Abstract
G-protein–coupled signal transduction mediates most cellular responses to hormones and neurotransmitters; this signaling system transduces a large variety of extracellular stimuli into neurons and is the most widely used mechanism for cell communication at the synaptic level. The heterotrimeric G-proteins have been well established as key regulators of neuronal growth, differentiation, and function. More recently, the heterotrimeric G-protein genes group was associated with general cognitive ability. Although heterotrimeric G-proteins are linked to both cognitive ability aond neuron signaling, it is unknown whether heterotrimeric G-proteins are also important for brain structure. We tested for association between local cerebral gray matter volume and the heterotrimeric G-protein genes group in 294 subjects; a replication analysis was performed in an independent sample of 238 subjects. Voxel-based morphometry revealed a strong replicated association between 2 genes encoding heterotrimeric G-proteins with specific local increase in medial frontal cortex volume, an area known to be involved in cognitive control and negative affect. This finding suggests that heterotrimeric G-proteins might modulate medial frontal cortex gray matter volume. The differences in gray matter volume due to variations in genes encoding G-proteins may be explained by the role of G-proteins in prenatal and postnatal neocortex development.
Keywords: brain volume, functional gene group, imaging genetics, MRI, VBM
Introduction
Heterotrimeric G-proteins (G-proteins) are the molecular switches that turn on intracellular signaling cascades in response to the activation of G-protein–coupled receptors (GPCRs) by extracellular stimuli (hormones and neurotransmitters) (Oldham and Hamm 2008). The G-proteins are abundantly expressed in the brain and are necessary for normal neuronal growth, cortical development, and brain function (Tanaka et al. 2007; Bromberg et al. 2008; Moers et al. 2008; Ma'ayan et al. 2009; Sanno et al. 2010; Tahirovic et al. 2010).
We previously reported a strong replicated association of the group of genes encoding synaptic G-proteins with cognitive ability, using a functional gene group analysis (Ruano et al. 2010). It is unknown, however, how G-proteins may affect cognitive ability.
Studies using magnetic resonance imaging (MRI) have consistently reported associations between brain morphometry and cognitive ability; the majority of these studies used voxel-based morphometry (VBM) to measure local gray and white matter volume across the entire brain. Large interindividual variation in total and regional brain volume exists, which is mainly due to differences at the genetic level. Twin studies have shown that many aspects of brain structure and function are highly heritable; genetic factors account for most of interindividual differences, with heritability estimates ranging from 82% for gray matter to 88% for white matter (Baare et al. 2001; Thompson et al. 2001). Even from early age on, MRI studies on neonates have shown high heritability of local and total brain morphology (Gilmore et al. 2010). Despite the generally high heritability of brain morphology, even the most highly associated common genetic polymorphisms explain less than 1–5% of the variation in most brain measurements (Thompson et al. 2010).
It is well known that the correlation between local brain volume and cognitive ability (Witelson et al. 2006; Rushton and Ankney 2009) is largely explained by correlations at a genetic level (Posthuma et al. 2002), genes that are important for cognitive ability may also be important for local brain volume. The group of genes encoding G-proteins poses an excellent candidate gene group for association with local brain volumes due to their known expression in the brain and their role in cortical development and neuronal growth. We thus set out to test whether genes encoding G-proteins explain individual differences in local brain volume using 2 relatively large data sets.
Materials and Methods
Subjects
A total of 532 healthy individuals were included from the Brain Imaging Genetics (BIG) study at the Donders Institute for Brain, Cognition and Behavior of the Radboud University Nijmegen Medical Center, the Netherlands. All participants included in the study were right handed and of European Caucasian descendent. The regional medical ethics committee approved the study, and all participants gave written informed consent.
In order to avoid the well-known interscanner confounding (Focke et al. 2011), the sample set was divided into 2 groups according to the MRI scanner used; 294 subjects were scanned in a 3-T Siemens scanner (Erlangen, Germany) and 238 subjects were scanned in a 1.5-T Siemens scanner. To increase the power to detect association, the larger sample was used as a discovery sample and the smaller sample was used as an independent sample for replication. We only included subjects aged 18–36 years as for this age group the effects of brain development or aging is negligible. The demographics of the 2 samples are described in Supplementary Table S1.
Genotyping
DNA was extracted from saliva using the Oragene DNA sample collection kit (DNA Genotek, Kanata, Canada). Genotypes were obtained using an Affymetrix GeneChip SNP 6.0 (AFFY6) array (Santa Clara, CA). The call rate threshold was set at 90% for the arrays, with an average call rate of 96.3%.
Single Nucleotide Polymorphisms Selection
All known single nucleotide polymorphisms (SNPs) within the heterotrimeric G-protein genes (Supplementary Table S2) that are expressed in the brain were retrieved in batch from the SNP database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/projects/SNP/) and were subsequently filtered for SNPs that were genotyped on the AFFY6 platform. The G-protein genes group expressed in the brain is composed of 27 genes (Supplementary Table S2), of which 25 (including 677 SNPs) were represented on the AFFY6 platform (no SNPs were available on this platform for the GNB2 and GNG3 genes). We excluded all genotyped SNPs that showed a minor allele frequency (MAF) less than 5% as well as genotyped SNPs that were in complete linkage disequilibrium (LD) with another SNP within the same gene. MAF and LD were calculated using PLINK-v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/). In total, we included 502 SNPs within 25 genes in the final analysis (Supplementary Table S2).
Brain Imaging
Image Acquisition
T1-weighted structural MRI data were acquired at either 3-T (n = 294) or 1.5-T (n = 238). All 1.5-T images were acquired at 1.5-T Siemens Sonata and Avanto scanners (Siemens), using small variations to a standard T1-weighted 3D magnetization prepared rapid gradient echo (MP-RAGE) sequence (time repetition [TR] 2300 ms, time to inversion [TI] 1100 ms, time echo [TE] 3.03 ms, 192 sagittal slices, field of view 256 mm). These variations included a TR/TI/TE/slices of 2730/1000/2.95/176, 2250/850/2.95/176, 2250/850/3.93/176, 2250/850/3.68/176, and the use of generalized autocalibrating partially parallel acquisitions (GRAPPA) parallel imaging with an acceleration factor of 2. All scans covered the entire brain and had a voxel size of 1 × 1 × 1 mm3. All 3-T images were acquired at 3-T Siemens Trio and TrioTim scanners (Siemens), using small variations to a standard T1-weighted 3D MP-RAGE sequence (TR 2300 ms, TI 1100 ms, TE 3.93 ms, 192 sagittal slices, field of view 256 mm). These variations included TR/TI/TE/slices of 2300/1100/3.03/192, 2300/1100/2.92/192, 2300/1100/2.96/192, 2300/1100/2.99/192, 1940/1100/3.93/176, 1960/1100/4.58/176, and the use of GRAPPA parallel imaging with an acceleration factor of 2. All scans covered the entire brain and had a voxel size of 1 × 1 × 1 mm3.
Conversion
Raw digital imaging and communications in medicine MR imaging data were converted to NIFTI format using the conversion as implemented in SPM5.
Brain Volume
Normalization, bias correction, and segmentation into gray matter, white matter, and cerebrospinal fluid were performed using the VBM toolbox in SPM using priors (default settings). This method uses an optimized VBM protocol (Ashburner and Friston 2000; Good et al. 2001) as well as a model based on hidden Markov random fields developed to increase signal-to-noise ratio (Cuadra et al. 2005). Total volume of gray matter, white matter, and cerebrospinal fluid were calculated by adding the resulting tissue probabilities. Brain volume was defined as the sum of white matter and gray matter volume.
VBM Preprocessing
Diffeomorphic image registration was performed using the DARTEL toolbox in SPM (Ashburner 2007). First, all images were realigned to templates created from 556 in-house data sets. Second, Jacobian scaled (“modulated”) images were calculated and subsequently transformed to Montreal Neurological Institute space using affine transformation. Finally, all data were smoothed with an 8-mm full-width at half-maximum Gaussian smoothing kernel.
Statistics
A full-factorial analysis of covariance was applied using genotype as factor, and participants' age, gender, total brain volume, and scan protocol were added to the model as covariates. Statistics were first done using an F-contrast and corrected for nonstationarity for cluster statistics (Hayasaka et al. 2004) at P < 0.05 (familywise error [FWE] corrected for whole brain). For the most significant findings, a T-statistic was applied to determine the direction of the effect. For the association analysis, we used a mass univariate model for pathway-based approaches to imaging genetics association studies as previously described by Inkster et al. (2010). In summary, for each of the 502 SNPs, we performed a separate VBM analysis for gray matter using standard tools in SPM5. This approach was used because the G-proteins genes are expressed ubiquitously in the whole brain and might show associations with many different brain regions. Two or 3 level group factor was used for genotype status: 1) for SNPs with an MAF >0.31 (more than 10% of subjects with the minor allele), a 2 degrees of freedom genotypic model was applied using an additive parameterization with a centered covariate; 2) for SNPs with MAF <0.31 (less than 10% of subjects with the minor allele), a recessive model merging the rare homozygous and heterozygous groups was used.
In order to find associations between the G-proteins genes group and variations in gray matter volume, we tested for associations between individual SNPs and gray matter variations. Cluster size of voxels was inferred using the toolbox “NS” implemented on SPM5 (Hayasaka et al. 2004). Results were corrected for nonstationarity and assessed initially at P < 0.001 uncorrected for the whole-brain volume. Subsequently, cluster statistics were applied using FWE correction, and results were considered significant at P-(FWE) < 5 × 10−4, which is the Bonferroni adjusted P-(FWE) value for the total number of SNPs tested in the whole analysis (502 SNPs).
In order to replicate the findings of the discovery sample, we performed the same analysis in our replication sample, using only the significant findings from the discovery sample as regions of interest (ROIs) to apply a small volume correction (SVC) to the results using the MarsBar toolbox implemented in SPM5 (http://marsbar.sourceforge.net/). This allows to specifically test the same brain regions in both samples, selecting only the cluster found to be significant in the discovery sample and testing it for association in the replication sample. Results were considered significant at P-(SVC) < 0.007, this is the adjusted P-(SVC) value for the total number of SNPs tested in the replication analysis (7 SNPs).
In summary, multiple testing corrections were applied at different levels: first, adjusting for testing multiple voxels in the whole brain was implemented using FWE correction; second, adjusting for testing multiple SNPs using Bonferroni correction, and finally, the gold standard of replication analysis was carried out to rule out any remaining false positive associations.
Results
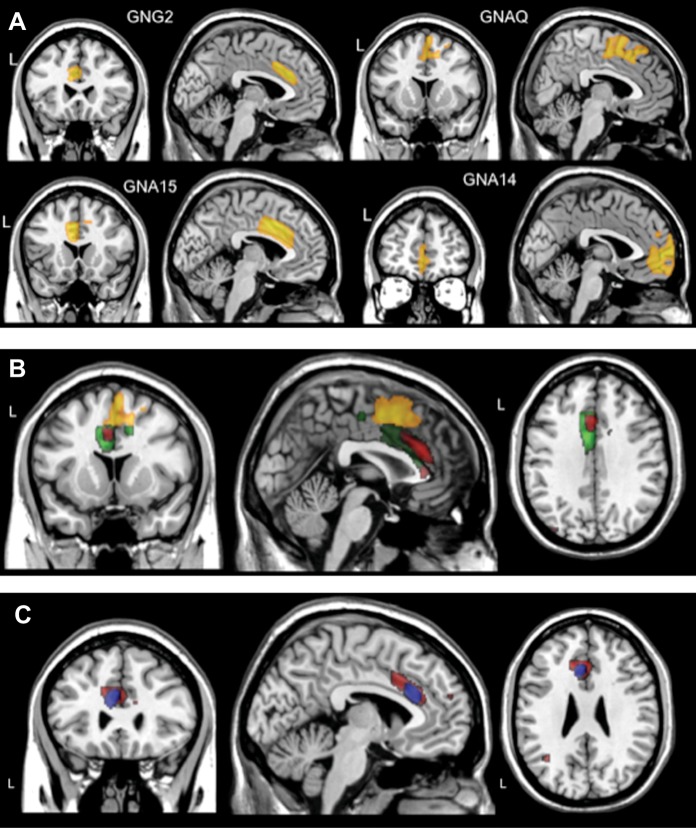
Using VBM of T1-weighted structural MRI measurements, we tested the association between local cerebral gray matter volume variations and 502 SNPs spanning 25 G-proteins genes. We identified 7 genes (GNG2, GNAQ, GNA14, GNA15, GNAO1, GNAL, GNB5) in which (after Bonferroni correction for all the 502 SNPs tested) at least one SNP was significantly associated with gray volume variations (denoted as P value Groupcorrected in Table 1). Four of these 7 genes (GNG2, GNAQ, GNA15, GNA14) mapped predominantly to gray matter differences in the same area, namely the medial frontal cortex (Fig. 1A); the associated areas in the medial frontal cortex overlapped in Brodmann area 32 for 3 of these genes: GNG2, GNAQ, and GNA15 (Fig. 1B). Two genes (GNAO1 and GNAL) mapped to significant differences in gray matter in the temporal lobe (Brodmann areas 20, 21, 28, 34) and 1 gene (GNB5) showed association for gray matter differences in a region in the occipital lobe (Brodmann area 18).
Table 1.
Association analysis of G-proteins genes with gray matter volume
| Discovery sample (n = 294) |
Replication sample (n = 238) |
||||||||||
| SNP* |
Cluster |
P values |
MAF | SVC P valuesd FWE corrected | |||||||
| rs number | MAF | Gene(a) | Brain region (Brodmann area) | Size | Local maximum |
FWE correctedb (α = 5 × 10−04) | Groupcorrectedc (α = 0.05) | ||||
| x | y | z | |||||||||
| rs11851703 | 0.0514 | GNG2(40) | Medial frontal cortex (24, 32) | 1302 | −2 | 30 | 25 | 5.05 × 10−05 | 0.02534 | 0.0506 | 0.002 |
| rs4745679 | 0.4851 | GNAQ(47) | Medial frontal cortex (8, 32) | 2047 | 15 | 9 | 51 | 9.27 × 10−07 | 0.00046 | 0.4838 | 0.034 |
| rs2238628 | 0.0864 | GNA15(8) | Medial frontal cortex (24, 32) | 2001 | −8 | 4 | 34 | 1.15 × 10−05 | 0.00575 | 0.0860 | NS |
| rs4745639 | 0.1476 | GNA14(85) | Medial frontal cortex (10, 11) | 3369 | −10 | 61 | 3 | 1.36 × 10−06 | 0.00682 | 0.1579 | NS |
| rs1382362 | 0.4012 | GNAO1(43) | Temporal lobe (28, 34) | 2574 | 25 | 0 | −21 | 7.60 × 10−07 | 0.00038 | 0.3801 | NS |
| rs17515178 | 0.1583 | GNAL(48) | Temporal lobe (20, 21) | 866 | −65 | −13 | −24 | 4.25 × 10−05 | 0.02132 | 0.1565 | NS |
| rs12396 | 0.3835 | GNB5(9) | Occipital lobe (18) | 902 | 31 | −85 | −15 | 5.77 × 10−05 | 0.02897 | 0.4013 | NS |
Only SNPs with significant associations with brain structural variation are shown, for cases in which multiple SNPs related to the same gene were significantly associated, the SNP that had the lowest structural association P-value is shown. MAF = minimum allele frequency.
In parenthesis the number of SNPs tested for that specific gene.
FWE-corrected p-values for the map-wise cluster based association test.
FWE-corrected p-values after applying Bonferroni correction based on 502 SNPs tested in the whole gene group analysis.
SVC-corrected p-values for the association test in the replication sample.
NS = No Suprathreshold Cluster association.
Figure 1.
Anatomical locations of SNP-association clusters of G-proteins genes that showed significant FWE corrected P values of local gray matter differences in the medial frontal cortex. (A) The anatomical cluster for each gene corresponds to the SNP that had the lowest significant association P values after Bonferroni correction in the discovery sample: GNG2 (rs1185703), GNAQ (rs4745679), GNA15 (rs2238628), GNA14 (rs4745639). (B) Overlap of 3 clusters of SNP-associated G-proteins genes that showed significant association with local gray matter differences in the medial frontal cortex (Brodmann areas 24 and 32). Red Cluster: GNG2 (rs11851703); yellow cluster: GNAQ (rs4745678); green cluster: GNA15 (rs2238628). (C) Anatomical location of the overlapping cluster of SNP rs11851703 (GNG2 gene) in the discovery sample (red cluster) and the replication sample (blue cluster). “L”: left hemisphere.
In order to replicate these findings, we tested the SNP that had the lowest structural association P value in the discovery sample for each of the 7 genes that were significantly associated with gray matter volume variation in an independent sample of 238 subjects. We found replicated regional association of 2 genes: GNG2 and GNAQ gene with differences in local gray matter volume in the medial frontal cortex (denoted as SVC P value in Table 1). No significant replications were observed for the temporal or the occipital lobe associations found in the discovery sample. If an additional Bonferroni correction is applied for the 7-replication tests, only the SNP rs11851703 on GNG2 gene would survive the α = 0.007 for this additional correction.
Using a t-test analysis (Supplementary Fig. 1) of the most significant replicated finding, we identified that the rare allele of the SNP rs11851703 (MAF = 0.051) is associated with an increased volume of gray matter in the medial frontal cortex in both the discovery and the replication sample. For the replication sample, this SNP explains almost 10% of the variance observed in the gray matter volume for the ROI obtained from the discovery analysis (T = 5.10; DF = 237; Cohen’s d = 0.6625; Effect-size r = 0.3144; r2 = 0.098).
Discussion
In this study, we investigated whether common genetic variants in the group of genes encoding G-proteins are related to local gray matter volume variations. We found that SNPs in 7 G-proteins genes (GNG2, GNAQ, GNA14, GNA15, GNAO1, GNAL, GNB5) were significantly associated with differences in 3 brain regions: the medial frontal cortex, the temporal lobe, and the occipital lobe. Interestingly, variants in 4 of the significantly associated G-proteins genes (GNG2, GNAQ, GNA15, GNA14) were associated with gray matter changes in an overlapping region of the medial frontal cortex. We conducted a second study to verify our initial result in an independent sample and were able to replicate the association for 2 of the 4 genes in exactly the same brain region of the medial frontal cortex as the discovery sample.
When interpreting results from an imaging genetics’ study, 2 main issues are important. The first issue is to show how likely a reported significant association is and to determine whether it might simply be due to chance. Lately, concerns have risen (Meyer-Lindenberg et al. 2008; Silver et al. 2010) about multiple testing issues that could lead to high false positives rates in imaging genetics (in our case, a whole-brain study of 502 SNPs across 600 000 voxels requires about 3 × 108 separate tests). Silver et al. (2010) recently compared different statistical approaches in neuroimaging genetics using VBM data and defined clear guidelines in order to reduce the probability of obtaining false positives results. In our study, we followed these guidelines and used a nonparametric nonstationary cluster size inference with a relatively high cluster-forming threshold (α = 0.001) on images smoothed an 8-mm Gaussian kernel (instead of the 12-mm Gaussian kernel smoothing recommended by Silver et al. 2010; we used an 8-mm Gaussian kernel because the smoothing kernel is always a trade-off between spatial accuracy [with as much as 12 mm you cannot claim that some effect is in a small structure] and statistical validity). However, the likelihood of false positive findings is reduced by the discovery–replication design that we used; with these parameters, rejection rates (at a nominal significance level of 5%) are expected to be well controlled as suggested by Silver et al. (2010). In addition to the statistical parameters used to determine corrected significance while accounting for spatial dependences between voxels (FWE whole-brain corrected), we only considered a finding to be statistically significant if the nominal P value was still significant after Bonferroni correction for 502 SNPs tested (Table 1), which is known to be overly conservative, especially when tests are nonindependent. Even after rigorous multiple testing correction in our discovery sample, associations remained significant for 4 G-proteins genes with the medial frontal cortex, a brain region that is known to be involved in cognitive control and negative affect (Shackman et al. 2011).
In genetic association studies, replication has become the gold standard to show that significant results, even after multiple corrections, are genuine. Most neuroimaging studies to date have been severely underpowered to detect associations; replicating association results require even larger samples, rendering the need for phenotyping standardization and data sharing even greater (Congdon et al. 2010). For imaging studies, this is not yet common practice mainly because of time and financial investments. However, more, large scaled MRI data sets are becoming available allowing for independent replication testing. We used both a discovery data set and an independent replication data set and showed replicated association with medial frontal cortex volume for 2 genes encoding G-proteins.
A second issue that is important when interpreting results from an imaging genetics study is to understand how common the discovered genetic association with a brain region is. That is, if a given number of randomly drawn genes (i.e., 100 groups of 500 SNPs) would be tested against the same number of voxels as used here, how likely is it that such genes would also show significant association? Such an approach will require performing 50 000 individual VBM analyses on 600 000 voxels. If this is likely, it means that besides the original genetic association is, although still bona vida, multiple other effects might be just as important: just one effect amongst very many other contributing effects. If not, it seems justified to conclude that the variation in the associated genes is one of the major genetic factors influencing this trait. Determine whether other genetic effects might be as important as the effect of the G-proteins would fit in an exploratory study. However, our current study was a hypothesis-driven study focusing on a functional group of genes with high prior validity.
It is important to notice that the rare allele of SNP rs11851703 has a relatively large effect size (r2 = 0.098) in our replication analysis. This could be explained by the fact that our model was limited to test univariate main effects, and it is only adequately powered to detect relatively large effects. Even though higher-order modeling (multivariate, nonadditive) is outside the scope of this paper, the field of imaging genetics should work toward developing reliable statistical methods to allow relating the combine effects of large number of genetic variants on equally multidimensional neuroimaging phenotypes (Meyer-Lindernberg 2011). The complexity of the high-volume data sets generated in imaging genetics is prone to produce too many false positives or false negatives. Even when our approach allowed us to prevent high rate of false positive findings, this approach might also increase the risk of type II errors, which at the end might represent the mayor limitation of this study.
The multiple layers of statistical protection and the independent replication analysis strengthen the conviction that the observed associations of gray matter volume variation in the medial frontal cortex with markers in GNG2 and GNAQ genes are genuine. For the most significant finding, in GNG2, we found that carriers of the rare allele of SNP rs11851703 in both the discovery and the replication sample had an increased gray matter volume of the medial frontal cortex, specifically in the anterior cingulate cortex (Brodmann areas 24 and 32), compared with noncarriers. The cingulate cortex has been implicated in both cognitive control and negative affect (Bush et al. 2000; Shackman et al. 2011). The anterior cingulate cortex has also been implicated in cognitive processes such as attention and executive functions, including performance monitoring (Botvinick et al. 2001; Braver et al. 2001). An increased gray matter volume as we found in the dorsal medial frontal cortex may explain differences in cognitive processes. During normal brain development, the maturation of the medial frontal cortex facilitates improvements in performance monitoring, an ability that is necessary to learn from experience, a critical feature of human cognition (Fitzgerald et al. 2010); this maturation could be modulated genetically by genes that regulate brain development and at the end creates differences in gray matter volume in this region.
The differences in gray matter volume due to variation in genes encoding G-proteins may be explained by the role of G-proteins in prenatal and postnatal neocortex development (Tanaka et al. 2007; Bromberg et al. 2008; Moers et al. 2008; Ma'ayan et al. 2009; Sanno et al. 2010; Tahirovic et al. 2010). Multiple GPCRs regulate the migratory activity of neurons by modulating the activity of Rac and/or Rho (Sah et al. 2000). The activation of RhoA via GPCRs is primarily mediated by the ubiquitously expressed G-proteins GNA12 and GNA13; these 2 G-proteins were found to be required for the proper positioning of migrating cortical plate neurons and Purkinje cells during brain development (Moers et al. 2008). The RhoA subfamily within the Ras superfamily of GTP-binding proteins was found to determine neuronal density during postnatal neocortex development by regulating the G-proteins’ activity (Sanno et al. 2010). Several signaling cascades activated by the G-proteins have also been implicated in genetic disorders that show different levels of cognitive ability impairment. Mutations in some of the genes encoding proteins of the signaling cascades activated by the G-proteins have been associated with developmental disorders that feature different levels of cognitive deficits (Cesarini et al. 2009).
In conclusion, our study shows that 2 members of the G-proteins genes explain individual differences in medial frontal cortex volume, a brain region known to be involved in cognitive control and negative affect. This suggests that G-proteins may also be important for disorders such as schizophrenia or attention deficit hyperactivity disorder where cognitive dysfunction is a prominent feature and that G-proteins may be responsible for at least part of the reported genetic association between brain volume and cognitive ability (Posthuma et al. 2002).
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
We gratefully acknowledge the financial support from the Netherlands Organization for Scientific Research (NWO) (grants NWO/VIDI 452-05-318, NWO 400-08-206, TOP) and (grant ZonMW 40-00812-98-07–032).
Supplementary Material
Acknowledgments
We thank all the participants. We thank Sabine Kooijman, Angelien Heister, Remco Makkinje, Marlies Naber, Marloes Steehouwer, and Terry Vrijenhoek for their assistance with the recruitment of participants, sample collection, and genotyping. Conflict of Interest : None declared.
References
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Baare WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Botvinick MM, Braver TS, Barch DM, Carter CS, Cohen JD. Conflict monitoring and cognitive control. Psychol Rev. 2001;108:624–652. doi: 10.1037/0033-295x.108.3.624. [DOI] [PubMed] [Google Scholar]
- Braver TS, Barch DM, Gray JR, Molfese DL, Snyder A. Anterior cingulate cortex and response conflict: effects of frequency, inhibition and errors. Cereb Cortex. 2001;11:825–836. doi: 10.1093/cercor/11.9.825. [DOI] [PubMed] [Google Scholar]
- Bromberg KD, Iyengar R, He JC. Regulation of neurite outgrowth by G(i/o) signaling pathways. Front Biosci. 2008;13:4544–4557. doi: 10.2741/3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush G, Luu P, Posner MI. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci. 2000;4:215–222. doi: 10.1016/s1364-6613(00)01483-2. [DOI] [PubMed] [Google Scholar]
- Cesarini L, Alfieri P, Pantaleoni F, Vasta I, Cerutti M, Petrangeli V, Mariotti P, Leoni C, Ricci D, Vicari S, et al. Cognitive profile of disorders associated with dysregulation of the RAS/MAPK signaling cascade. Am J Med Genet A. 2009;149:140–146. doi: 10.1002/ajmg.a.32488. [DOI] [PubMed] [Google Scholar]
- Congdon E, Poldrack RA, Freimer NB. Neurocognitive phenotypes and genetic dissection of disorders of brain and behavior. Neuron. 2010;68:218–230. doi: 10.1016/j.neuron.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadra MB, Cammoun L, Butz T, Cuisenaire O, Thiran JP. Comparison and validation of tissue modelization and statistical classification methods in T1-weighted MR brain images. IEEE Trans Med Imaging. 2005;24:1548–1565. doi: 10.1109/TMI.2005.857652. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KD, Perkins SC, Angstadt M, Johnson T, Stern ER, Welsh RC, Taylor SF. The development of performance-monitoring function in the posterior medial frontal cortex. Neuroimage. 2010;49:3463–3473. doi: 10.1016/j.neuroimage.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focke NK, Helms G, Kaspar S, Diederich C, Tóth V, Dechent P, Mohr A, Paulus W. Multi-site voxel-based morphometry—not quite there yet. Neuroimage. 2011;56:1164–1170. doi: 10.1016/j.neuroimage.2011.02.029. [DOI] [PubMed] [Google Scholar]
- Gilmore JH, Schmitt JE, Knickmeyer RC, Smith JK, Lin W, Styner M, Gerig G, Neale MC. Genetic and environmental contributions to neonatal brain structure: a twin study. Hum Brain Mapp. 2010;31:1174–1182. doi: 10.1002/hbm.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;22:676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Inkster B, Nichols TE, Saemann PG, Auer DP, Holsboer F, Muglia P, Matthews PM. Pathway-based approaches to imaging genetics association studies: Wnt signaling, GSK3beta substrates and major depression. Neuroimage. 2010;53:908–917. doi: 10.1016/j.neuroimage.2010.02.065. [DOI] [PubMed] [Google Scholar]
- Ma'ayan A, Jenkins SL, Barash A, Iyengar R. Neuro2A differentiation by Galphai/o pathway. Sci Signal. 2009;2:cm1. doi: 10.1126/scisignal.254cm1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. The future of fMRI and genetics research. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2011.10.063. http://dx.doi.org/10.1016/j.neuroimage.2011.10.063. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. Neuroimage. 2008;40:655–661. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Moers A, Nurnberg A, Goebbels S, Wettschureck N, Offermanns S. Galpha12/Galpha13 deficiency causes localized overmigration of neurons in the developing cerebral and cerebellar cortices. Mol Cell Biol. 2008;28:1480–1488. doi: 10.1128/MCB.00651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJ, Baare WF, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 2002;5:83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- Ruano D, Abecasis GR, Glaser B, Lips ES, Cornelisse LN, de Jong AP, Evans DM, Davey Smith G, Timpson NJ, Smit AB, et al. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am J Hum Genet. 2010;86:113–125. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton JP, Ankney CD. Whole brain size and general mental ability: a review. Int J Neurosci. 2009;119:691–731. doi: 10.1080/00207450802325843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah VP, Seasholtz TM, Sagi SA, Brown JH. The role of Rho in G protein-coupled receptor signal transduction. Annu Rev Pharmacol Toxicol. 2000;40:459–489. doi: 10.1146/annurev.pharmtox.40.1.459. [DOI] [PubMed] [Google Scholar]
- Sanno H, Shen X, Kuru N, Bormuth I, Bobsin K, Gardner HA, Komljenovic D, Tarabykin V, Erzurumlu RS, Tucker KL. Control of postnatal apoptosis in the neocortex by RhoA-subfamily GTPases determines neuronal density. J Neurosci. 2010;30:4221–4231. doi: 10.1523/JNEUROSCI.3318-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackman AJ, Salomons TV, Slagter HA, Fox AS, Winter JJ, Davidson RJ. The integration of negative affect, pain and cognitive control in the cingulate cortex. Nat Rev Neurosci. 2011;12:154–167. doi: 10.1038/nrn2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M, Montana G, Nichols TE. False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 2010;54:992–1000. doi: 10.1016/j.neuroimage.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, Hellal F, Neukirchen D, Hindges R, Garvalov BK, Flynn KC, Stradal TE, Chrostek-Grashoff A, Brakebusch C, Bradke F. Rac1 regulates neuronal polarization through the WAVE complex. J Neurosci. 2010;30:6930–6943. doi: 10.1523/JNEUROSCI.5395-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Ishii K, Kasai K, Yoon SO, Saeki Y. Neural expression of G protein-coupled receptors GPR3, GPR6, and GPR12 up-regulates cyclic AMP levels and promotes neurite outgrowth. J Biol Chem. 2007;282:10506–10515. doi: 10.1074/jbc.M700911200. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen ET, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M, et al. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Martin NG, Wright MJ. Imaging genomics. Curr Opin Neurol. 2010;23:368–373. doi: 10.1097/WCO.0b013e32833b764c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witelson SF, Beresh H, Kigar DL. Intelligence and brain size in 100 postmortem brains: sex, lateralization and age factors. Brain. 2006;129:386–398. doi: 10.1093/brain/awh696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.