcp.20589. [DOI] [PubMed] [Google Scholar]
- 29.Lindstrom MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem. 2008;283:15568–76. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–12. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multifunctional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair. 2009;8:1215–24. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–9. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and downregulates rRNA synthesis. Genes Dev. 2005;19:933–41. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–9. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol. 2001;3:445–52. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- 36.Llanos S, Cuadrado A, Serrano M. MSK2 inhibits p53 activity in the absence of stress. Sci Signal. 2009;2:57. doi: 10.1126/scisignal.2000205. [DOI] [PMC free article] [PubMed] [Google Scholar]