Abstract
Nanog levels in pluripotent stem cells are heterogeneous and reflect two different and interchangeable cell states, respectively poised to self-renew (Nanog-high subpopulation) or to differentiate (Nanog-low subpopulation). However, little is known about the mechanisms responsible for this pattern of Nanog expression. Here, we have examined the impact of the histone methyltransferase Ezh2 on pluripotent stem cells and on Nanog expression. Interestingly, induced pluripotent stem (iPS) cells lacking Ezh2 presented higher levels of Nanog due to a relative expansion of the Nanog-high subpopulation, and this was associated to severe defects in differentiation. Moreover, we found that the Nanog promoter in embryonic stem (ES) cells and iPS cells coexists in two univalent chromatin configurations, one characterized by H3K4me3 and the other by H3K27me3, being the latter dependent on the presence of functional Ezh2. Finally, the levels of expression of Ezh2, as well as the amount of H3K27me3 present at the Nanog promoter, were higher in the Nanoglow subpopulation of ES/iPS cells. Together, these data indicate that Ezh2 directly regulates the epigenetic status of the Nanog promoter affecting the balance of Nanog expression in pluripotent stem cells and, therefore, the equilibrium between self-renewal and differentiation.
Keywords: Ezh2, Polycomb, Nanog, iPS, epigenetics, stem cells
Introduction
Polycomb repressive complex 2 (PRC2) is one of several protein complexes that participate in the establishment and maintenance of specific chromatin configurations, being its most characteristic mark the tri-methylation of histone 3 at residue lysine 27 (H3K27me3).1 The core of PRC2 is formed by three protein subunits, namely, Eed, Suz12 and Ezh2, where Ezh2 is the catalytic subunit responsible for H3K27 tri-methylation. Ezh2 is expressed during development, including embryonic stem (ES) cells, whereas its paralog Ezh1 is preferentially expressed in adult differentiated tissues.2 In agreement with this, recent data in ES cells have indicated that Ezh2 is responsible for the bulk of H3K27me3, while Ezh1 plays a detectable and complementary role in determining the levels of H3K27me3 at a subset of PRC2 regulated genes.3
PRC2 is known to play a critical role during differentiation. In particular, ES cells deficient in Eed,4-6 Suz127 or Ezh23 present aberrant patterns of expression of key differentiation regulators. However, despite the accumulated knowledge on ES differentiation, little is known about the putative impact of PRC2 on the maintenance of stemness by self-renewal. Current models of self-renewal assign a critical role to three stemness factors, namely, Nanog, Oct4 and Sox2, which together form an intricate network of interactions and feed-forward loops.8 Particular attention has been given to Nanog due, among other observations, to the fact that its expression is the final and key step in the establishment of self-renewal and pluripotency in a variety of reprogramming settings.9 An intriguing observation regarding Nanog consists on its heterogeneous levels within the inner cell mass at the blastocyst stage,10 as well as, within a given cell population of in vitro cultured ES cells.11,12 Indeed, it is possible to separate Nanog-high and Nanog-low ES cell subpopulations, and each of them, in time, recreates the original mixture of Nanog-high and Nanog-low states, indicating that these states are in a dynamic interchange.11 This in-built heterogeneity of Nanog has been proposed to reflect a complex interplay between Oct4, and probably also Sox2, with Nanog.13 According to the known roles of Nanog in promoting self-renewal and preventing differentiation, cells in the Nanog-high state are prone to self-renew, whereas cells in the Nanog-low state are prone to differentiate.11,12,14 Recently, the Satb1 and Satb2 proteins involved in the regulation of the high-order structure of chromatin have been shown to bind to the Nanog locus, including its promoter region, and to play antagonistic roles in the balance between Nanog-high and Nanog-low states.15 Here, we have addressed the role of Polycomb on self-renewal and differentiation of iPS, and we have uncovered an unexpected role of Polycomb in the regulation of Nanog expression and in the balance between Nanog-high and Nanog-low states.
Results
Generation and validation of Ezh2-null iPS
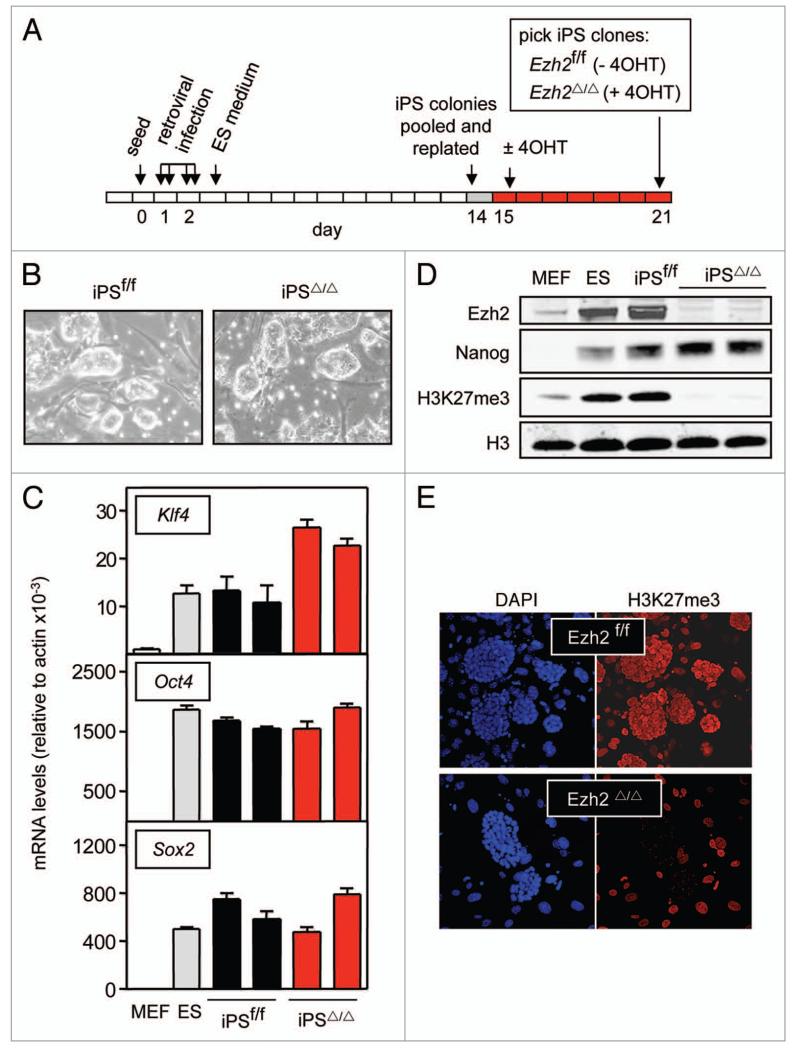
The successful establishment of Ezh2-null ES cell cultures has been achieved by use of a conditional loxP-flanked (floxed) Ezh2 allele and excision after previous establishment of the ES cultures.3 However, at the time we initiated this project, it had been reported that it was not possible to derive viable embryonic stem (ES) cells from Ezh2-null blastocysts.16 To circumvent this reported inability to obtain Ezh2-null ES cells, we took advantage of the possibility to reprogram somatic cells into induced pluripotent stem cells (iPS).17 For this, we used primary mouse embryo fibroblasts (MEFs) carrying a conditional loxP-flanked Ezh2 allele18 and an inducible Cre allele (ROSA26:CreERT).19 Reprogramming with Oct4, Klf4 and Sox2 (3-factors reprogramming) produced iPS colonies carrying the floxed allele (iPSf/f) that were replated in the absence or presence of 4-hydroxytamoxifen (4OHT). As expected, clones grown in the absence of 4OHT retained intact the floxed alleles (iPSf/f, Ezh2-proficient; n = 6 clones) and those grown in the presence of 4OHT had deleted the floxed alleles (iPSΔ/Δ, Ezh2-null; n = 21 clones) (Fig. 1A). The resulting Ezh2-null iPSΔ/Δ cells did not differentiate spontaneously and colonies had a normal morphology (Fig. 1B). Moreover, all the iPS clones analyzed expressed the endogenous stemness factors Klf4, Oct4 and Sox2, regardless of the status of Ezh2 (Fig. 1C, two clones of each genotype are shown as example). Protein analyses by immunoblotting confirmed expression of Ezh2 in iPSf/f cells at levels similar to those in ES cells, and significantly higher than in MEFs; in contrast, as expected, iPSΔ/Δ cells completely lacked Ezh2 (Fig. 1D). Paralleling these results, immunoblotting and immunofluorescence analyses indicated that the total levels of histone 3 tri-methylated at lysine 27 (H3K27me3) were dramatically decreased in iPSΔ/Δ cells to levels that were essentially undetectable (Fig. 1D and E) (note that in Fig. 1E the immunofluorescence signal in the iPSΔ/Δ preparations corresponds to the feeder cells and not to the iPS colonies). In keeping with the observations reported in ES cells lacking Suz12 or Eed,6,7 differentiation genes Gata3 and Gata4 were de-repressed in the Ezh2-null iPS, while the differentiation gene Gata1 remained unchanged (Sup. Fig. 1a). We wondered whether the elimination of Ezh2 had an effect on the mRNA levels of Jmjd3 and Utx, which are the two demethylases that reverse the mark introduced by Ezh2 at H3K27,20 but we did not observe any alteration (Sup. Fig. 1a). Also, Sirt1 is bound to Ezh2-containing complexes in ES cells,21 but no significant changes in Sirt1 mRNA or protein were observed in iPSΔ/Δ cells (Sup. Fig. 1a and 1b). As expected, the stemness protein Nanog was expressed in all the analyzed iPS clones (Fig. 1D). Interestingly, we noticed that the protein levels of Nanog were consistently increased in Ezh2-null iPS (iPSΔ/Δ) compared to Ezh2-proficient iPS (iPSf/f) cells (Fig. 1D and see more clones in Sup. Fig. 1c). In summary, we conclude that Ezh2 is not essential for the maintenance of iPS cells and, at the same time, is responsible for the bulk levels of H3K27me3 in these cells.
Figure 1.
Generation and validation of Ezh2-null ipS. (A) Strategy to obtain Ezh2-null ipS. Ezh2-floxed MeFs were infected with the three reprogramming factors, oct4, Klf4 and Sox2, and two weeks later ipSf/f colonies were pooled and replated in the absence or presence of 4OHT. Colonies of iPSf/f (−4OHt) or iPSΔ/Δ (+4OHt) cells were picked one week later. (B) Morphology of iPSf/f and iPSΔ/Δ colonies. Cells were cultured on feeder fibroblasts. Both genotypes presented a typical morphology and did not differentiate spontaneously. (C) qRT-pCR analysis of stemness markers. Relative endogenous expression of each gene was normalized to actin and error bars represent standard deviation. The figure shows the data for two independent clones. A total of six clones were analyzed per genotype yielding similar results. (D) Protein levels of Ezh2, H3K27me3 and Nanog in iPSf/f and iPSΔ/Δ cells. The figure shows the data for two independent iPSΔ/Δ clones. Additional clones (5 clones of iPSf/f and 8 clones of iPSΔ/Δ) are shown in Supplemental Figure 1c. (E) Confocal immunofluorescence of H3K27me3. The figure is representative of a total analysis of six iPSf/f clones and five iPSΔ/Δ clones. Note that the positive H3K27me3 signal in the iPSΔ/Δ panel corresponds to the feeder fibroblasts.
Impaired differentiation of Ezh2-null cells
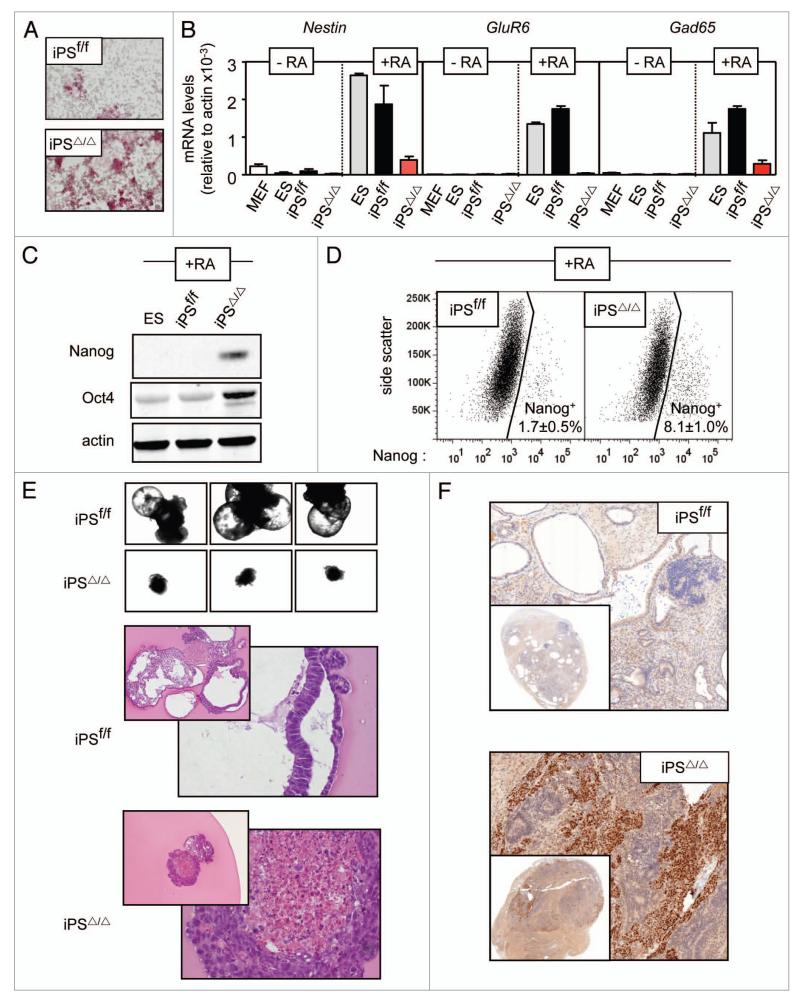
Previous investigators have reported that Ezh2-null ES cells have a defective activation of mesoendodermal differentiation markers when cells are deprived of leukemia inhibitory factor (LIF).3 Here, we have further extended this concept by studying neuroectodermal differentiation, embryoid body formation and teratoma formation. We began by examining neuroectodermal differentiation upon treatment of iPS cells with retinoic acid (RA) in the absence of LIF.22 These differentiation conditions produced, after 4 days, a drastic reduction in the expression of alkaline phosphatase in iPSf/f cells, whereas iPSΔ/Δ cells still retained abundant expression of this stemness marker (Fig. 2A). Examination of three neural differentiation markers, namely, Nestin, GluR6 and Gad65 mRNAs, indicated a clear failure of iPSΔ/Δ cells to upregulate these markers when compared with iPSf/f or ES cultures (Fig. 2B). Further supporting an impaired differentiation in Ezh2-null iPS, the protein levels of Nanog and Oct4 were not completely ablated, as was the case in Ezh2-proficient iPS (Fig. 2C and D). Similar results on defective activation of differentiation markers and defective repression of stemness factors have been reported in ES cells lacking Ezh2 3 or Suz12.7 To further support these data, we performed microarray analyses of gene expression in RA-treated iPS cells lacking or having Ezh2 (n = 3, per genotype). The induction of a number of genes involved in neural differentiation was significantly impaired in RA-treated Ezh2-null iPS cells (see heatmap of 25 neural differentiation genes in Sup. Fig. 2a). Conversely, the repression of genes characteristic of stem cells was also impaired in RA-treated Ezh2-null iPS cells (see heatmap of 12 stem cell genes in Sup. Fig. 2b). A complete list of all the significant changes in this microarray analysis is provided in Supplemental Tables 1 and 2. In summary, absence of Ezh2 reduces the ability of iPS cells to differentiate in vitro in response to retinoic acid.
Figure 2.
Impaired differentiation of Ezh2-null iPS cells. (A) Cultures of iPS cells were treated with retinoic acid (RA) in the absence of LIF for four days and differentiation was assessed by the loss of the stemness marker alkaline phosphatase detected by a histochemical reaction. The figure is representative of a total analysis of three clones of each genotype. (B) Expression by qRT-PCR of the indicated neural markers before (−RA) or after (+RA) RA-induced differentiation. Values correspond to the average and standard deviation of a single clone per genotype. The figure is representative of a total analysis of six clones of each genotype. (C) Levels of stemness proteins Nanog and Oct4 upon RA-induced differentiation. The figure is representative of a total of three clones of each genotype. (D) FACS analysis of Nanog four days after RA-induced differentiation. The figure is representative of a total of three clones of each genotype. Values correspond to the average and standard deviation of the percentage of Nanog-positive cells in a total of three clones per genotype. (E) Embryoid bodies formed by ipSf/f and ipSΔ/Δ cells. Top panels, low magnification images of three embryoid bodies of each genotype. Bottom panels, sections of embryoid bodies stained with hematoxylin and eosin at two different magnifications. Images are representative of a total of six clones of each genotype. (F) Teratomas formed by iPSf/f and iPSΔ/Δ cells. Sections of teratomas were stained for Nanog by immunohistochemistry and counterstained with hematoxylin. Insets show low magnification views of the entire teratomas. Images are representative of a total of two clones of each genotype, and each clone was used to generate six independent teratomas.
For the formation of embryoid bodies, cells were put under non-adherent conditions for 2 weeks and in the absence of LIF. As expected, iPSf/f cells formed mature embryoid bodies with cavities (Fig. 2E) and rhythmic contractions (not shown). In contrast, iPSΔ/Δ cells could not progress beyond small aggregates lacking cavities (Fig. 2E). Finally, injection of iPS cells into nude mice generated visible teratomas after 6 weeks regardless of the presence or absence of Ezh2. Histological analyses indicated that Ezh2-proficient teratomas were largely devoid of Nanog expression (Fig. 2F) and consisted mostly in well-differentiated structures with representation of the three primary germ layers (Sup. Fig. 3a). In contrast, Ezh2-null teratomas were formed by large undifferentiated masses positive for Nanog (Fig. 2F) and Oct4 (Sup. Fig. 3b), surrounded by areas of ectodermal differentiation (Sup. Fig. 3a). Upon inspection of multiple sections, it was possible to identify rare areas within Ezh2-null teratomas showing endodermal or mesodermal differentiation (Sup. Fig. 3a). Together, these results demonstrate the importance of Ezh2 for the differentiation of pluripotent stem cells into the three germ layers.
Role of Ezh2 in the balance between Nanog-high and Nanog-low states
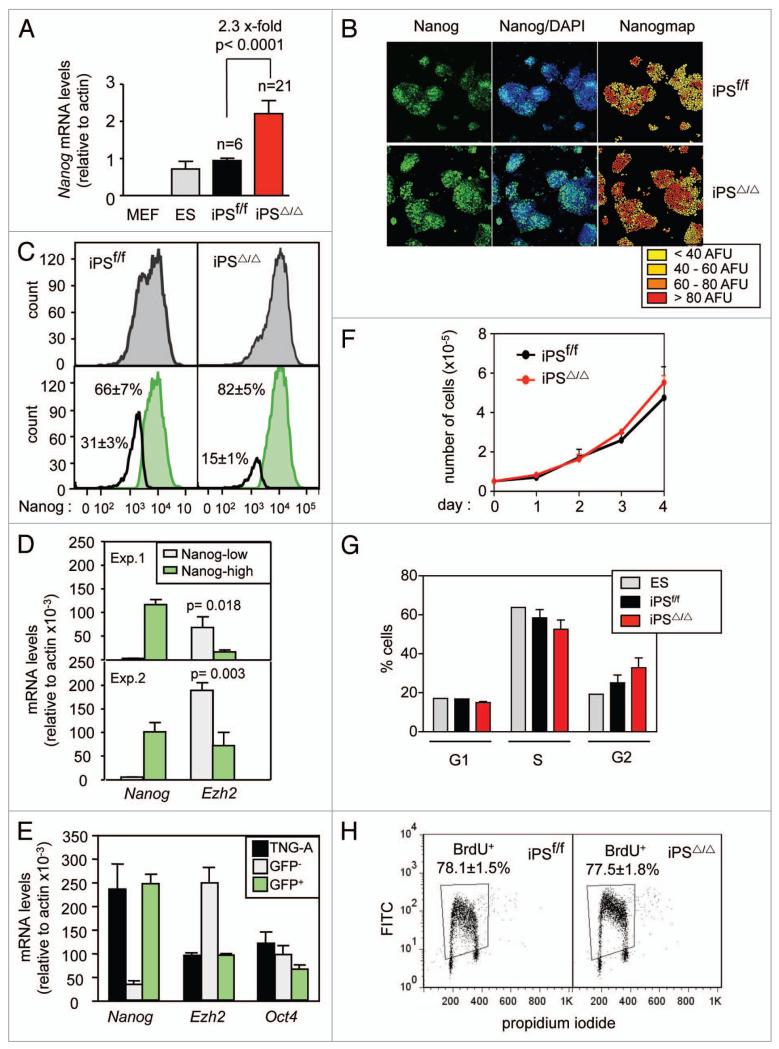
As mentioned above, the protein levels of Nanog are moderately higher in Ezh2-null iPS compared to wild-type iPS or ES cells (Fig. 1D and Sup. Fig. 1c). To further explore this, we measured Nanog mRNA levels and observed that they were increased about 2.3x-fold in iPSΔ/Δ cells compared to iPSf/f cells (Fig. 3A). This observation, together with the above-shown differentiation defect of Ezh2-null iPS cells, suggests that these cells are partially locked in the self-renewal state. This concept was supported by microarray gene expression data comparing wt and Ezh2-null iPS cells under normal culture conditions (n = 3 per genotype). Indeed, this analysis revealed very few changes between Ezh2-null and wt iPS cells (88 downregulated genes and 43 upregulated genes, all with a false discovery rate lower than 0.15 and a fold change higher than two-fold; Sup. Tables 3 and 4). Interestingly, among the upregulated genes there were many genes associated to stemness (see heatmap of 7 stemness genes in Sup. Fig. 4). Notably, many of these genes are effectors of the Wnt pathway, such as Lef1, Pitx2 and Porcn.
Figure 3.
Ratio between Nanog-high and Nanog-low sub-populations in Ezh2-null iPS cells. (A) Expression by qRT-PCR of Nanog. Values are relative to iPSf/f and correspond to the average and standard deviation. Student’s t-test was used to determine statistical significance. (B) Levels of Nanog protein by confocal immunofluorescence. Fluorescence was quantified per cell in arbitrary fluorescence units (AFUs) and colored according to the indicated ranges. The figure is representative of a total analysis of three iPS clones of each genotype. (C) FACS analysis of Nanog. The upper part shows the direct Nanog profile, and the lower part the deconvolution of the profile into two peaks. Deconvoluted peaks were quantified in six clones of each genotype and values correspond to the average and standard deviation. Student’s t-test indicated that the two Nanog peaks (low and high) in iPSΔ/Δ cells were significantly different from the corresponding Nanog peaks in iPSf/f cells (p < 0.001). (D) Expression by qRT-PCR of Nanog and Ezh2 in Nanog-low and Nanog-high subpopulations of wild-type ES cells sorted by cytometry using anti-Nanog staining. Two independent experiments were performed. Each qRT-PCR determination was done in triplicate. Student’s t-test was used to determine statistical significance. (E) Expression by qRT-PCR of Nanog, Ezh2 and Oct4 in Nanog-low and Nanog-high subpopulations of TNG-A ES cells sorted by cytometry according to the GFP fluorescence signal. Student’s t-test indicated that the levels of Ezh2 in GFP-cells were significantly different from the parental TNG-A cells and from GFP+ cells. (F) Proliferation of iPS in gelatin-coated plates. Values correspond to the average and standard deviation of a total of three clones per genotype. (G) Analysis of cell cycle phases measured by the incorporation of propidium iodide. Values correspond to the average and standard deviation of a total of three clones per genotype. (H) Quantification of DNA replication by incorporation of BrdU for 30 min. The figure is representative of a total of three clones per genotype and values correspond the average and standard deviation of the three clones.
The levels of Nanog in ES cells are known to fluctuate between Nanog-high and Nanog-low states, which are related, respectively, to their proneness to self-renew or to differentiate.11,12,14 We wondered whether the higher amounts of Nanog in Ezh2-null iPS had an impact on the balance between the high and low Nanog states. For this, we performed quantitative confocal immunofluorescence of Nanog (which we call “Nanogmap”). These analyses confirmed the heterogeneous nature of Nanog levels in control wild-type iPS cells (Fig. 3B). Interestingly, immunofluorescence images showed that the fraction of Nanog-high cells (colored red in the figure) was higher in Ezh2-deficient iPS compared to Ezh2-proficient iPS cells (Fig. 3B). To validate and confirm this observation with an independent method, we performed cell cytometry with cell populations stained with a fluorescently-labeled antibody against Nanog. This method allowed the detection of two peaks of Nanog expression in control iPS cells (Fig. 3C). Deconvolution of these peaks into Nanog-low and Nanog-high populations indicated that the absence of Ezh2 results in the expansion of the Nanog-high subpopulation at the expense of a decrease in the Nanog-low subpopulation (Fig. 3C). Since the absence of Ezh2 reduces the size of the Nanog-low compartment, we wondered whether the expression levels of Ezh2 itself were higher in this compartment. In fact, quantification of Ezh2 mRNA by qRT-PCR showed significantly higher levels of Ezh2 expression in the Nanog-low population, compared to Nanog-high cells (Fig. 3D, note the presence of two independent assays). In an effort to further associate high Ezh2 levels with Nanog-low cells, we used TNG-A ES cells, which carry a GFP reporter knocked-in at the Nanog locus.11 Purification of GFP-negative (Nanog-low) and GFP-positive (Nanog-high) populations was achieved by fluorescence-activated cell sorting. Interestingly, GFP-negative cells had significantly higher levels of Ezh2 mRNA levels compared to GFP-positive cells (Fig. 3E). Equal levels of Oct4 confirmed the pluripotent status of these cells, in agreement with previous reports.11–13 Finally, we asked whether the observed imbalance between Nanog-low and Nanog-high sub-populations is associated or not to an altered proliferation rate of the cultures. However, analyses of cell number (Fig. 3F), cell cycle phase distribution (Fig. 3G) and BrdU-labelling (Fig. 3H), indicated that the presence or absence of Ezh2 does not affect the proliferation of iPS cells. These results indicate that the observed effects of Ezh2 on differentiation and Nanog expression are not associated to changes in the proliferation rate of cells. Together, these results are compatible with Ezh2 exerting a direct control on the balance between Nanog-low and Nanog-high populations.
In an effort to further reinforce the above conclusions, we inactivated Nanog mRNA by transfection of specific siRNAs into wt and Ezh2-null iPS. This resulted in dramatic and sudden differentiation of the cells regardless of their Ezh2 status (data not shown). In fact, previous reports using siRNAs against Nanog in ES cells have also observed sudden differentiation.23
Epigenetic regulation of Nanog expression by Ezh2
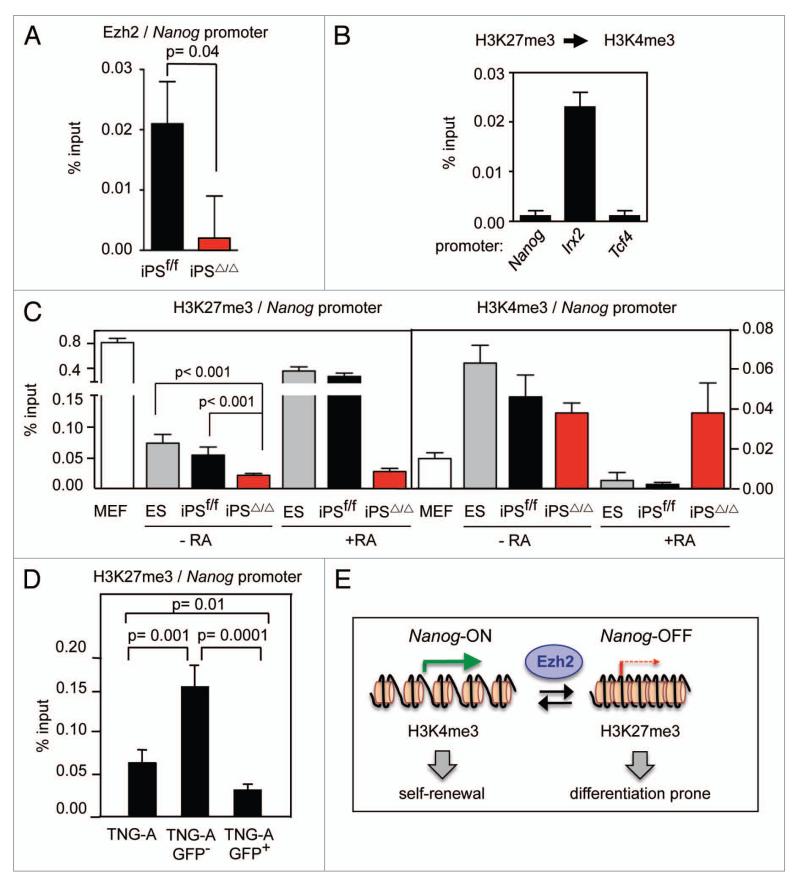
In an attempt to test whether Ezh2 could be directly implicated in the expression of Nanog, we examined by chromatin immunoprecipitation (ChIP) the presence of Ezh2 at the Nanog promoter. Interestingly, we observed detectable binding of Ezh2 in ES/iPS cells (Fig. 4A). As expected, Ezh2-null iPS had background levels of chromatin immunoprecipitation (Fig. 4A). Next, we asked whether the Nanog promoter could be bivalent, that is, carrying simultaneously and on the same DNA molecules the epigenetic marks H3K4me3 and H3K27me3.24 Sequential ChIP of H3K27me3, followed by ChIP of H3K4me3 indicated that the Nanog promoter is not bivalent in murine ES cells (Fig. 4B). In this experiment, the Irx2 promoter served as a positive bivalent control and the promoter of Tcf4 as a negative control. Having observed that the Nanog promoter binds Ezh2, but is not bivalent, we continued by examining the presence of H3K27me3 at the Nanog promoter. Interestingly, H3K27me3 was detected at the Nanog promoter in ES and iPSf/f under standard growing conditions (Fig. 4C). The levels of this mark were clearly above the background and moreover, were significantly decreased in iPSΔ/Δ cells compared to iPSf/f cells (Fig. 4C), thus indicating that Ezh2 is directly responsible, at least to a large extent, for the presence of H3K27me3 at the Nanog promoter. The remaining levels of H3K27me3 are likely maintained by Ezh1.3 On the other hand, H3K4me3 was also present at the Nanog promoter and was not affected by the absence of Ezh2 under normal growing conditions (Fig. 4C). Upon exposure of ES or wild-type iPS to RA-differentiation, the levels of H3K27me3 at the Nanog promoter were dramatically increased and those of H3K4me3 essentially disappeared (Fig. 4C), a pattern consistent with the full repression of Nanog expression upon differentiation. In contrast, iPSΔ/Δ cells retained high levels H3K4me3 and low levels H3K27me3 upon treatment with RA (Fig. 4C), confirming once more their defective differentiation. Finally, to directly demonstrate an association between Nanog levels and H3K27me3 at the Nanog promoter, we took advantage of the TNG-A cells.11 Purified GFP-negative (Nanog-low) TNG-A ES cells had significantly higher levels of H3K27me3 at the Nanog promoter compared to GFP-positive (Nanog-high) cells (Fig. 4D). Together, these results are consistent with a model in which the Nanog promoter exists in two univalent epigenetic configurations, one characterized by H3K4me3 and associated to high levels of expression of Nanog, and another one characterized by H3K27me3 where Nanog expression is reduced (Fig. 4E).
Figure 4.
Epigenetic marks at the Nanog promoter in Ezh2-null iPS cells. (A) Chromatin immunoprecipitation (ChIP) of Ezh2 and qPCR of the Nanog promoter in the indicated cells under standard culture conditions. Values correspond to the average and standard deviation of triplicate qPCR reactions for each immunoprecipitation. Student’s t-test was used to determine statistical significance. The data are representative of a total of two clones per genotype. (B) Sequential ChIP, first of H3K27me3 and then of H3K4me3, at the Nanog promoter in wild-type ES cells. Irx2 and Tcf4 promoters were used as positive and negative controls of bivalency, respectively. Values correspond to the average and standard deviation of triplicate qPCR reactions for each immunoprecipitation. (C) ChIP of H3K4me3 and H3K27me3 and q-PCR of the Nanog promoter in the indicated cells in standard growth conditions (−RA) or four days after differentiation (+RA). Values correspond to the average and standard deviation of triplicate q-PCR reactions of a single clone per genotype. Student’s t-test was used to determine statistical significance. The data are representative of a total of three clones per genotype. (D) ChIP of H3K27me3 and q-PCR of the Nanog promoter in TNG-A ES cells sorted by cytometry according to the GFP fluorescence signal. Values correspond to the average and standard deviation of triplicate q-PCR reactions. Student’s t-test was used to determine statistical significance. (E) Model of Nanog regulation by Ezh2 protein.
Discussion
Here, we have examined the impact of Polycomb repressive complex 2 (PRC2) on the biology of pluripotent stem cells, with a particular focus on the critical stemness factor Nanog. The PRC2 complex is responsible for the epigenetic modification of chromatin characterized by the H3K27me3 mark.
Previous investigators had already implicated PRC2 in the differentiation of ES cells lacking Eed, Suz12 or Ezh2.3-7 The mechanisms by which PRC2 participates in differentiation programs are complex, including repression of stemness genes, but also activation of differentiation genes through mechanisms that are still poorly understood.7 It is important to remark that under normal self-renewal conditions, i.e., in the absence of differentiation cues, PRC2-deficient ES cells already present numerous gene expression changes, as previously determined by microarray analyses.3,6,25 These alterations include de-repression of differentiation genes, such as Gata4,3,6,25 (and our own data in Sup. Fig. 1a), as well as, upregulation of stemness genes, as reported here by us, particularly in relation to the Wnt pathway (Sup. Fig. 4). The final balance of these alterations is that Ezh2-null iPS cells preserve their self-renewal growth, but are severely impaired in differentiation.
During the course of these studies, we noticed that Nanog levels are moderately increased in Ezh2-null iPS. It is well established that the levels of Nanog in ES cells are heterogeneous and dynamic, transitioning from low to high in a cell autonomous manner.11,12 A large body of evidence indicates that the balance between Nanog-high and Nanog-low states is associated with the capacity of cells to self-renew or to differentiate, respectively.14 At present, however, very little is known about the factors that control the equilibrium between these two states. The proteins Satb1 and Satb2 implicated in the higher-order organization of chromatin constitute a notable exception and have been reported to modify both the levels of expression of Nanog and the ratio between Nanog-high and Nanog-low subpopulations.15 Interestingly, when we performed single cell quantifications of Nanog, both by immunofluorescence and by cytometry, we observed that the absence of Ezh2 expanded the Nanog-high compartment at the expense of the Nanog-low subpopulation.
Moreover, we have observed that the Nanog-low compartment is associated with high levels of Ezh2 expression, and conversely the Nanog-high population has low levels of Ezh2 expression. Even more, by taking advantage of a GFP reported knocked-in at the Nanog locus,11 we have observed that Nanog-low cells have high levels of expression of Ezh2 and high levels of its associated epigenetic mark H3K27me3 at the Nanog promoter. In contrast, Nanog-high cells have low expression of Ezh2 and low levels of H3K27me3 at the Nanog promoter. Together, these observations strongly suggest that Ezh2 is a direct regulator of Nanog expression.
To reinforce the concept that Ezh2 could be a direct regulator of the expression of Nanog, we examined in further detail the epigenetic configuration of the Nanog promoter. Chromatin immunoprecipitation analyses confirmed the presence of Ezh2 and its associated epigenetic mark H3K27me3 at the Nanog promoter under standard growing conditions. At the same time, and in agreement with the high levels of expression of Nanog in ES and iPS cells, the activation-associated mark H3K4me3 was also present at the Nanog promoter. The coexistence of H3K27me3 and H3K4me3 at the same promoter is suggestive of a bivalent configuration, although bivalent genes are, in general, poorly expressed.24,28 We have directly tested the possibility that the Nanog promoter could be bivalent. Interestingly, sequential ChIP of H3K27me3 and H3K4me3 clearly indicated that the Nanog promoter is not bivalent, but rather consists in a mixture of two univalent populations, one with the repressive mark H3K27me3 and the other with the active mark H3K4me3.
Fluctuations in Nanog expression are at the basis of the balance between self-renewal and differentiation in pluripotent stem cells.11,12,14 The results reported here provide strong evidence that Ezh2 contributes to the balance between Nanog-high and Nanog-low states through direct epigenetic modification of the Nanog promoter with H3K27me3 (Fig. 4E).
Methods
Cell culture
ES cells were isolated from C57BL/6 blastocysts at the Transgenic Unit of the CNIO. TNG-A ES cells carry a GFP reporter knocked-in at the Nanog locus11 and were a kind gift of Dr. Austin Smith (Cambridge, UK). ES/iPS cells were maintained on feeders using standard culture conditions, namely, DMEM (high glucose) supplemented with serum replacement (KSR, 15%, Invitrogen), LIF 1,000 u/ml, non-essential amino acids, glutamax and beta-mercaptoethanol (we refer to this as “complete ES/iPS medium”). For protein and RNA analyses, as well as, for chromatin immunoprecipitation (ChIP), ES/iPS cells were previously transferred to gelatinized plates (0.1% gelatin) and grown in these plates for two or three passages before preparation of cell extracts.
Reprogramming
Reprogramming of primary mouse embryo fibroblasts (MEFs) was performed following a protocol previously described by us.27 For the generation of Ezh2-null iPS, MEFs carrying both CreERT at the ROSA26 locus19 (Jackson Laboratory #004847) and a floxed Ezh2 allele18 were reprogrammed with 3F. The resulting iPS cells were plated at single cell density and expanded either in the absence or presence of 4-hydroxytamoxifen (4OHT) to obtain, respectively, iPSf/f (Ezh2-proficient) or iPSΔ/Δ (Ezh2-deficient) clones. For this study, a total of 6 iPSf/f clones and of 21 iPSΔ/Δ clones were used. After expansion and confirmation of the excision, the iPSΔ/Δ clones were cultured as their wild-type controls, that is, in the absence of 4OHT. Excision of the floxed Ezh2 region was achieved by treating iPSf/f cells plated at low density with 4OHT at a final concentration of 0.2 μM. Complete ES/iPS medium with 4OHT was replaced every day. Individual colonies were formed after one week, picked and amplified. Upon confirmation of Ezh2 complete loss, these clones were named iPSΔ/Δ.
Global gene expression analyses
Total cellular RNA samples were extracted by combining the Trizol reagent with the RNeasy Mini kit from Qiagen and were analyzed on Mouse Whole Genome DNA microarrays (Agilent P/N G4122F) by following the manufacturer’s instructions. Images were quantified using Agilent Feature Extraction Software (v10.1.1). Microarray background subtraction was carried out using normexp method. To normalize the dataset, we performed loess within arrays normalization and quantiles between arrays normalization. Differentially expressed genes were obtained by applying linear models with R limma package (Bioconductor project, www.bioconductor.org).28 To account for multiple hypotheses testing, the estimated significance level (p value) was adjusted using Benjamini & Hochberg False Discovery Rate (FDR) correction.
Quantitative real-time PCR
Total RNA was obtained using Trizol (Life Technologies) following the manufacturer’s instructions. Total RNA (2 μg) preparations were treated with “Ready-to-go you-prime first-strand beads” (GE Healthcare) to generate cDNA according to the manufacturer’s protocol. Quantitative real-time PCR was performed using DNA Master SYBR Green I mix (Applied Biosystems) in an ABI PRISM 7700 thermocycler (Applied Biosystems). All the primers used for qPCR are listed in Supplemental Table 5. Quantifications were made applying the ΔCt method:29 when determining mRNA expression levels, ΔCt = (Ct of gene of interest - Ct of actin); when determining the recovery of chromatin immunoprecipitations, ΔCt = (Ct of immunoprecipitated promoter - Ct of input promoter) taking into account the corresponding correction for the input dilution factor.
Chromatin immunoprecipitation
ES/iPS cells from a densely grown 150 mm dishes were crosslinked with 1% formaldehyde for 15 min at r.t. Crosslinking was stopped by addition of glycine to a final concentration of 0.125 M. Fixed cells were lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0) and sonicated. Protein concentration was measured and, for each condition, 60 μg of protein were reserved as input and 600 μg were processed for immunoprecipiation. Specifically, protein extracts (600 μg) were diluted in dilution buffer (1% Triton-X100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris-HCl pH 8.0 containing protease inhibitors) and precleared with 60 μl of A/G plus-agarose beads (SantaCruz, sc-2003). The antibodies used for immunoprecipitation were Ezh2 (1:100; Cell Signalling, 4905), histone-3 trimethyl Lys4 (dilution 1:100; Abcam, 8580) and histone-3 trimethyl Lys27 (dilution 1:100; Upstate 07-449). Immune complexes were precipitated with A/G plus-agarose beads and washed sequentially with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 150 mM NaCl), followed by high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 500 mM NaCl), then with LiCl immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0), and finally, washed twice with TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA). Immunoprecipitates were then eluted in 500 μl of elution buffer (1% SDS, 0.1 M NaHCO3). To reverse crosslinking, a volume of 30 μl of NaCl 5 M was added to all samples, including inputs, and were incubated o.n. at 65°C. After this, samples were treated with a total amount of 40 μg of proteinase-K and 20 μg of RNaseA for 1 h at 65°C. Finally, DNA was extracted with phenol-chloroform, resuspended in TE and used for quantitative PCR. Sequential ChIP experiments were carried out essentially as above with modifications to elute the first immunoprecitate, as previously reported.24
Protein analyses
Cells were lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0) and sonicated. Extracts were centrifuged to eliminate cellular debris, diluted in Laemmli buffer and boiled. Protein extracts (30 μg) were loaded on NuPAGE 4–12% gradient Bis-Tris gels, transferred to nitrocellulose and incubated with antibodies against Nanog (dilution 1:500; Chemicon, Ab5731), Ezh2 (1:500; Cell Signalling, 4905), H3K27me3 (1:1,000; Upstate, 07-449), histone H3 (1:1,000; Upstate 05-499), Oct4 (1:500; SantaCruz, H-134) and Sox2 (1:500; Santa Cruz, Y-17).
Immunofluorescence
ES/iPS cells were cultured on gelatin-coated chamber slides with feeders, fixed in 4% PFA and permeabilized with PBS 1x + 0.1% Triton. For H3K27me3 immunofluorescence, cells were blocked (5% BSA, 0.2% Tween-20 in PBS 1x, 1 h at r.t.) before an o.n. incubation at 4°C with α-H3K27me3 (1:500; Upstate 07-449) in blocking buffer. Slides were then stained with Cy3-conjugated secondary antibody (Invitrogen, in 2% BSA-PBS 1x, 1 h at r.t.) prior mounting in Vectashield antifade medium-DAPI (4′,6-Diamidino-2-phenylindole, Vector Laboratories). For Nanog immunofluorescence, cells were blocked (5% BSA/1% Australian FBS/0.2% Tween-20 in PBS 1x, 1 h at r.t.) before an o.n. incubation at 4°C with α-Nanog antibody (1:50, Novus Biologicals, NB100-58842) in Dako Antibody Diluent with Background Reducing Components. Slides were then stained with Alexa Fluor488-conjugated secondary antibody (Invitrogen, in Dako REAL buffer, 1 h at r.t.) followed by DAPI staining to visualize the nuclei. Slides were mounted with Vectashield (Vector Laboratories) before confocal analysis.
Confocal microscopy was performed with a TCS SP5 laser scanning spectral microscope (Leica Microsystems) equipped with a Plan-Apochromat 20x/0.70 NA oil objective. 8-bits images were acquired using the Leica LAS AF v.2.1 software (Leica Microsystems). The pictures show the maximum projection of Z-stacks. For quantitative measurement of Nanog immunofluorescence, the Argon-488 laser was held at a constant intensity to capture all the images.
For the generation of Nanog fluorescence intensity maps (Nanogmap) the images were analyzed with Definiens XD software package. The DAPI image was used to define the nuclear area and the Alexa Fluor488 image was used to quantify the average Nanog fluorescence for each nucleus. The nuclei were then classified in four categories according to their average Nanog fluorescence.
Cytometry
For the analysis of the cell cycle phases, ES/iPS cells (1 × 106) were washed twice with PBS 1x and then resuspended in 330 μl of cold PBS 1x. Fixation was performed vortexing and adding drop by drop 660 μl of ice cold 100% ethanol and left for 2 h at 4°C. Then, cells were pelleted at 800 rpm for 5 min and resuspended in 500 μl of PBS, 1 μl RNaseA (from a stock of 100 mg/ml) and 10 μl propidium iodide (from a stock of 1 mg/ml) to determine DNA content.
For measuring DNA replication, cultures were incubated for 30 min with a final concentration of 10 μM BrdU (Sigma, B5002). After this, cells (1 × 106) were fixed as above, pelleted and resuspended in denaturing solution (2 N HCl) for 20 min at r.t. Then, cells were washed twice with PBS containing 1% BSA and 0.5% Tween-20, and incubated 20 min with mouse anti-BrdU (dilution 1:20; Dako M0744) in PBS containing 0.5% BSA and 0.5% Tween-20. Subsequently, cells were washed and incubated 20 min with secondary antibody rabbit anti-mouse FITC (Dako). Finally, cells were washed twice, pelleted and resuspended in 0.5 ml propidium iodide (final concentration 50 μg/ml in PBS) for FACS analysis.
For cytometric detection of Nanog, cells were fixed and permeabilized using “Foxp3 Staining Buffer Set” (eBioscience 00-5523) according to the manufacturer’s instructions. Staining was carried out by adding Alexa Fluor 647 anti-mouse Nanog antibody (eBioscience, Cat. 51-5761), 0.1 μg of antibody per sample, in 200 μl of permeabilization buffer for 1 h at 4°C in the dark. In all cases, cytometry was performed in a FACSCanto II (BD Biosciences). Deconvolution into peaks was done using the software “Flowjo.”
Two strategies were used to sort Nanog-high and Nanog-low subpopulations. When using wild-type ES populations (Fig. 3D), cells were stained with Nanog as above and were sorted in a BDFacs Aria IIu equipment (BD Biosciences). When using TNG-A ES cells (Figs. 3E and 4D), cells were sorted live based on their GFP fluorescence and immediately after processed for RNA or ChIP analyses.
In vitro differentiation
Differentiation of iPS in vitro was performed following a variant of a previously reported protocol.32 Briefly, cells (1 × 106) were transferred to gelatinized 100 mm dish plates in “complete ES/iPS medium” (see above), the following day (day 1) cells were incubated in ES/iPS medium without LIF, and for the subsequent days, cells were incubated in ES/iPS medium without LIF and with retinoic acid (RA) at a final concentration of 1 μM. This culture medium (−LIF and +RA) was replaced daily during a four days. Alkaline phosphatase staining was performed using a kit according to the manufacturer’s instructions (Millipore, SCR004).
Embryoid body formation and teratomas
iPS cells grown on feeders were tripsinized and 5,000 cells in 20 μl were plated in hanging drops on Petri dish lids in “complete ES/iPS medium” without LIF. Three days later, embryoid bodies were transferred to Petri dishes and kept growing in “ES/iPS complete medium” without LIF for 2 weeks. Embryoid bodies (20) were collected in Bioscience. an 1.5 ml Eppendorf tube, washed with PBS and fixed with 25 μl of formalin for 1 h, at r.t. After fixation, gelatin was added to a final concentration of 5% and, after gelatinized, 1 ml of formalin was added and kept o.n. at 4°C. Each gelatin block was included in paraffin and subsequently sectioned and stained with haematoxylin and eosin.
Teratomas were produced by subcutaneous injection of iPS cells (1 × 106) in irradiated nude mice (4 Gy, injections were performed 1 day post-irradiation) until tumors were clearly detectable.
Acknowledgments
We are indebted to Austin Smith for the TNG-A ES cells. We are grateful to Maribel Muñoz, Marta Cañamero, Lola Martinez, Arancha Garcia, Orlando Dominguez and Diego Megias for their expert technical support. A.V. is supported by the FPI Program of the Spanish Ministry of Science. D.P. is supported by a Long Term EMBO Fellowship. H.L. is supported by the Juan de la Cierva Program of the Spanish Ministry of Science. Work in the laboratory of M.S. is funded by the CNIO and by grants from the Spanish Ministry of Science (SAF and CONSOLIDER), the Regional Government of Madrid (GsSTEM), the European Union (PROTEOMAGE, ERC Advanced Grant), and the “Marcelino Botin” Foundation.
Footnotes
Note Supplemental materials can be found at: www.landesbioscience.com/journals/cc/article/15658
References
- 1.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 2.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–32. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–7. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136:2311–22. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–37. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–24. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–4. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 12.Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–42. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 13.Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–6. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savarese F, Davila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, et al. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–38. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–6. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–31. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 19.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–22. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–64. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stavridis MP, Collins BJ, Storey KG. Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development. 137:881–90. doi: 10.1242/dev.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–48. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 25.Pasini D, Bracken AP, Agger K, Christensen J, Hansen K, Cloos PA, et al. Regulation of stem cell differentiation by histone methyltransferases and demethylases. Cold Spring Harb Symp Quant Biol. 2008;73:253–63. doi: 10.1101/sqb.2008.73.009. [DOI] [PubMed] [Google Scholar]
- 26.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–9. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–75. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor upregulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–22. [PubMed] [Google Scholar]